Abstract
Our aim was to comprehensively analyze promoter hypermethylation of a panel of novel and known methylation markers for thyroid neoplasms and to establish their relationship with BRAF mutation and clinicopathologic parameters of thyroid cancer. A cohort of thyroid tumors, consisting of 44 cancers and 44 benign thyroid lesions, as well as 15 samples of adjacent normal thyroid tissue, was evaluated for BRAF mutation and promoter hypermethylation. Genes for quantitative methylation specific PCR (QMSP) were selected by a candidate gene approach. Twenty-two genes were tested: TSHR, RASSF1A, RARβ2, DAPK, hMLH1, ATM, S100, p16, CTNNB1, GSTP1, CALCA, TIMP3, TGFßR2, THBS1, MINT1, CTNNB1, MT1G, PAK3, NISCH, DCC, AIM1 and KIF1A. The PCR-based “mutector assay” was used to detect BRAF mutation. All p values reported are two sided. Considerable overlap was seen in the methylation markers among the different tissue groups. Significantly higher methylation frequency and level were observed for KIF1A and RARß2 in cancer samples compared with benign tumors. A negative correlation between BRAF mutation and RASSF1A methylation, and a positive correlation with RARß2 methylation were observed in accordance with previous results. In addition, positive correlation with TIMP3 and a marginal correlation with DCC methylation were observed. The present study constitutes a comprehensive promoter methylation profile of thyroid neoplasia and shows that results must be analyzed in a tissue-specific manner to identify clinically useful methylation markers. Integration of genetic and epigenetic changes in thyroid cancer will help identify relevant biologic pathways that drive its development.
Keywords: BRAF, RARβ2, RASSF1A, TIMP3, biomarkers, hypermethylation, thyroid cancer, thyroid tissue
Introduction
Genomic research has been able to identify cancer-specific genetic and epigenetic alterations. The field of classic genetics largely concentrates on the DNA sequence. Important mutations that lead to human cancer have been identified by genetics research. In contrast, the field of epigenetics refers to changes in the genome that alter gene expression without altering the DNA sequence itself. DNA methylation constitutes the most studied epigenetic event in cancer. The methylation of cytosine residues in the promoter region of genes inhibits transcriptional binding and hence gene expression.1,2 Evidence points to a complex succession of critical molecular events that activates proto-oncogenes, and/or silences tumor suppressor genes (TSGs), leading to the development of cancer. Molecular biology techniques that detect these alterations could be powerful tools to potentially enhance diagnosis, understand tumor biology as well as characterize tumor’s behavior, thereby providing new tools for cancer management.
The present study concentrates on two molecular phenomena in thyroid cancer: BRAF mutation and promoter DNA methylation. The BRAF activating mutation V600E constitutes a common oncogenic mechanism in up to 69% of papillary thyroid cancers (PTC).3,4 Activation of the RAF/MEK/MAPK signaling pathway interferes with proliferation, differentiation, and apoptosis.5 Furthermore, BRAF mutation has been associated with poor prognosis in PTC patients.6 DNA methylation in promoter regions of TSGs is a well-established event that has been described in virtually all tumor types. However, considerable variation exists between individual methylated genes among different tumor types.7-10 In an effort to expand our knowledge of DNA methylation in thyroid cancer, a total of 22 cancer related genes were selected for methylation analysis in adjacent normal thyroid, benign thyroid tumors, and thyroid cancer. The genes studied were selected based on previous reported association with thyroid cancer as well as genes never evaluated in thyroid cancer with known tumor suppressor properties or promoter methylation in other cancer types.
The present study seeks to examine the methylation signatures of a panel of novel and known genes and to integrate methylation profiling with the most important genetic alteration (BRAF mutation) identified for thyroid cancer to date. Some studies have tested the methylation status only in thyroid cancer tissues and not in benign or normal controls.11 We decided to test a comprehensive cohort of tissue samples that included normal tissues as well as benign neoplasias and thyroid cancers in an effort to molecularly differentiate these three groups. We have also tested BRAF mutations in all our thyroid cancer samples, and correlated BRAF mutation status with methylation profiling.
Results
We examined a cohort of 15 normal thyroid tissue samples, 44 benign thyroid lesions (6 hyperplastic nodules, 12 follicular adenomas, 6 adenomatoid nodules, 1 adenomatoid hyperplasia, 6 multinodular goiters, 1 multinodular hyperplasia, 12 Hϋrthle adenomas) and 44 thyroid cancers [27 papillary (10 of which were of the follicular variant of papillary thyroid cancer), 7 follicular, 2 Hϋrthle cell and 8 medullary carcinomas]. Demographic and clinicopathological characteristics are detailed in Table 1. No significant differences in demographic characteristic between sample groups were observed (data not shown). Staging for thyroid cancer was done according to the American Joint Committee on Cancer (AJCC) TNM system.
Table 1. Demographic and clinical characteristics of study subjects (n = 103).
| Characteristic |
No. (%) Cancer Patients (Total = 44) |
No. (%) Benign thyroid pathology patients (Total = 44) |
No. (%) Normal thyroid subjects (Total = 15) |
|---|---|---|---|
|
Gender |
|
|
|
| Female |
33 (75%) |
29 (65.9%) |
12 (80%) |
| Male Unknown |
10 (22.7%) 1(2.3%) |
11 (25%) 4 (9.1%) |
1 (6.7%) 2 (13.3%) |
|
Median Age (range) |
47 (16–74) |
50 (25–92) |
50 (26–92) |
|
Stage |
|
|
|
| I |
25 (56.8%) |
|
|
| II |
7 (15.9%) |
|
|
| III |
9 (20.5%) |
|
|
| IV |
2 (4.5%) |
|
|
| Unknown | 1 (2.3%) |
Frequency of methylation in different types of thyroid tissues
We examined 22 genes of diverse function, including cell cycle regulation, tumor suppression, and DNA repair in thyroid tissues by QMSP. One would expect to see a trend of increasing methylation, across the three categories of samples: normal, benign and cancer. We did not find significant trends for the majority of the genes tested for promoter methylation, in either binary data (Cochran-Armitage tests) or continuous data (Cuzick tests) analyses.
The frequencies of individual gene methylation per tissue group are shown in Table 2. In our analysis of trends of increasing methylation across categories, KIF1A was the only marker with an increased probability of methylation in the tumor samples (14% in the cancer tissue and 0% in the normal and benign; Cochran-Armitage p value = 0.02). While the trend test across three categories for RARβ2 was not significant, a standard Wilcoxon rank sum test directly comparing benign and malignant tumors confirmed a previously reported6 difference in methylation levels (p = 0.05, borderline significant). The frequencies of methylation for normal tissues, benign tissues, and thyroid cancer were CTNNB1 20%, 3% and 16%; GSTP1 0%, 7%, 7%; TIMP3 27%, 42%, and 51%. AIM1 was methylated in one of the tumors but not in any other tissue (0%, 0%, and 3%). No significant differences in methylation were seen for genes DAPK (71%, 64% and 65%), CDH1 (67%, 66%, and 56%), and RARβ2 (7%, 2%, and 14%). NISCH was methylated in all normal samples but not in all tumors (100% and 86%). To create a panel of genes that could distinguish the different categories of samples, we combined genes with high specificity: GSTP1, P16, RARβ2 and KIF1A and analyzed the frequencies based on any one of these markers being methylated. This combination marker (COMBO) was positive in 2 (13%) normal samples, 5 (11%) benign samples, and 12 (27%) thyroid cancers.
Table 2. Frequencies of individual gene methylation per tissue group (Cochran-Armitage and Cuzick tests of trend across sample classes).
| Marker | Missing n |
Normal n (%) |
Benign Adenoma n (%) |
Carcinoma n (%) |
C-A p value |
Cuzick p value |
|---|---|---|---|---|---|---|
|
RASSF1A |
1 |
13 (87) |
43 (98) |
38 (88) |
0.67 |
0.19 |
|
TSHR
1
|
1 |
9 (60) |
24 (55) |
20 (45) |
0.27 |
0.47 |
|
AIM1 |
19 |
0 |
0 |
1 (3) |
0.32 |
0.85 |
|
ATM |
19 |
8 (80) |
19 (53) |
27 (71) |
0.74 |
0.63 |
|
CALCA |
2 |
13 (87) |
37 (84) |
39 (93) |
0.33 |
0.11 |
|
CDH1 |
1 |
10 (67) |
29 (66) |
24 (56) |
0.34 |
0.36 |
|
DAPK |
2 |
10 (71) |
28 (64) |
28 (65) |
0.77 |
0.85 |
|
DCC |
19 |
3 (30) |
9 (25) |
11 (29) |
0.90 |
0.84 |
|
GSTP1 |
1 |
0 |
3 (7) |
3 (7) |
0.42 |
0.73 |
|
MINT1 |
19 |
7 (70) |
24 (67) |
25 (66) |
0.82 |
0.47 |
|
hMLH1 |
19 |
7 (70) |
19 (53) |
27 (71) |
0.44 |
0.22 |
|
MT1G |
19 |
1 (10) |
10 (28) |
8 (21) |
0.80 |
0.77 |
|
P16 |
1 |
1 (7) |
1 (2) |
2 (5) |
0.94 |
0.98 |
|
RAR-ß2 |
1 |
1 (7) |
1 (2) |
6 (14) |
0.14 |
0.50 |
|
S100A2 |
1 |
4 (27) |
24 (55) |
21 (49) |
0.32 |
0.77 |
|
TGFßR2 |
1 |
9 (60) |
33 (75) |
31 (72) |
0.54 |
0.65 |
|
Thbs1 |
19 |
4 (40) |
10 (28) |
6 (16) |
0.08 |
0.17 |
|
Timp3 |
2 |
4 (27) |
18 (42) |
22 (51) |
0.10 |
0.21 |
|
Pak3 |
23 |
3 (33) |
10 (29) |
16 (44) |
0.26 |
0.40 |
|
Nisch |
22 |
8 (100) |
31 (86) |
32 (86) |
0.46 |
0.12 |
|
Kif1a |
23 |
0 |
0 |
5 (14) |
0.02* |
0.35 |
|
CTNNB1 |
19 |
2 (20) |
1 (3) |
6 (16) |
0.60 |
0.80 |
| COMBO 2 | 0 | 2 (13) | 5(11) | 12 (27) | 0.09 | NA |
1TSHR is median dichotomized. All other markers are zero dichotomized. 2COMBO is positive if any one of: GSTP1, P16, RARβ2 or KIF1A is positive. No continuous data. Cochran-Armitage tests for trend were used for dichotomized methylation variables. The frequency and percent positive are shown for normal, benign and malignant categories. The Cuzick test for trend is a non-parametric test for the continuous data values (data not shown).
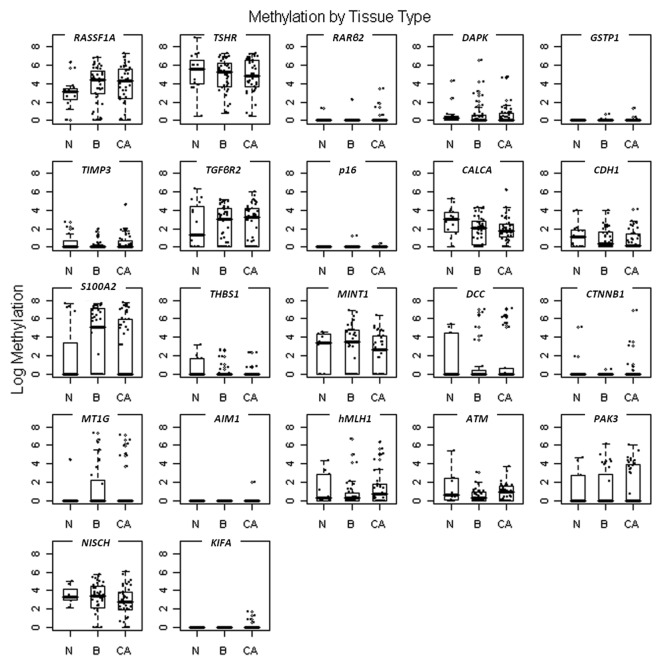
Figure 1 shows box plots of all the genes for the continuous methylation data in the three major sample categories (normal, benign and cancer). The box plots show overlap between the methylation ratios of almost all the genes in the different tissue groups. KIF1A methylation was detected in five cancer samples (14%) and in none of the normal or benign samples. Although very low frequency of methylation was observed for KIF1A, it can be used with other markers in the diagnosis of thyroid nodules due to its 100% specificity.
Figure 1. Promoter methylation levels for the different markers in the cancer patients (n = 44) [CA], the benign pathology patients (n = 44) [B], and the normal thyroid tissues (n = 15) [N]. The quantity of methylation is expressed as the ratio of the PCR product for the gene of interest to that of β-actin multiplied by 1,000. Boxplots show the middle 50% of the data, the median with a bar in the center, and bars extending the median by 1.5 times the interquartile range.
When making binary determinations of the presence or absence of methylation based on different cutoffs for methylation, such as zero, the 75th percentile or the 90th percentile of methylation level in the normal tissue group, there were no significant differences in the methylation status in the different groups. Due to the limited amounts of DNA not all the genes were tested for all samples (identified as missing in Table 2). Similarly we were not able to test the methylation status of all the potential relevant genes in these samples due to limited amount of DNA.
We performed a correlation analysis for all pairs of markers (Spearman correlation shown in Table 3). The strongest correlations (r ≥ 0.7) were between TGFβR2 and TSHR in normal and tumor samples. A strong correlation between CDH1 and TSHR was observed in normal, while a moderate correlation was observed in tumors (r ≥ 0.5). A moderate correlation between TIMP3 and THSR was observed exclusively in the tumor samples’ group.
Table 3. Summary of Spearman correlations for pair of genes (promoter methylation) in thyroid samples.
| THYROID CANCER | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
RASSF1A |
0.56 * |
0.41 |
0.06 |
0.23 |
0.48 |
0.22 |
-0.11 |
0 |
0.08 |
-0.18 |
-0.15 |
0.14 |
0.33 |
-0.1 |
0.08 |
-0.13 |
0.24 |
0.11 |
0.04 |
-0.11 |
-0.06 |
| 0.61 * |
TSHR |
0.63 * |
0.6 * |
0.52 |
0.77 * |
0.47 |
0.15 |
-0.03 |
0.31 |
0.21 |
-0.04 |
0.04 |
0.36 |
-0.07 |
0.15 |
-0.18 |
0.1 |
0.26 |
0.1 |
0.15 |
-0.23 |
| 0.36 |
0.5 * |
CALCA |
0.68 |
0.37 |
0.45 |
0.25 |
0.27 |
0.34 |
0.33 |
0.36 |
-0.26 |
-0.1 |
0.36 |
0.06 |
0.04 |
-0.17 |
0.09 |
0.08 |
-0.21 |
0.02 |
-0.24 |
| 0.41 |
0.74 * |
0.48 |
CDH1 |
0.39 |
0.41 |
0.45 |
0.36 |
0.23 |
0.42 |
0.58 |
-0.03 |
0 |
0.4 |
-0.03 |
0.19 |
0.09 |
0.15 |
0.14 |
-0.18 |
0.33 |
-0.18 |
| 0.23 |
0.35 |
0.23 |
0.49 |
TIMP3 |
0.36 |
0.26 |
0.19 |
0.25 |
0.22 |
0.05 |
-0.04 |
-0.01 |
0.29 |
-0.16 |
0.21 |
-0.23 |
0.18 |
0.18 |
-0.1 |
0.24 |
-0.16 |
| 0.6 |
0.84 * |
0.49 |
0.66 |
0.27 |
TGFβR2 |
0.34 |
-0.07 |
-0.1 |
0.17 |
0.03 |
-0.08 |
0.06 |
0.32 |
-0.14 |
0.14 |
-0.27 |
0.02 |
0.51 |
0.06 |
0 |
-0.13 |
| 0.22 |
-0.05 |
-0.14 |
-0.16 |
0.1 |
-0.05 |
MINT1 |
-0.01 |
0.17 |
0.37 |
0.29 |
-0.02 |
0.05 |
0.37 |
0.1 |
0.43 |
-0.01 |
0 |
0.01 |
0.22 |
0.17 |
0.08 |
| 0.1 |
0.15 |
0.35 |
0.34 |
0.34 |
0.27 |
0.07 |
DCC |
0.37 |
0.25 |
0.25 |
-0.01 |
0.18 |
0.2 |
0.17 |
-0.1 |
-0.13 |
0.08 |
-0.07 |
-0.09 |
0.09 |
-0.1 |
| 0.22 |
0.23 |
0.01 |
0.27 |
0.17 |
0.22 |
0.02 |
0.14 |
MT1G |
0.12 |
0.22 |
-0.14 |
0.15 |
0.25 |
0.05 |
-0.04 |
-0.2 |
0.33 |
0.14 |
-0.04 |
0.18 |
-0.08 |
| -0.07 |
-0.05 |
0.03 |
0.05 |
0.1 |
0.02 |
0.14 |
-0.09 |
-0.08 |
RARβ2 |
0.25 |
-0.11 |
-0.09 |
0.15 |
0.15 |
0.18 |
-0.08 |
0.08 |
0.05 |
-0.03 |
0.26 |
-0.07 |
| -0.07 |
0.08 |
-0.01 |
0.16 |
0.29 |
-0.08 |
-0.35 |
-0.07 |
-0.04 |
-0.09 |
DAPK |
0.07 |
0.1 |
0.19 |
0.18 |
0.09 |
0.07 |
-0.02 |
-0.06 |
-0.15 |
0.27 |
-0.18 |
| -0.13 |
-0.19 |
-0.15 |
-0.14 |
-0.04 |
-0.24 |
-0.18 |
-0.09 |
0.32 |
-0.04 |
-0.12 |
GSTP1 |
-0.06 |
-0.13 |
-0.05 |
-0.13 |
0.15 |
0.08 |
0.11 |
0.15 |
0.17 |
-0.05 |
| 0.12 |
0.22 |
0.14 |
0.09 |
0.08 |
0.26 |
-0.18 |
-0.09 |
-0.08 |
-0.04 |
0.19 |
-0.04 |
p16 |
0.2 |
0.21 |
-0.1 |
-0.07 |
0.36 |
-0.01 |
-0.18 |
0.21 |
-0.04 |
| 0.09 |
0.16 |
0.04 |
0.12 |
0.13 |
0.21 |
0.13 |
-0.03 |
0.01 |
-0.1 |
0.09 |
-0.1 |
-0.1 |
THBS1 |
0.04 |
0.16 |
0.07 |
0.08 |
0.15 |
-0.17 |
0.03 |
-0.07 |
| 0.03 |
-0.11 |
-0.06 |
0.05 |
0.11 |
-0.08 |
0.16 |
0.19 |
0.06 |
-0.17 |
-0.16 |
0.01 |
-0.17 |
0.2 |
S100A2 |
0.06 |
-0.04 |
0.05 |
-0.1 |
-0.02 |
0.18 |
-0.14 |
| -0.05 |
0.01 |
-0.01 |
-0.02 |
-0.22 |
0.06 |
-0.22 |
-0.15 |
-0.15 |
-0.04 |
-0.1 |
-0.04 |
-0.04 |
0.25 |
-0.24 |
CTNNB1 |
0.06 |
0.01 |
-0.05 |
0.09 |
0.03 |
-0.07 |
| 0.1 |
0.22 |
0.19 |
0.21 |
0 |
0.22 |
-0.22 |
-0.18 |
-0.04 |
-0.01 |
0.1 |
-0.15 |
0.24 |
0.18 |
-0.18 |
0.36 |
hMLH1 |
-0.07 |
-0.14 |
0.1 |
-0.04 |
-0.08 |
| -0.03 |
0 |
0.1 |
-0.02 |
0.09 |
-0.03 |
0.1 |
-0.09 |
0.05 |
0.16 |
-0.17 |
-0.16 |
-0.16 |
0.03 |
0 |
0.32 |
0.01 |
ATM |
0.04 |
-0.04 |
0.43 |
-0.2 |
| -0.04 |
0.05 |
0.12 |
0.01 |
-0.21 |
0.04 |
-0.05 |
0.16 |
0.16 |
-0.1 |
-0.1 |
-0.1 |
. |
-0.14 |
-0.15 |
0.01 |
0 |
0.24 |
PAK3 |
0.16 |
0.11 |
0.04 |
| 0.19 |
0.07 |
-0.13 |
-0.13 |
0.06 |
0.06 |
0.26 |
-0.25 |
0.1 |
0.2 |
-0.11 |
-0.2 |
. |
0.23 |
0.2 |
0.05 |
0.17 |
0.16 |
0.13 |
NISCH |
-0.06 |
-0.25 |
| . |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
KIF1A |
-0.07 |
| . |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
. |
AIM1 |
| |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
|
THYROID NORMAL + BENIGN PATHOLOGY |
|
|
|
|
|
|
|
|
|||||||||||
The diagonal row of boxes lists the markers analyzed; correlations are indicated at the horizontal and vertical intersections of the markers. Light gray shading represents R > 0.5; dark gray shading represents R > 0.7. The samples were split into two groups: correlation between the pair of genes in tumors (n = 44) indicated in the upper half of the plot and correlations between normal (benign pathology and normal thyroid tissue, n = 59) indicated in the lower half of the plot. Black asterisk (*) indicates similar correlations observed in both groups. Black dot (.) indicates that the correlation could not be calculated due to values being mainly zero.
The frequency of DCC methylation was 36% (8/22) in papillary and 18% (5/28) in follicular neoplasms (adenomas and carcinomas). None of the other markers showed significant correlation with demographic and clinicopathologic data such as age, sex, or stage. Thyroid cancer patients were divided into two age groups, those older than 45 y and those younger than 45 y, to analyze the status of methylation given that AJCC determined that 45 y of age is a cut-off point for decreased prognosis. No significant correlation was found with methylation values and age groups by non-parametric Wilcoxon testing.
Frequency of BRAF mutation in thyroid tissues:
We analyzed the BRAF mutation status in all thyroid cancer samples. Consistent with previous results, we found that 15 of the 25 papillary thyroid cancers were positive for the mutation (60%), and the mutation was not detectable in the other carcinomas tested, in agreement with previous findings showing that BRAF mutation is present in papillary thyroid cancers but not the other subtypes.3 To our knowledge, there is no published report about germline mutation of BRAF in normal thyroid tissue. We therefore coded all nonmalignant samples as negative for the BRAF mutation for the purpose of the correlation analyses.
Correlation between BRAF mutation and methylation
In papillary thyroid cancer, we found significant correlations between the methylation status of four genes (TIMP3, RASSF1A, RARβ2 and DCC) and the V600E BRAF mutation. The mutation was present in 15 of the papillary thyroid cancer tumors in our sample set. RASSF1A methylation decreased the probability of BRAF mutation, OR = 0.74 (95% CI: 0.56, 0.97), p = 0.035, while methylation in other genes increased the probability of BRAF mutation: RARβ2 OR = 2.63 (95% CI: 1.00, 6.89), p = 0.05, TIMP3, OR = 2.04 (95% CI: 1.00, 4.12), p = 0.05, and DCC, OR = 1.32 (95% CI: 1.07, 1.63), p = 0.01. Multivariate logistic regression confirmed the negative association with RASSF1A, and the positive association with RARβ2, TIMP3 as well as a marginal positive correlation with DCC. Table 4 shows the odds ratios as well as the confidence intervals for these associations, in univariate and multivariate analyses.
Table 4. Univariate and multivariate analyses of BRAF mutation and promoter methylation of the genes that showed statistically significant correlations.
|
BRAF mutation vs. promoter methylation |
Chi Square p value |
Odds Ratio |
95% CI |
|||
|---|---|---|---|---|---|---|
| Univariate/Multivariate | Univariate/Multivariate | Univariate/ Multivariate | ||||
|
DCC* RARβ2* RASSF1A* TIMP3* |
0.01 0.05 0.035 0.05 |
0.09 0.01 0.005 0.01 |
1.32 2.63 0.74 2.04 |
1.29 4.35 0.53 3.04 |
(1.07–1.63) (1.00–6.89) (0.56–0.97) (1.0–4.12) |
(0.97–1.72) (1.47–12.79) (0.34–0.82) (1.26–7.34) |
Log transformed
Discussion
Dissecting all the genetic and epigenetic alterations involved in thyroid cancer is essential for our understanding of the pathogenesis of this disease, and hence, for more precise diagnosis, accurate prognosis prediction and appropriate management of patients. In this study, we evaluated the most common mutation found in papillary thyroid cancer as well as a comprehensive panel of candidate cancer methylation markers that include markers tested previously in thyroid cancer and cancer specific methylation markers that had not yet been tested in thyroid cancer. Nineteen of the genes we have tested in this study have been previously analyzed in thyroid cancer (TSHR, RASSF1A, RARß2, DAPK, TIMP3, hMLH1, p16, ATM, TGFßR2, PAK3, NISCH, KIF1A, CALCA, CDH1, S100A2, THBS1, GSTP1, CTNNB1 and MT1G).6,8,11-18 Among these 19 genes, PAK3, NISCH, and KIF1A were previously only tested in a small set of thyroid cancer samples by our group as a part of our comprehensive approach to discover methylated genes in cancer.8 Three of the 22 genes, MINT1, DCC, and AIM1, have not been tested in thyroid cancer to date.
In general, our analysis revealed considerable overlap between promoter hypermethylation of normal thyroid tissue, benign hyperplastic states and tumors, and malignant tumors. Although with very low frequency (14%), KIF1A methylation was found to be 100% cancer specific and this frequency is consistent with our previous findings.8 The analysis of BRAF mutations confirmed the previously reported inverse relationship with RASSF1A methylation,6,17,19 as well as the direct relationship with RARβ2 methylation6 and with TIMP3.16 Additionally we describe a positive correlation between DCC methylation and BRAF mutation. To our knowledge this is the first evidence indicating a relationship between these two genes and thyroid cancer. Interestingly, DCC was only methylated in the papillary subtype.
We were unable to confirm previously reported promising results for several potential methylation markers, possibly because the scope of previous methylation studies was too narrowly defined, particularly regarding samples tested and methylation detection methods used. Our group previously reported differential methylation in cancer and benign samples for the genes TSHR and RARß2. Using conventional MSP we reported TSHR methylation was 59% (23/39) for papillary thyroid tumors and 47% (7/15) for follicular tumors and 0% (0/8) for the normal and benign tumors.13 The current study used QMSP, a more sensitive and automated method that shows 45% (20/44) methylation in tumors, 55% (24/44) in benign tumors, and 60% (9/15) in normal tissues. Although methylation levels are generally low in normal and benign tissues, the overlap between benign and malignant tumors precluded reaching statistical significance in this cohort, and will complicate the use of this marker in a diagnostic and/or prognostic setting. RARß2 had shown significantly higher methylation in thyroid cancer in comparison to benign thyroid (22% of the papillary and follicular thyroid carcinomas, and only 4% of the adenomas) in our prior QMSP study.6 However, in this study we found methylation in only 14% of thyroid cancers, compared with 2% in the benign tumors, and 7% of the normal. The source of RARß2 methylation in the normal thyroid is unclear at this time, and also has the potential to confound diagnostic testing, although the statistically significant difference between benign lesions and malignant tumors found in our previous study was confirmed. Larger sample sizes are needed to confirm the frequency of methylation of RARβ2 in thyroid cancer and various benign thyroid neoplasms and normal thyroid tissue. Hu et al. reported the methylation of DAPK, RARβ2, and TIMP3 in papillary thyroid cancer through the use of QMSP.16 Their frequency of methylation for DAPK and TIMP3 in thyroid cancer is similar to ours (64 vs. 65% for DAPK and 55 vs. 51% for TIMP3), while different for RARβ2 (58 vs. 14%). Unfortunately, in the present study as well as a previous one from our group, we have shown that normal thyroid samples and cancer samples have overlapping frequencies of methylation for DAPK, TIMP3, and RARβ2 (DAPK 65% tumors vs. 63% benign vs. 71% in normal; TIMP3 51% tumors vs. 42% benign vs. 27% normal; RARβ2 mentioned above).6 These findings limit the use of these particular genes as diagnostic markers for cancer in thyroid specimens and question the neoplastic relevance of the hypermethylation present in the tumors.
The present results raise concerns regarding the laboratory techniques and tissue samples selection in the study of methylation. Quantitative Methylation Specific PCR (known as QMSP or methylight) is based on a real time PCR and is more sensitive than conventional methylation specific PCR (MSP). Eads et al.20 reported a sensitivity of detection of 1 methylated allele in a background of 10,000 unmethylated alleles (1:10,000). Herman et al.9 reported when first reporting conventional MSP a sensitivity of 1:1,000, which represents a 10-fold difference. QMSP, being a real time PCR assay is more objective to be analyzed than conventional PCR, with reduced cross-contamination, as there is no post-PCR analysis (gel electrophoresis). The quantitative type of assay is more specific due to the incorporation of the methylation specific probe, in addition to the pair of primers. In the other hand, QMSP is able to detect only fully methylated molecules, while MSP can also detect partially methylated molecules. Virmani et al.21 compared their results of APC promoter methylation in lung cancer cell lines using conventional MSP and QMSP, and observed higher percentages when using the quantitative method. MSP was used to test the majority of the genes previously reported as hypermethylated in thyroid cancer (MT1G, TSHR, CTNNB1, DAPK, ATM, p16, and hMLH1).12-15 For most of the genes, we were unable to establish cutoff levels that could reliably distinguish methylation frequencies in the normal and benign groups from the cancer group. Overall, the high levels of methylation in normal and benign thyroid tumors reported in this study are consistent with our previous QMSP study in thyroid tissues.6 An additional difference among various studies is the methodology to process tissues, which is also a factor that can change the detected levels of methylation. Furthermore, some of the previous studies did not use appropriate normal controls for the determination of thyroid cancer specific methylated genes.11 Of significant concern to our group is the evaluation of the methylation markers on comprehensive sample sets that not only include cancer cases, but also normal and benign tumors that are a common part of the clinical differential diagnosis.
Methylation of TSHR often occurred together with TGFβR2 and CDH1 in normal, benign and tumor samples. These associations could have happened by chance alone and should be interpreted with caution. A possible interaction of these genes in thyroid physiology will need to be further assessed. There was only one interaction, between TIMP3 and THSR, which was observed exclusively in cancer. None of these genes showed a cancer specific hypermethylation pattern in our study. Although both were frequently methylated in all samples, further studies should be conducted to elucidate the role of this coordinated methylation. A lack of inhibition of TSHR by TIMPs has been previously reported.22 It is possible that pathways related to both TIMP3 and TSHR signaling need to be altered for developing subsets of thyroid cancers.
Genes studied here are putative TSGs that are active in a wide variety of normal tissues and have been reported to be hypermethylated across many tumor types (some examples are p16, CTNNB1, and DAPK). Reddy et al. demonstrated, however, that DAPK promoter methylation was present in lymphocytes from normal individuals, specifically in B lymphocytes.23 This is an example of a tissue specific methylation of a gene that has been reported as a cancer specific methylated gene. Similarly, we have recently reported RARβ2 methylation presence in cancer-free patients (29%; 46/157).24 In the later study, RARβ2 promoter methylation was observed in 45% of subjects who had a high-fat diet. Many groups are investigating how exogenous as well as endogenous factors (like hormones) participate in promoter methylation.25,26 Thyroid being an endocrine organ, exposure to variations of hormone levels could lead to specific promoter methylation patterns like RARβ2. In contrast, there are TSGs that are specifically affected in particular tumor types like GSTP1 in prostate cancer.27 We believe each potential TSG needs to be evaluated in the context of the tissue type being examined. In this context, it should come as no surprise that an endocrine organ such as the thyroid may show significant differences in pathway regulation and TSG-dependency compared with non-endocrine epithelial organs, from which most currently known cancer methylation markers have been developed.
The relevance for the RAF/MEK/MAPK kinase pathway in thyroid cancer being upregulated either by a BRAF activating mutation or RASSF1A methylation silencing is further confirmed by our study. RASSF1A methylation has been reported in several cancer types.6,28-30 This gene contains a Ras-binding domain, and its association to Ras activation has been demonstrated in vitro, as well as its ability to induce apoptosis.31-33 Accumulated data suggested its direct correlation to RAF/MEK/MAPK oncogenic pathway. It would be interesting to see how DCC plays a role in this pathway for papillary tumors, in a larger cohort of samples. DCC is a postulated TSG initially identified in colon cancer that mediates apoptosis by a mechanism requiring receptor ligand activity.34,35 TIMP3 proteins are inhibitors of the matrix metalloproteinases, a group of peptidases involved in degradation of the extracellular matrix. TIMP3 is of particular interest for therapeutic purposes due to its characteristics of inhibiting different aspects of tumor development, mainly because it is a potent angiogenesis inhibitor. The positive association of TIMP3 methylation with BRAF mutations suggests that methylation of this metastasis suppressor gene may play a role in the aggressiveness of PTC conferred by BRAF mutation. Further studies are necessary to understand the biological relevance of the positive correlation between BRAF mutation and promoter methylation of TIMP3. After establishing biological relevance, specific therapy may be developed for BRAF mutated PTC patients with or without TIMP3 methylation. The methylation markers associated with BRAF mutations may well have specific roles in thyroid carcinomas, but since only a minority of thyroid carcinomas in this cohort were mutated in BRAF, it is likely that our cohort lacked the statistical power to show their association with specific clinical behavior of thyroid carcinoma.
The present study constitutes a comprehensive profile of hypermethylated genes previously reported as altered in thyroid cancer as well as an extended panel of genes never evaluated in thyroid tissues. Moreover, we also analyzed BRAF mutations, a common genetic event in papillary thyroid cancer. Methylation profiling of thyroid tissues to date has concentrated on a candidate gene approach for known TSGs in epithelial cancers. To our knowledge, no genes have been identified as thyroid cancer specific methylated genes using appropriate controls and sufficient numbers of benign tumor samples for comparison. It is possible that methylation is not a common event for thyroid cancer, but since epigenetic control seems otherwise ubiquitous, it is more likely that the relevant methylation markers remain unidentified. Alternatively, it is possible that the identification of combinations of markers, both genetic and epigenetic, reflecting alterations in several regulatory and metabolic pathways may be required to achieve acceptable positive and negative predictive values in a clinical diagnostic test. Correct diagnosis of thyroid nodules from FNAs specimens and prognostic profiling of thyroid cancers constitute pressing challenges in the current medical practice. Additional systematic genome-wide approaches using primary tumors and appropriate controls will be necessary to develop new panels of biomarkers for thyroid cancer. The planned comprehensive effort by the National Cancer Institute’s The Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov/)-thyroid consortium should provide valuable insights in thyroid cancer-specific methylome changes.
Materials and Methods
Tissue samples
We selected 109 patients who underwent surgical resection for a thyroid tumor from 2000–2003 at The Johns Hopkins Hospital (Baltimore, MD) from whom a frozen tumor sample was available for DNA extraction. Collection of tissue and demographic data was performed in accordance to the guidelines of The Johns Hopkins University Institutional Review Board under protocol NA_00018307. The molecular studies were performed under protocol 03–11–12–06e. Tissue was routinely obtained from the center of the lesions and from uninvolved adjacent thyroid tissue and snap frozen in liquid nitrogen. To avoid field-cancerization effects (the increased risk of the adjacent epithelial surface of the tumor for the development of malignant lesions due to the potential presence of multiple molecular alterations in the entire region), only normal thyroid tissue sampled adjacent to benign nodules was used in this study. All cases were classified according to the clinical pathology report, and cryosections were obtained from experimental tissue samples to verify the presence of tumor tissue and scored for the presence of inflammatory cell infiltrates suggestive of thyroiditis. Cases with chronic lymphocytic (Hashimoto) thyroiditis as a secondary diagnosis were excluded from the study, since extensive inflammation has been shown to induce aberrant DNA methylation.36
DNA extraction and Sodium Bisulfite Treatment
DNA was extracted from frozen thyroid tissue, and subsequently subjected to bisulfite treatment, as described previously.6
Methylation analysis
Bisulfite treated DNA was used as a template for the fluorogenic gene-specific QMSP reactions. Quantitative PCR was performed in a TaqMan 7900HT Applied Biosystem and analyzed by a sequence detector system (SDS 2.3; Applied Biosystems), as previously described.37Table 5 shows the primer and probe sequences used. Each plate included studied DNA samples, positive (in-vitro methylated leukocyte DNA) and negative (normal leukocyte DNA or DNA from a known unmethylated cell line) controls and multiple water blanks (molecular grade water was used as a non-template control). Leukocyte DNA from a healthy individual was methylated in vitro with excess SssI methyltransferase (New England Biolabs Inc.) to generate completely methylated DNA, and serial dilutions (90–0.009 ng) of this DNA were used to construct a calibration curve for each plate. The β-actin gene was used to normalize the fluorescence emission as well as an internal loading control, β-actin primers and probe were designed to amplify a region that is devoid of CpG nucleotides which allows amplification independent of its methylation status. The methylation ratio is defined as the ratio of the fluorescence emission intensity values for the gene-specific PCR products to those of the β-actin (reference gene) and then multiplied by 1,000 for easier tabulation. All samples were within the assay’s range of sensitivity and reproducibility, based on the amplification of the internal reference standard [threshold cycle (CT) value for β-actin of 40].
Table 5. Primers and probes used in QMSP assay.
| Gene | Forward primer 5’-3’ | Probe 5’-3’ (6-FAM-5’- 3’-6-TAMRA) | Reverse primer 5’-3’ | Amplicon Location (Genbank numbering) | Accession Number |
|---|---|---|---|---|---|
|
β-actin |
TGGTGATGGAGGAGGTTTAGTAAGT |
ACCACCACCCAACACACAATAACAAACACA |
AACCAATAAAACCTACTCCTCCCTTAA |
390-522 |
Y00474 |
|
TSHR |
GGTGTAGAGTTGAGAATGAGGTGATTTC |
ACAACACCAACTACAACAAATCCGCCGA |
GCCCAAATCCCTAAACAAATCG |
188-310 |
BC024205 |
|
RASSF1A |
GCGTTGAAGTCGGGGTTC |
ACAAACGCGAACCGAACGAAACCA |
CCCGTACTTCGCTAACTTTAAACG |
45-119 |
NM_007182 |
|
RARβ2 |
GGGATTAGAATTTTTTATGCGAGTTGT |
TGTCGAGAACGCGAGCGATTCG |
TACCCCGACGATACCCAAAC |
907-999 |
X56849 |
|
DAPK |
GGATAGTCGGATCGAGTTAACGTC |
TTCGGTAATTCGTAGCGGTAGGGTTTGG |
CCCTCCCAAACGCCGA |
4-102 |
X76104 |
|
hMLH1 |
CGTTATATATCGTTCGTAGTATTCGTGTTT |
CGCGACGTCAAACGCCACTACG |
CTATCGCCGCCTCATCGT |
254-341 |
U26559 |
|
ATM |
CGGGTCGAATGTTTTGGGG |
ATCCAATATCACGCGATCTCCGC |
GCAAAACACGATATACCCATAC |
82-160 |
NM_000051.3 |
|
S100A2 |
TGGTTTCGATTTTTTGATTTCG |
CGACCGAACGCGATAACTTACTCCTA |
CGACCGAACGCGATAACTTACTCCTA |
5075-5317 |
Y07755 |
|
p16 |
TTATTAGAGGGTGGGGCGGATCGC |
AGTAGTATGGAGTCGGCGGCGGG |
GACCCCGAACCGCGACCGTAA |
25-174 |
U12818 |
|
CDH1 |
AATTTTAGGTTAGAGGGTTATCGCGT |
CGCCCACCCGACCTCGCAT |
TCCCCAAAACGAAACTAACGAC |
842-911 |
L34545 |
|
GSTP1 |
AGTTGCGCGGCGATTTC |
CGGTCGACGTTCGGGGTGTAGCG |
GCCCCAATACTAAATCACGACG |
1033-1172 |
M24485 |
|
CALCA |
GTTTTGGAAGTATGAGGGTGACG |
ATTCCGCCAATACACAACAACCAATAAACG |
TTCCCGCCGCTATAAATCG |
1706-1806 |
X15943 |
|
TIMP3 |
GCGTCGGAGGTTAAGGTTGTT |
AACTCGCTCGCCCGCCGAA |
CTCTCCAAAATTACCGTACGCG |
1051-1143 |
U33110 |
|
TGFβR2 |
GAGGGGAGGCGGTAGAT |
CGACGTCCAACCCCTAACTCTC |
CAACTTCAACTCAACGCTACG |
(-)224-91 |
NM_001024847.2 |
|
THBS1 |
CGACGCACCAACCTACCG |
ACGCCGCGCTCACCTCCCT |
GTTTTGAGTTGGTTTTACGTTCGTT |
1642-1716 |
J04835 |
|
MINT1 |
ATTTTCGAAGCGTTTGTTTGGC |
GCGAAACTCCCCTACTCTCCAAC |
ACAAAAAACCTCAACCCCGC |
63970-64053 |
AC026774.7 |
|
CTNNB1 |
GGAAAGGCGCGTCGAGT |
CGCGCGTTTCCCGAACCG |
TCCCCTATCCCAAACCCG |
583-664 |
X89448 |
|
MT1G |
TGCGAAAGGGGTCGTTTTGC |
GCGATCCCGACCTAAACTATACG |
AACCCGCTAAATCCGCACC |
120-235 |
J03910 |
|
PAK3 |
TTACGGTCGTCGTTATTATCG |
AACCAAAAAAAATAAAAAATCACAACCG |
ACCGAAAATTCTACCCTTCG |
943-1065 |
NM_002578 |
|
NISCH |
TTTTTTTCGTATAGA GTTCGT |
CGCGACCCAACACGCAATAATACTC |
CTAAACCTCTCTAAAATTCG |
361-517 |
NM_007184 |
|
DCC |
TTGTTCGCGATTTTTGGTTTC |
GCGCTAAACAAAAAAACTCCGAAAA |
ACCGATTACTTAAAAATACGCG |
549-680 |
NM_005215 |
|
AIM1 |
CGCGGGTATTGGATGTTAGT |
GGGAGCGTTGCGGATTATTCGTAG |
CCGACCCACCTATACGAAAA |
6862-6982 |
AL359292.12 |
| KIF1A | GCGCGATAAATTAGTTGGCGATT | CCTCCCGAAACGCTAATTAACTACGCG | CTCGACGACTACTCTACGCTAT | 870-1010 | NM_004321 |
Gene Selection
The thyrotropin receptor (TSHR) gene plays an important role in thyroid function by initiating thyroid hormone synthesis. TSHR has been reported as hypermethylated in thyroid cancers and unmethylated in benign and normal thyroid tissues. 6,12,13 RASSF1A is a direct player in the RAF/MEK/MAPK pathway and is considered a negative regulator of the cell cycle progression.38 RARß2 is a retinoic acid receptor commonly silenced in cancer that was found by our group to be hypermethylated in thyroid cancer and not in benign thyroid tissues.6,19 p16, DAPK, hMLH1, ATM, MT1G, TIMP3, TGFßR2 and CTNNB1 have been previously found to be hypermethylated in thyroid cancers. 6,11,12,14,19,39 The role of p16 as an important cell cycle regulator has been widely described.40 DAPK is involved in apoptosis.41 hMLH1 is involved in DNA repair42 while ATM regulates cell cycle in DNA damage scenarios.43 S100A2, GSTP1, CALCA, THBS1, MINT1, CTNNB1, PAK3, DCC, AIM1, and KIF1A are other genes known to be methylated in cancer. 8,27,37,44
BRAF mutation detection
Samples were analyzed for the thymine (T) - adenine (A) miss-sense mutation at nucleotide 1796 in the BRAF gene. Briefly, PCR primer sequences were designed to amplify a 102-bp fragment of exon 15 (5′- GAAGACCTCACAGTAAAAATAGGTGA-3′ and 5′-CCACAAAATGGATCCAGACA-3′). PCR amplification was performed using 100 ng of genomic DNA as template. Cycling conditions were as follows: a denaturation step at 95°C for 5 min was followed by two cycles of denaturation at 95°C for 1 min, annealing at 60°C for 1 min, primer extension at 72°C for 1 min, two cycles of denaturation at 95°C for 1 min, annealing at 58°C for 1 min, primer extension at 72°C for 1 min, 35 cycles of denaturation at 95°C for 1 min, annealing at 56°C for 1 min, primer extension at 72°C for 1 min, and one final extension at 72°C for 5 min. Amplified fragments were separated on an agarose gel and visualized by ethidium bromide staining. Analysis of the products was performed using the colorimetric Mutector assay according to the manufacturer’s instructions (TrimGen). A detection primer was designed that does not permit primer extension when the target base is wild-type. When the target base is mutated, primer extension continues and a color reaction is observed. The assay was preformed according to the manufacturer’s instructions. The melanoma cell line HTB 72 was used as positive control for the BRAF T1796A mutation, and the cervical cancer cell line ME180, known to be wild type for BRAF at T1796, served as a negative control. This assay is reported to have sensitivity and specificity nearly of 100%, and it is capable of detecting as low as 1% mutated DNA in a wild-type background.45
Statistical analysis
The primary objective of this study was to describe the methylation patterns of 22 genes in normal, benign and cancer thyroid tissue. Continuous distributions of QMSP ratios are often distinctly non-normal with a clump of zeros in the lower tail of a distribution of continuous values. To evaluate the two parts of these distributions for a trend of increasing methylation across the categories of samples: normal, benign and cancer, two types of analyses were performed. In the first, methylation was considered binary and the presence or absence of methylation was analyzed with an exact version of the Cochran-Armitage trend test. In the second, the continuous methylation distributions of each gene were evaluated across the tissue categories with the non-parametric Cuzick test for trend.
A secondary objective of this study was to determine if methylation of these genes was associated with an increased probability of BRAF mutation. Genes associated with this outcome were selected based on univariate and multivariate logistic regression modeling. All statistical computations were performed using the SAS system46 StatXact or R. All p values reported are two sided. p values of < 0.05 were considered statistically significant.
Disclosure of Potential Conflicts of Interest
This work was supported by National Cancer Institute Grant U01-CA84986, P50 DE019032 and R01 CA107247–04) and a grant from the American Cancer Society (RSG-08–003–01-CCE). The funding agencies had no role in the design of the study, data collection, analysis, interpretation of the results, preparation of the manuscript, or the decision to submit the manuscript for publication. Under a licensing agreement between Oncomethylome Sciences, SA and the Johns Hopkins University, D.S. is entitled to a share of royalty received by the University upon sales of diagnostic products described in this article. D.S. owns Oncomethylome Sciences, SA stock, which is subject to certain restrictions under University policy. D.S. is a paid consultant to Oncomethylome Sciences, SA and is a paid member of the company’s Scientific Advisory Board. The Johns Hopkins University in accordance with its conflict of interest policies is managing the terms of this agreement. Dr. Hoque is a recipient of Young Clinical Scientist Award from the Flight Attendant Medical Research Institute and the Young Investigator Award from the International Association for the Study of Lung Cancer. The other authors have nothing to disclose.
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/20524
References
- 1.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–59. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 2.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–54. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 3.Cohen Y, Xing M, Mambo E, Guo Z, Wu G, Trink B, et al. BRAF mutation in papillary thyroid carcinoma. J Natl Cancer Inst. 2003;95:625–7. doi: 10.1093/jnci/95.8.625. [DOI] [PubMed] [Google Scholar]
- 4.Kimura ET, Nikiforova MN, Zhu Z, Knauf JA, Nikiforov YE, Fagin JA. High prevalence of BRAF mutations in thyroid cancer: genetic evidence for constitutive activation of the RET/PTC-RAS-BRAF signaling pathway in papillary thyroid carcinoma. Cancer Res. 2003;63:1454–7. [PubMed] [Google Scholar]
- 5.Wan PT, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, et al. Cancer Genome Project Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–67. doi: 10.1016/S0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 6.Hoque MO, Rosenbaum E, Westra WH, Xing M, Ladenson P, Zeiger MA, et al. Quantitative assessment of promoter methylation profiles in thyroid neoplasms. J Clin Endocrinol Metab. 2005;90:4011–8. doi: 10.1210/jc.2005-0313. [DOI] [PubMed] [Google Scholar]
- 7.Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–9. [PubMed] [Google Scholar]
- 8.Hoque MO, Kim MS, Ostrow KL, Liu J, Wisman GB, Park HL, et al. Genome-wide promoter analysis uncovers portions of the cancer methylome. Cancer Res. 2008;68:2661–70. doi: 10.1158/0008-5472.CAN-07-5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–6. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suzuki H, Gabrielson E, Chen W, Anbazhagan R, van Engeland M, Weijenberg MP, et al. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat Genet. 2002;31:141–9. doi: 10.1038/ng892. [DOI] [PubMed] [Google Scholar]
- 11.Ishida E, Nakamura M, Shimada K, Higuchi T, Takatsu K, Yane K, et al. DNA hypermethylation status of multiple genes in papillary thyroid carcinomas. Pathobiology. 2007;74:344–52. doi: 10.1159/000110028. [DOI] [PubMed] [Google Scholar]
- 12.Smith JA, Fan CY, Zou C, Bodenner D, Kokoska MS. Methylation status of genes in papillary thyroid carcinoma. Arch Otolaryngol Head Neck Surg. 2007;133:1006–11. doi: 10.1001/archotol.133.10.1006. [DOI] [PubMed] [Google Scholar]
- 13.Xing M, Usadel H, Cohen Y, Tokumaru Y, Guo Z, Westra WB, et al. Methylation of the thyroid-stimulating hormone receptor gene in epithelial thyroid tumors: a marker of malignancy and a cause of gene silencing. Cancer Res. 2003;63:2316–21. [PubMed] [Google Scholar]
- 14.Guan H, Ji M, Hou P, Liu Z, Wang C, Shan Z, et al. Hypermethylation of the DNA mismatch repair gene hMLH1 and its association with lymph node metastasis and T1799A BRAF mutation in patients with papillary thyroid cancer. Cancer. 2008;113:247–55. doi: 10.1002/cncr.23548. [DOI] [PubMed] [Google Scholar]
- 15.Huang Y, de la Chapelle A, Pellegata NS. Hypermethylation, but not LOH, is associated with the low expression of MT1G and CRABP1 in papillary thyroid carcinoma. Int J Cancer. 2003;104:735–44. doi: 10.1002/ijc.11006. [DOI] [PubMed] [Google Scholar]
- 16.Hu S, Liu D, Tufano RP, Carson KA, Rosenbaum E, Cohen Y, et al. Association of aberrant methylation of tumor suppressor genes with tumor aggressiveness and BRAF mutation in papillary thyroid cancer. Int J Cancer. 2006;119:2322–9. doi: 10.1002/ijc.22110. [DOI] [PubMed] [Google Scholar]
- 17.Nakamura N, Carney JA, Jin L, Kajita S, Pallares J, Zhang H, et al. RASSF1A and NORE1A methylation and BRAFV600E mutations in thyroid tumors. Lab Invest. 2005;85:1065–75. doi: 10.1038/labinvest.3700306. [DOI] [PubMed] [Google Scholar]
- 18.Bello MJ, De Campos JM, Isla A, Casartelli C, Rey JA. Promoter CpG methylation of multiple genes in pituitary adenomas: frequent involvement of caspase-8. Oncol Rep. 2006;15:443–8. [PubMed] [Google Scholar]
- 19.Xing M, Cohen Y, Mambo E, Tallini G, Udelsman R, Ladenson PW, et al. Early occurrence of RASSF1A hypermethylation and its mutual exclusion with BRAF mutation in thyroid tumorigenesis. Cancer Res. 2004;64:1664–8. doi: 10.1158/0008-5472.CAN-03-3242. [DOI] [PubMed] [Google Scholar]
- 20.Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, et al. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Virmani AK, Tsou JA, Siegmund KD, Shen LY, Long TI, Laird PW, et al. Hierarchical clustering of lung cancer cell lines using DNA methylation markers. Cancer Epidemiol Biomarkers Prev. 2002;11:291–7. [PubMed] [Google Scholar]
- 22.de Bernard S, Misrahi M, Huet JC, Beau I, Desroches A, Loosfelt H, et al. Sequential cleavage and excision of a segment of the thyrotropin receptor ectodomain. J Biol Chem. 1999;274:101–7. doi: 10.1074/jbc.274.1.101. [DOI] [PubMed] [Google Scholar]
- 23.Reddy AN, Jiang WW, Kim M, Benoit N, Taylor R, Clinger J, et al. Death-associated protein kinase promoter hypermethylation in normal human lymphocytes. Cancer Res. 2003;63:7694–8. [PubMed] [Google Scholar]
- 24.Brait M, Ford JG, Papaiahgari S, Garza MA, Lee JI, Loyo M, et al. Association between lifestyle factors and CpG island methylation in a cancer-free population. Cancer Epidemiol Biomarkers Prev. 2009;18:2984–91. doi: 10.1158/1055-9965.EPI-08-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feil R. Environmental and nutritional effects on the epigenetic regulation of genes. Mutat Res. 2006;600:46–57. doi: 10.1016/j.mrfmmm.2006.05.029. [DOI] [PubMed] [Google Scholar]
- 26.Herceg Z. Epigenetics and cancer: towards an evaluation of the impact of environmental and dietary factors. Mutagenesis. 2007;22:91–103. doi: 10.1093/mutage/gel068. [DOI] [PubMed] [Google Scholar]
- 27.Jerónimo C, Henrique R, Hoque MO, Mambo E, Ribeiro FR, Varzim G, et al. A quantitative promoter methylation profile of prostate cancer. Clin Cancer Res. 2004;10:8472–8. doi: 10.1158/1078-0432.CCR-04-0894. [DOI] [PubMed] [Google Scholar]
- 28.Burbee DG, Forgacs E, Zöchbauer-Müller S, Shivakumar L, Fong K, Gao B, et al. Epigenetic inactivation of RASSF1A in lung and breast cancers and malignant phenotype suppression. J Natl Cancer Inst. 2001;93:691–9. doi: 10.1093/jnci/93.9.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ibanez de Caceres I, Battagli C, Esteller M, Herman JG, Dulaimi E, Edelson MI, et al. Tumor cell-specific BRCA1 and RASSF1A hypermethylation in serum, plasma, and peritoneal fluid from ovarian cancer patients. Cancer Res. 2004;64:6476–81. doi: 10.1158/0008-5472.CAN-04-1529. [DOI] [PubMed] [Google Scholar]
- 30.Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet. 2000;25:315–9. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- 31.Khokhlatchev A, Rabizadeh S, Xavier R, Nedwidek M, Chen T, Zhang XF, et al. Identification of a novel Ras-regulated proapoptotic pathway. Curr Biol. 2002;12:253–65. doi: 10.1016/S0960-9822(02)00683-8. [DOI] [PubMed] [Google Scholar]
- 32.Ortiz-Vega S, Khokhlatchev A, Nedwidek M, Zhang XF, Dammann R, Pfeifer GP, et al. The putative tumor suppressor RASSF1A homodimerizes and heterodimerizes with the Ras-GTP binding protein Nore1. Oncogene. 2002;21:1381–90. doi: 10.1038/sj.onc.1205192. [DOI] [PubMed] [Google Scholar]
- 33.Vos MD, Ellis CA, Bell A, Birrer MJ, Clark GJ. Ras uses the novel tumor suppressor RASSF1 as an effector to mediate apoptosis. J Biol Chem. 2000;275:35669–72. doi: 10.1074/jbc.C000463200. [DOI] [PubMed] [Google Scholar]
- 34.Mehlen P, Rabizadeh S, Snipas SJ, Assa-Munt N, Salvesen GS, Bredesen DE. The DCC gene product induces apoptosis by a mechanism requiring receptor proteolysis. Nature. 1998;395:801–4. doi: 10.1038/27441. [DOI] [PubMed] [Google Scholar]
- 35.Lo KW, Kwong J, Hui AB, Chan SY, To KF, Chan AS, et al. High frequency of promoter hypermethylation of RASSF1A in nasopharyngeal carcinoma. Cancer Res. 2001;61:3877–81. [PubMed] [Google Scholar]
- 36.Niwa T, Tsukamoto T, Toyoda T, Mori A, Tanaka H, Maekita T, et al. Inflammatory processes triggered by Helicobacter pylori infection cause aberrant DNA methylation in gastric epithelial cells. Cancer Res. 2010;70:1430–40. doi: 10.1158/0008-5472.CAN-09-2755. [DOI] [PubMed] [Google Scholar]
- 37.Brait M, Begum S, Carvalho AL, Dasgupta S, Vettore AL, Czerniak B, et al. Aberrant promoter methylation of multiple genes during pathogenesis of bladder cancer. Cancer Epidemiol Biomarkers Prev. 2008;17:2786–94. doi: 10.1158/1055-9965.EPI-08-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shivakumar L, Minna J, Sakamaki T, Pestell R, White MA. The RASSF1A tumor suppressor blocks cell cycle progression and inhibits cyclin D1 accumulation. Mol Cell Biol. 2002;22:4309–18. doi: 10.1128/MCB.22.12.4309-4318.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu S, Ewertz M, Tufano RP, Brait M, Carvalho AL, Liu D, et al. Detection of serum deoxyribonucleic acid methylation markers: a novel diagnostic tool for thyroid cancer. J Clin Endocrinol Metab. 2006;91:98–104. doi: 10.1210/jc.2005-1810. [DOI] [PubMed] [Google Scholar]
- 40.Jen J, Harper JW, Bigner SH, Bigner DD, Papadopoulos N, Markowitz S, et al. Deletion of p16 and p15 genes in brain tumors. Cancer Res. 1994;54:6353–8. [PubMed] [Google Scholar]
- 41.Gozuacik D, Bialik S, Raveh T, Mitou G, Shohat G, Sabanay H, et al. DAP-kinase is a mediator of endoplasmic reticulum stress-induced caspase activation and autophagic cell death. Cell Death Differ. 2008;15:1875–86. doi: 10.1038/cdd.2008.121. [DOI] [PubMed] [Google Scholar]
- 42.Li GM, Modrich P. Restoration of mismatch repair to nuclear extracts of H6 colorectal tumor cells by a heterodimer of human MutL homologs. Proc Natl Acad Sci U S A. 1995;92:1950–4. doi: 10.1073/pnas.92.6.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bolderson E, Scorah J, Helleday T, Smythe C, Meuth M. ATM is required for the cellular response to thymidine induced replication fork stress. Hum Mol Genet. 2004;13:2937–45. doi: 10.1093/hmg/ddh316. [DOI] [PubMed] [Google Scholar]
- 44.Carvalho AL, Jeronimo C, Kim MM, Henrique R, Zhang Z, Hoque MO, et al. Evaluation of promoter hypermethylation detection in body fluids as a screening/diagnosis tool for head and neck squamous cell carcinoma. Clin Cancer Res. 2008;14:97–107. doi: 10.1158/1078-0432.CCR-07-0722. [DOI] [PubMed] [Google Scholar]
- 45.Benoit NE, Goldenberg D, Deng SX, Rosenbaum E, Cohen Y, Califano JA, et al. Colorimetric approach to high-throughput mutation analysis. Biotechniques. 2005;38:635–9. doi: 10.2144/05384PF01. [DOI] [PubMed] [Google Scholar]
- 46.SAS/STAT Users Guide. Cary: SAS Institute Inc., 1999. [Google Scholar]