Abstract
In the title compound, C20H22O5, the tetrahydropyran, cyclohexene and cyclohexane rings of the xanthene ring system adopt half-chair, half-boat and chair conformations, respectively. The mean plane of the four roughly planar atoms of the tetrahydropyran ring (r.m.s. deviation = 0.111 Å) forms a dihedral angle of 82.91 (4)° with the methoxybenzene group. In the crystal, molecules are linked via O—H⋯O and C—H⋯O hydrogen bonds into sheets lying parallel to the ac plane. The crystal is further consolidated by weak C—H⋯π interactions.
Related literature
For background to the applications of xanthene, see: Menchen et al. (2003 ▶); Knight & Stephens (1989 ▶). For our previous studies in this area, see: Palakshi Reddy et al. (2010 ▶); Reddy et al. (2009 ▶). For ring conformations, see: Cremer & Pople (1975 ▶). For a related structure, see: Loh et al. (2011 ▶). For bond length data, see: Allen et al. (1987 ▶). For the stability of the temperature controller used for data collection, see: Cosier & Glazer (1986 ▶).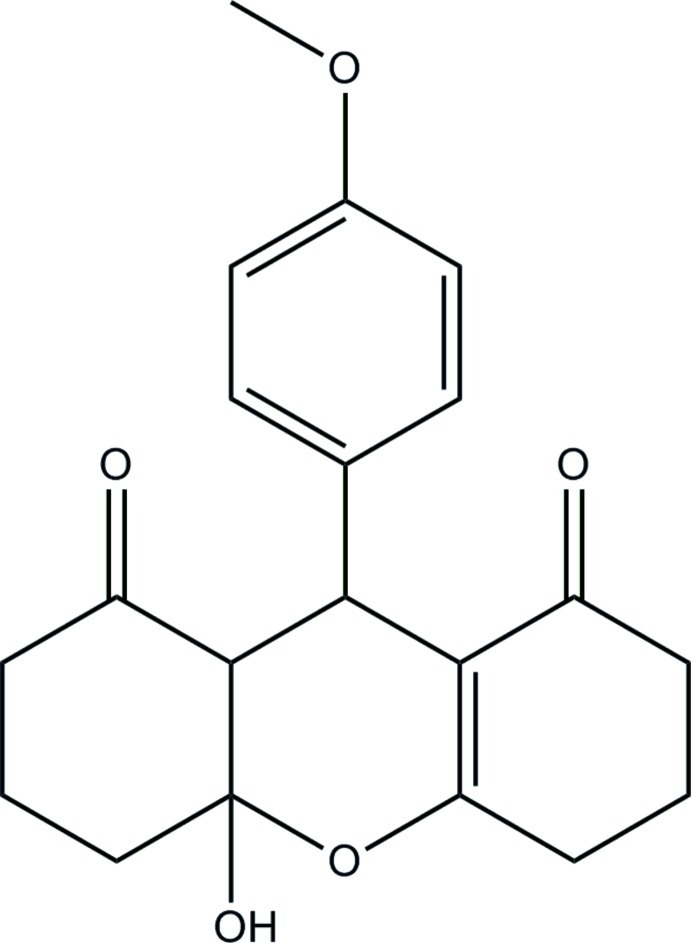
Experimental
Crystal data
C20H22O5
M r = 342.38
Orthorhombic,
a = 15.7611 (9) Å
b = 18.0089 (11) Å
c = 11.6451 (7) Å
V = 3305.3 (3) Å3
Z = 8
Mo Kα radiation
μ = 0.10 mm−1
T = 100 K
0.48 × 0.23 × 0.11 mm
Data collection
Bruker APEX DUO CCD diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2009 ▶) T min = 0.954, T max = 0.990
97212 measured reflections
7190 independent reflections
6068 reflections with I > 2σ(I)
R int = 0.045
Refinement
R[F 2 > 2σ(F 2)] = 0.037
wR(F 2) = 0.108
S = 1.05
7190 reflections
231 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.55 e Å−3
Δρmin = −0.24 e Å−3
Data collection: APEX2 (Bruker, 2009 ▶); cell refinement: SAINT (Bruker, 2009 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXTL (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXTL; molecular graphics: SHELXTL; software used to prepare material for publication: SHELXTL and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S160053681203005X/hb6880sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S160053681203005X/hb6880Isup2.hkl
Supplementary material file. DOI: 10.1107/S160053681203005X/hb6880Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
Cg1 is the centroid of the C2–C7 ring.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O4—H1O4⋯O3i | 0.886 (17) | 1.935 (17) | 2.8156 (8) | 172.0 (16) |
| C12—H12B⋯O4ii | 0.99 | 2.50 | 3.1879 (9) | 126 |
| C12—H12A⋯Cg1iii | 0.99 | 2.78 | 3.6557 (8) | 147 |
| C16—H16A⋯Cg1iv | 0.99 | 2.78 | 3.7467 (8) | 165 |
Symmetry codes: (i) 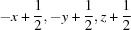 ; (ii)
; (ii) 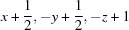 ; (iii)
; (iii) 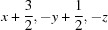 ; (iv)
; (iv) 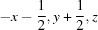 .
.
Acknowledgments
HKF and CWO thank Universiti Sains Malaysia (USM) for the Research University Grant (No. 1001/PFIZIK/811160). CWO also thanks the Malaysian Goverment and USM for the award of the post of Research Officer under Research University Grant No. 1001/PFIZIK/811160. VV, SS and BPR are grateful to VIT University for providing facilities to carry out research work.
supplementary crystallographic information
Comment
Xanthene derivatives are important heterocyclic compounds: their uses vary from dyes (Menchen et al., 2003) to agricultural bactericides (Knight et al., 1989). In continuation of our earlier interest in 1,4-DHP's and piperidones (Palakshi Reddy et al. 2009; Palakshi Reddy et al. 2010), herein we report the crystal structure of the title compound.
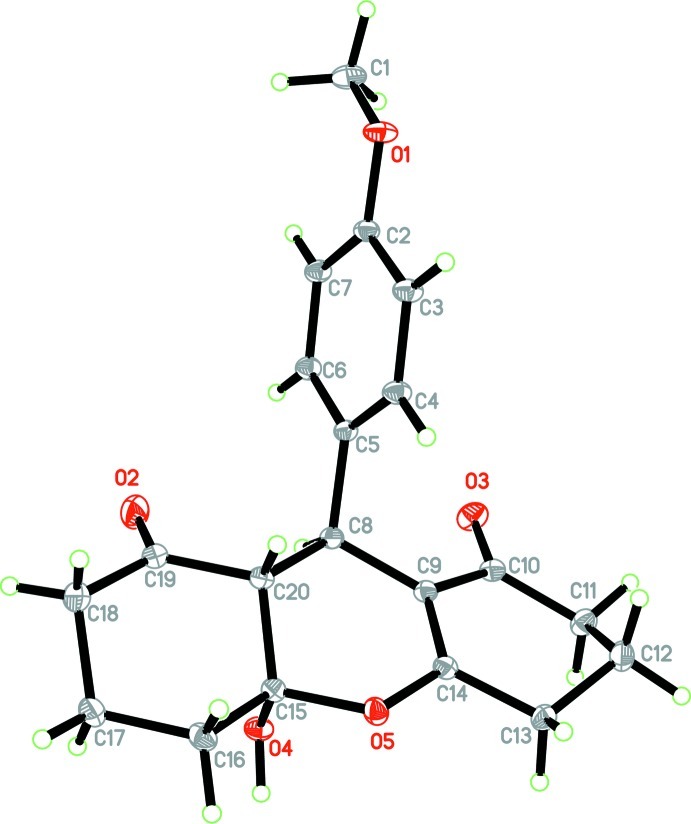
In the title compound (Fig. 1), the xanthene ring system consists of three rings which adopt different conformations. The tetrahydropyran ring (O5/C8/C9/C14/C15/C20) adopts a half chair conformation with the puckering parameters Q = 0.4980 (7) Å, θ = 122.73 (8)°, φ = 104.23 (9)° (Cremer & Pople, 1975). The cyclohexene (C9–C14) and cyclohexane (C15–C20) rings adopt half boat and chair conformations with the puckering parameters Q = 0.4905 (8) Å, θ = 117.37 (9)°, φ = 349.74 (10)° and Q = 0.5575 (8) Å, θ = 176.39 (8)°, φ = 192.6 (13)° (Cremer & Pople, 1975), respectively. The mean plane of the tetrahydropyran ring [r.m.s deviation = 0.111 Å] forms a dihedral angle of 82.91 (4)° with the methoxyphenyl group (C1–C7/O1). The bond lengths and angles are comparable to those in a related structure (Loh et al., 2011).
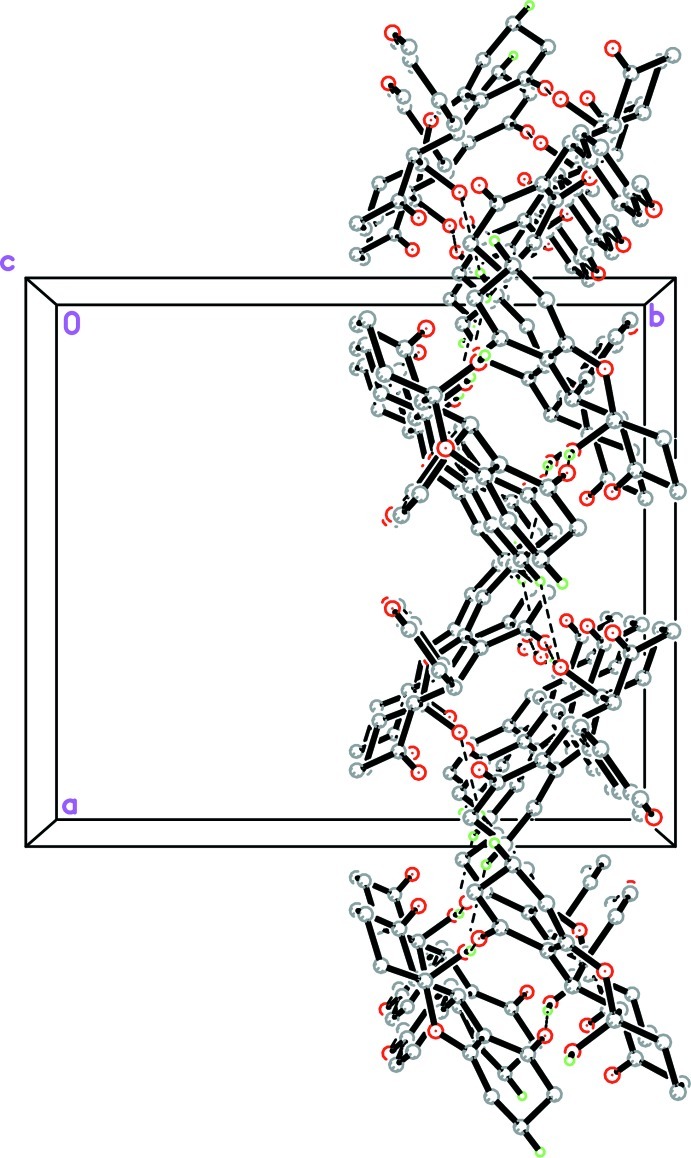
In the crystal structure (Fig. 2), the molecules are linked via intermolecular O4—H1O4···O3 and C12—H12B···O4 hydrogen bonds (Table 1) into two-dimensional networks parallel to the ac plane. The crystal structure is further consolidated by weak C—H···π interactions (Table 1), involving the centroid of the benzene ring (C2–C7; Cg1).
Experimental
A mixture of 4-methoxybenzaldehyde (1 mol) and 1,3-cyclohexanedione (2 mol) was refluxed in acetonitrile for 3 h. The progress of the reaction was monitored by TLC. After completion of the reaction, it was kept for 2 days for solid formation. The pure product was obtained by recrystallization from acetonitrile in the form of colourless blocks. M.p.: 194–196°C; Yield 70%.
Refinement
Atom H1O4 was located from the difference map and was refined freely [O–H = 0.887 (17) Å]. The remaining H atoms were positioned geometrically and refined using a riding model with Uiso(H) = 1.2 or 1.5Ueq(C) (C–H = 0.95, 0.98, 0.99 and 1.00 Å). A rotating group model was applied to the methyl group. In the final refinement, one outlier (1 0 4) was omitted.
Figures
Fig. 1.
The molecular structure of the title compound, showing 50% probability displacement ellipsoids.
Fig. 2.
The crystal packing of the title compound, viewed along the c axis. H atoms not involved in the intermolecular interactions (dashed lines) have been omitted for clarity.
Crystal data
| C20H22O5 | F(000) = 1456 |
| Mr = 342.38 | Dx = 1.376 Mg m−3 |
| Orthorhombic, Pbcn | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2n 2ab | Cell parameters from 9871 reflections |
| a = 15.7611 (9) Å | θ = 2.6–34.8° |
| b = 18.0089 (11) Å | µ = 0.10 mm−1 |
| c = 11.6451 (7) Å | T = 100 K |
| V = 3305.3 (3) Å3 | Block, colourless |
| Z = 8 | 0.48 × 0.23 × 0.11 mm |
Data collection
| Bruker APEX DUO CCD diffractometer | 7190 independent reflections |
| Radiation source: fine-focus sealed tube | 6068 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.045 |
| φ and ω scans | θmax = 34.8°, θmin = 1.7° |
| Absorption correction: multi-scan (SADABS; Bruker, 2009) | h = −25→25 |
| Tmin = 0.954, Tmax = 0.990 | k = −28→28 |
| 97212 measured reflections | l = −18→18 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.037 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.108 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.05 | w = 1/[σ2(Fo2) + (0.059P)2 + 0.8159P] where P = (Fo2 + 2Fc2)/3 |
| 7190 reflections | (Δ/σ)max = 0.001 |
| 231 parameters | Δρmax = 0.55 e Å−3 |
| 0 restraints | Δρmin = −0.24 e Å−3 |
Special details
| Experimental. The crystal was placed in the cold stream of an Oxford Cryosystems Cobra open-flow nitrogen cryostat (Cosier & Glazer, 1986) operating at 100 (1) K. |
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.41955 (4) | 0.43646 (3) | −0.11953 (4) | 0.01606 (10) | |
| O2 | 0.12092 (4) | 0.39264 (4) | 0.24042 (6) | 0.02378 (13) | |
| O3 | 0.35506 (4) | 0.19277 (3) | 0.24263 (5) | 0.01785 (11) | |
| O4 | 0.18491 (3) | 0.32355 (3) | 0.50833 (5) | 0.01370 (10) | |
| O5 | 0.32107 (3) | 0.36949 (3) | 0.53699 (4) | 0.01306 (10) | |
| C1 | 0.38478 (5) | 0.41214 (5) | −0.22594 (6) | 0.01835 (14) | |
| H1A | 0.4187 | 0.4321 | −0.2894 | 0.028* | |
| H1B | 0.3262 | 0.4299 | −0.2328 | 0.028* | |
| H1C | 0.3855 | 0.3578 | −0.2289 | 0.028* | |
| C2 | 0.38251 (4) | 0.40829 (4) | −0.02218 (5) | 0.01168 (11) | |
| C3 | 0.42230 (5) | 0.42661 (4) | 0.08114 (6) | 0.01375 (12) | |
| H3A | 0.4718 | 0.4567 | 0.0811 | 0.016* | |
| C4 | 0.38909 (4) | 0.40059 (4) | 0.18390 (6) | 0.01299 (11) | |
| H4A | 0.4165 | 0.4130 | 0.2540 | 0.016* | |
| C5 | 0.31611 (4) | 0.35644 (4) | 0.18628 (5) | 0.01067 (11) | |
| C6 | 0.27766 (4) | 0.33891 (4) | 0.08261 (6) | 0.01223 (11) | |
| H6A | 0.2279 | 0.3092 | 0.0828 | 0.015* | |
| C7 | 0.31024 (4) | 0.36386 (4) | −0.02199 (6) | 0.01273 (11) | |
| H7A | 0.2834 | 0.3507 | −0.0921 | 0.015* | |
| C8 | 0.27857 (4) | 0.33101 (4) | 0.30012 (5) | 0.01100 (11) | |
| H8A | 0.2310 | 0.2958 | 0.2841 | 0.013* | |
| C9 | 0.34387 (4) | 0.29187 (4) | 0.37349 (5) | 0.01071 (11) | |
| C10 | 0.37999 (4) | 0.22215 (4) | 0.33249 (6) | 0.01227 (11) | |
| C11 | 0.44481 (5) | 0.18319 (4) | 0.40660 (6) | 0.01532 (12) | |
| H11A | 0.4845 | 0.1553 | 0.3568 | 0.018* | |
| H11B | 0.4155 | 0.1470 | 0.4568 | 0.018* | |
| C12 | 0.49507 (5) | 0.23714 (4) | 0.48070 (6) | 0.01564 (12) | |
| H12A | 0.5300 | 0.2698 | 0.4312 | 0.019* | |
| H12B | 0.5337 | 0.2092 | 0.5320 | 0.019* | |
| C13 | 0.43452 (4) | 0.28408 (4) | 0.55218 (6) | 0.01375 (12) | |
| H13A | 0.4103 | 0.2532 | 0.6144 | 0.016* | |
| H13B | 0.4664 | 0.3253 | 0.5882 | 0.016* | |
| C14 | 0.36412 (4) | 0.31523 (4) | 0.48106 (6) | 0.01110 (11) | |
| C15 | 0.23657 (4) | 0.38542 (4) | 0.49304 (6) | 0.01137 (11) | |
| C16 | 0.20491 (5) | 0.45323 (4) | 0.55788 (6) | 0.01448 (12) | |
| H16A | 0.2459 | 0.4945 | 0.5488 | 0.017* | |
| H16B | 0.2004 | 0.4415 | 0.6407 | 0.017* | |
| C17 | 0.11806 (5) | 0.47705 (4) | 0.51201 (7) | 0.01725 (13) | |
| H17A | 0.1003 | 0.5234 | 0.5509 | 0.021* | |
| H17B | 0.0758 | 0.4382 | 0.5303 | 0.021* | |
| C18 | 0.11950 (5) | 0.48996 (5) | 0.38134 (7) | 0.02103 (15) | |
| H18A | 0.0610 | 0.4990 | 0.3537 | 0.025* | |
| H18B | 0.1538 | 0.5346 | 0.3640 | 0.025* | |
| C19 | 0.15647 (5) | 0.42367 (4) | 0.31926 (6) | 0.01632 (13) | |
| C20 | 0.24286 (4) | 0.39890 (4) | 0.36411 (6) | 0.01216 (11) | |
| H20A | 0.2833 | 0.4409 | 0.3521 | 0.015* | |
| H1O4 | 0.1773 (11) | 0.3167 (9) | 0.5830 (15) | 0.044 (4)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0198 (2) | 0.0200 (2) | 0.0083 (2) | −0.00409 (19) | 0.00179 (17) | 0.00088 (17) |
| O2 | 0.0187 (3) | 0.0334 (3) | 0.0192 (3) | 0.0069 (2) | −0.0042 (2) | −0.0018 (2) |
| O3 | 0.0242 (3) | 0.0162 (2) | 0.0131 (2) | 0.0013 (2) | −0.00347 (19) | −0.00439 (18) |
| O4 | 0.0146 (2) | 0.0138 (2) | 0.0126 (2) | −0.00304 (16) | 0.00192 (16) | 0.00182 (17) |
| O5 | 0.0119 (2) | 0.0149 (2) | 0.0124 (2) | 0.00174 (16) | −0.00052 (16) | −0.00358 (16) |
| C1 | 0.0258 (4) | 0.0206 (3) | 0.0086 (3) | −0.0017 (3) | 0.0007 (2) | −0.0008 (2) |
| C2 | 0.0138 (3) | 0.0127 (3) | 0.0085 (2) | 0.0004 (2) | 0.00115 (19) | 0.00019 (19) |
| C3 | 0.0145 (3) | 0.0166 (3) | 0.0101 (2) | −0.0037 (2) | 0.0003 (2) | 0.0000 (2) |
| C4 | 0.0137 (3) | 0.0161 (3) | 0.0092 (2) | −0.0031 (2) | −0.0005 (2) | −0.0003 (2) |
| C5 | 0.0116 (2) | 0.0115 (2) | 0.0089 (2) | 0.00015 (19) | 0.00017 (19) | 0.00024 (19) |
| C6 | 0.0130 (3) | 0.0134 (3) | 0.0103 (2) | −0.0016 (2) | −0.00051 (19) | −0.0007 (2) |
| C7 | 0.0144 (3) | 0.0146 (3) | 0.0092 (2) | −0.0012 (2) | −0.0010 (2) | −0.0008 (2) |
| C8 | 0.0113 (2) | 0.0122 (2) | 0.0095 (2) | −0.00026 (19) | 0.00045 (19) | 0.00068 (19) |
| C9 | 0.0119 (2) | 0.0110 (2) | 0.0093 (2) | 0.00007 (19) | 0.00030 (19) | −0.00021 (19) |
| C10 | 0.0143 (3) | 0.0119 (2) | 0.0106 (2) | −0.0004 (2) | 0.0003 (2) | −0.0001 (2) |
| C11 | 0.0182 (3) | 0.0133 (3) | 0.0145 (3) | 0.0032 (2) | −0.0021 (2) | −0.0012 (2) |
| C12 | 0.0130 (3) | 0.0175 (3) | 0.0164 (3) | 0.0025 (2) | −0.0017 (2) | −0.0023 (2) |
| C13 | 0.0128 (3) | 0.0165 (3) | 0.0119 (3) | 0.0010 (2) | −0.0021 (2) | −0.0016 (2) |
| C14 | 0.0110 (2) | 0.0116 (2) | 0.0107 (2) | −0.00030 (19) | 0.00070 (19) | −0.00083 (19) |
| C15 | 0.0111 (3) | 0.0117 (2) | 0.0114 (2) | −0.00005 (19) | 0.00047 (19) | −0.0001 (2) |
| C16 | 0.0159 (3) | 0.0133 (3) | 0.0143 (3) | 0.0013 (2) | 0.0031 (2) | −0.0016 (2) |
| C17 | 0.0165 (3) | 0.0179 (3) | 0.0173 (3) | 0.0049 (2) | 0.0041 (2) | 0.0010 (2) |
| C18 | 0.0232 (3) | 0.0223 (3) | 0.0176 (3) | 0.0103 (3) | 0.0031 (3) | 0.0032 (3) |
| C19 | 0.0157 (3) | 0.0199 (3) | 0.0133 (3) | 0.0047 (2) | 0.0021 (2) | 0.0039 (2) |
| C20 | 0.0130 (3) | 0.0132 (3) | 0.0103 (2) | 0.0011 (2) | 0.0019 (2) | 0.0011 (2) |
Geometric parameters (Å, º)
| O1—C2 | 1.3724 (8) | C9—C14 | 1.3595 (9) |
| O1—C1 | 1.4240 (9) | C9—C10 | 1.4590 (9) |
| O2—C19 | 1.2121 (10) | C10—C11 | 1.5102 (10) |
| O3—C10 | 1.2366 (8) | C11—C12 | 1.5220 (10) |
| O4—C15 | 1.3914 (8) | C11—H11A | 0.9900 |
| O4—H1O4 | 0.887 (17) | C11—H11B | 0.9900 |
| O5—C14 | 1.3561 (8) | C12—C13 | 1.5225 (10) |
| O5—C15 | 1.4552 (8) | C12—H12A | 0.9900 |
| C1—H1A | 0.9800 | C12—H12B | 0.9900 |
| C1—H1B | 0.9800 | C13—C14 | 1.4940 (10) |
| C1—H1C | 0.9800 | C13—H13A | 0.9900 |
| C2—C7 | 1.3919 (10) | C13—H13B | 0.9900 |
| C2—C3 | 1.3963 (9) | C15—C16 | 1.5201 (10) |
| C3—C4 | 1.3876 (9) | C15—C20 | 1.5241 (9) |
| C3—H3A | 0.9500 | C16—C17 | 1.5306 (11) |
| C4—C5 | 1.3986 (9) | C16—H16A | 0.9900 |
| C4—H4A | 0.9500 | C16—H16B | 0.9900 |
| C5—C6 | 1.3872 (9) | C17—C18 | 1.5395 (11) |
| C5—C8 | 1.5223 (9) | C17—H17A | 0.9900 |
| C6—C7 | 1.3961 (9) | C17—H17B | 0.9900 |
| C6—H6A | 0.9500 | C18—C19 | 1.5124 (11) |
| C7—H7A | 0.9500 | C18—H18A | 0.9900 |
| C8—C9 | 1.5120 (9) | C18—H18B | 0.9900 |
| C8—C20 | 1.5383 (9) | C19—C20 | 1.5250 (10) |
| C8—H8A | 1.0000 | C20—H20A | 1.0000 |
| C2—O1—C1 | 116.20 (6) | C13—C12—H12A | 109.7 |
| C15—O4—H1O4 | 108.5 (11) | C11—C12—H12B | 109.7 |
| C14—O5—C15 | 115.53 (5) | C13—C12—H12B | 109.7 |
| O1—C1—H1A | 109.5 | H12A—C12—H12B | 108.2 |
| O1—C1—H1B | 109.5 | C14—C13—C12 | 111.78 (6) |
| H1A—C1—H1B | 109.5 | C14—C13—H13A | 109.3 |
| O1—C1—H1C | 109.5 | C12—C13—H13A | 109.3 |
| H1A—C1—H1C | 109.5 | C14—C13—H13B | 109.3 |
| H1B—C1—H1C | 109.5 | C12—C13—H13B | 109.3 |
| O1—C2—C7 | 124.19 (6) | H13A—C13—H13B | 107.9 |
| O1—C2—C3 | 115.68 (6) | O5—C14—C9 | 123.23 (6) |
| C7—C2—C3 | 120.13 (6) | O5—C14—C13 | 112.08 (6) |
| C4—C3—C2 | 119.60 (6) | C9—C14—C13 | 124.67 (6) |
| C4—C3—H3A | 120.2 | O4—C15—O5 | 109.43 (5) |
| C2—C3—H3A | 120.2 | O4—C15—C16 | 112.80 (6) |
| C3—C4—C5 | 121.28 (6) | O5—C15—C16 | 106.51 (5) |
| C3—C4—H4A | 119.4 | O4—C15—C20 | 106.96 (5) |
| C5—C4—H4A | 119.4 | O5—C15—C20 | 108.57 (5) |
| C6—C5—C4 | 118.13 (6) | C16—C15—C20 | 112.50 (6) |
| C6—C5—C8 | 121.31 (6) | C15—C16—C17 | 110.21 (6) |
| C4—C5—C8 | 120.53 (6) | C15—C16—H16A | 109.6 |
| C5—C6—C7 | 121.69 (6) | C17—C16—H16A | 109.6 |
| C5—C6—H6A | 119.2 | C15—C16—H16B | 109.6 |
| C7—C6—H6A | 119.2 | C17—C16—H16B | 109.6 |
| C2—C7—C6 | 119.17 (6) | H16A—C16—H16B | 108.1 |
| C2—C7—H7A | 120.4 | C16—C17—C18 | 111.97 (6) |
| C6—C7—H7A | 120.4 | C16—C17—H17A | 109.2 |
| C9—C8—C5 | 111.58 (5) | C18—C17—H17A | 109.2 |
| C9—C8—C20 | 110.23 (5) | C16—C17—H17B | 109.2 |
| C5—C8—C20 | 108.97 (5) | C18—C17—H17B | 109.2 |
| C9—C8—H8A | 108.7 | H17A—C17—H17B | 107.9 |
| C5—C8—H8A | 108.7 | C19—C18—C17 | 111.03 (6) |
| C20—C8—H8A | 108.7 | C19—C18—H18A | 109.4 |
| C14—C9—C10 | 118.45 (6) | C17—C18—H18A | 109.4 |
| C14—C9—C8 | 122.42 (6) | C19—C18—H18B | 109.4 |
| C10—C9—C8 | 118.81 (6) | C17—C18—H18B | 109.4 |
| O3—C10—C9 | 121.41 (6) | H18A—C18—H18B | 108.0 |
| O3—C10—C11 | 119.98 (6) | O2—C19—C18 | 123.23 (7) |
| C9—C10—C11 | 118.47 (6) | O2—C19—C20 | 122.49 (7) |
| C10—C11—C12 | 112.31 (6) | C18—C19—C20 | 114.28 (6) |
| C10—C11—H11A | 109.1 | C15—C20—C19 | 109.02 (5) |
| C12—C11—H11A | 109.1 | C15—C20—C8 | 111.99 (5) |
| C10—C11—H11B | 109.1 | C19—C20—C8 | 113.16 (6) |
| C12—C11—H11B | 109.1 | C15—C20—H20A | 107.5 |
| H11A—C11—H11B | 107.9 | C19—C20—H20A | 107.5 |
| C11—C12—C13 | 109.76 (6) | C8—C20—H20A | 107.5 |
| C11—C12—H12A | 109.7 | ||
| C1—O1—C2—C7 | −5.83 (10) | C10—C9—C14—O5 | −165.29 (6) |
| C1—O1—C2—C3 | 173.89 (6) | C8—C9—C14—O5 | 8.13 (10) |
| O1—C2—C3—C4 | 179.92 (6) | C10—C9—C14—C13 | 13.68 (10) |
| C7—C2—C3—C4 | −0.35 (11) | C8—C9—C14—C13 | −172.89 (6) |
| C2—C3—C4—C5 | −0.24 (11) | C12—C13—C14—O5 | −166.48 (6) |
| C3—C4—C5—C6 | 0.28 (10) | C12—C13—C14—C9 | 14.45 (10) |
| C3—C4—C5—C8 | −177.51 (6) | C14—O5—C15—O4 | 64.53 (7) |
| C4—C5—C6—C7 | 0.28 (10) | C14—O5—C15—C16 | −173.25 (6) |
| C8—C5—C6—C7 | 178.05 (6) | C14—O5—C15—C20 | −51.88 (7) |
| O1—C2—C7—C6 | −179.41 (6) | O4—C15—C16—C17 | −63.35 (7) |
| C3—C2—C7—C6 | 0.89 (10) | O5—C15—C16—C17 | 176.59 (5) |
| C5—C6—C7—C2 | −0.86 (10) | C20—C15—C16—C17 | 57.76 (8) |
| C6—C5—C8—C9 | 128.25 (7) | C15—C16—C17—C18 | −54.64 (8) |
| C4—C5—C8—C9 | −54.04 (8) | C16—C17—C18—C19 | 51.97 (9) |
| C6—C5—C8—C20 | −109.80 (7) | C17—C18—C19—O2 | 127.53 (8) |
| C4—C5—C8—C20 | 67.91 (8) | C17—C18—C19—C20 | −52.57 (9) |
| C5—C8—C9—C14 | 122.43 (7) | O4—C15—C20—C19 | 67.95 (7) |
| C20—C8—C9—C14 | 1.21 (9) | O5—C15—C20—C19 | −174.06 (5) |
| C5—C8—C9—C10 | −64.17 (8) | C16—C15—C20—C19 | −56.44 (7) |
| C20—C8—C9—C10 | 174.62 (6) | O4—C15—C20—C8 | −58.05 (7) |
| C14—C9—C10—O3 | 169.77 (7) | O5—C15—C20—C8 | 59.94 (7) |
| C8—C9—C10—O3 | −3.90 (10) | C16—C15—C20—C8 | 177.56 (6) |
| C14—C9—C10—C11 | −6.00 (9) | O2—C19—C20—C15 | −125.90 (8) |
| C8—C9—C10—C11 | −179.66 (6) | C18—C19—C20—C15 | 54.19 (8) |
| O3—C10—C11—C12 | 155.40 (7) | O2—C19—C20—C8 | −0.58 (10) |
| C9—C10—C11—C12 | −28.78 (9) | C18—C19—C20—C8 | 179.51 (6) |
| C10—C11—C12—C13 | 55.12 (8) | C9—C8—C20—C15 | −34.60 (7) |
| C11—C12—C13—C14 | −47.83 (8) | C5—C8—C20—C15 | −157.37 (5) |
| C15—O5—C14—C9 | 18.89 (9) | C9—C8—C20—C19 | −158.31 (6) |
| C15—O5—C14—C13 | −160.19 (6) | C5—C8—C20—C19 | 78.92 (7) |
Hydrogen-bond geometry (Å, º)
Cg1 is the centroid of the C2–C7 ring.
| D—H···A | D—H | H···A | D···A | D—H···A |
| O4—H1O4···O3i | 0.886 (17) | 1.935 (17) | 2.8156 (8) | 172.0 (16) |
| C12—H12B···O4ii | 0.99 | 2.50 | 3.1879 (9) | 126 |
| C12—H12A···Cg1iii | 0.99 | 2.78 | 3.6557 (8) | 147 |
| C16—H16A···Cg1iv | 0.99 | 2.78 | 3.7467 (8) | 165 |
Symmetry codes: (i) −x+1/2, −y+1/2, z+1/2; (ii) x+1/2, −y+1/2, −z+1; (iii) x+3/2, −y+1/2, −z; (iv) −x−1/2, y+1/2, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HB6880).
References
- Allen, F. H., Kennard, O., Watson, D. G., Brammer, L., Orpen, A. G. & Taylor, R. (1987). J. Chem. Soc. Perkin Trans. 2, pp. S1–19.
- Bruker (2009). SADABS, APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Cosier, J. & Glazer, A. M. (1986). J. Appl. Cryst. 19, 105–107.
- Cremer, D. & Pople, J. A. (1975). J. Am. Chem. Soc. 97, 1354–1358.
- Knight, C. G. & Stephens, T. (1989). Biochem. J. 258, 683–689. [DOI] [PMC free article] [PubMed]
- Loh, W.-S., Fun, H.-K., Reddy, B. P., Vijayakumar, V. & Sarveswari, S. (2011). Acta Cryst. E67, o35–o36. [DOI] [PMC free article] [PubMed]
- Menchen, S. M., Benson, S. C., Lam, J. Y. L., Zhen, J. Y. L., Sun, W., Rosenblum, D., Khan, B. B. & Taing, S. H. (2003). US Patent No. 6583168.
- Palakshi Reddy, B., Vijayakumar, V., Sarveswari, S., Ng, S. W. & Tiekink, E. R. T. (2010). Acta Cryst. E66, o2806–o2807. [DOI] [PMC free article] [PubMed]
- Reddy, B. P., Vijayakumar, V., Narasimhamurthy, T., Suresh, J. & Lakshman, P. L. N. (2009). Acta Cryst. E65, o916. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S160053681203005X/hb6880sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S160053681203005X/hb6880Isup2.hkl
Supplementary material file. DOI: 10.1107/S160053681203005X/hb6880Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report