Abstract
Chronic infection with the HIV results in poor HIV-specific CD4 T cell proliferation, but more recent analyses using intracellular cytokine staining demonstrated that IFN-γ-producing, HIV-specific CD4 T cells can be detected for years in HIV-infected subjects. Because it is not known whether the majority of HIV-specific T cells are lost or become dysfunctional, we examined the kinetics of the T cell response over an extended period of time using a panel of 10 HLA-DR tetramers loaded with HIV p24 peptides. Tetramer+ CD4 T cells were present at a relatively high frequency during acute infection, but the size of these populations substantially contracted following suppression of viral replication. Short-term cessation of antiretroviral therapy resulted in a burst of viral replication and concomitant expansion of tetramer+ CD4 T cells, and these populations again contracted following reinitiation of therapy. The kinetics with which these cell populations contracted were characteristic of effector T cells, a conclusion that was supported by their phenotypic (CCR7−CD45RA−) and functional properties (IFN-γ+). Continued high-level viremia resulted in the physical loss of the majority of tetramer+ CD4 T cells, and the decline of HIV p24-specific CD4 T cells occurred more rapidly and was more substantial than the reduction of total CD4 T cell numbers. We conclude that the population of HIV p24-specific CD4 T cells is initially responsive to changes in the levels of viral Ags, but that the majority of these cells are lost in a setting of chronic viremia.
Infection with the HIV leads to destruction of CD4 T cells and a severe immunodeficiency state characterized by opportunistic infections (1, 2). The CD8 T cell response to the virus has been analyzed in great detail with the MHC class I tetramer technology introduced in 1996 and single-cell techniques for definition of T cell function (3, 2). However, although animal models and data in humans indicate that virus-specific CD4 T cells are critical for effective immune control, in part due to help provided to CD8 T cells (4 –10) and B cells (11), much less is currently known about the CD4 T cell response to the virus due to the lower frequency of HIV-specific CD4 T cells and technical difficulties in the generation of MHC class II tetramers. A precise understanding of the CD4 T cell response to the virus may be critical for the design of preventive or therapeutic vaccines.
Although HIV-specific lymphocyte proliferation is usually weak or undetectable in chronically infected subjects, CD4 T cell proliferation to HIV Ags is detectable in early HIV infection or in subjects with spontaneous control of viral replication (long-term nonprogressors) (12–18). The lack of a proliferative response during chronic infection could either be due to deletion of HIV-specific cells, e.g., by preferential infection of HIV-specific CD4 T cells during early stages of the disease (19, 20), or due to an impaired proliferative capacity of these cells. The prevailing notion that the majority of HIV-specific CD4 T cells are lost during early stages of the infection was challenged by the finding that CD4 T cells producing IFN-γ following stimulation with HIV Ags could be detected in the majority of chronically infected subjects even in the absence of HIV-specific proliferative responses (21). Resolution of this issue requires a quantitative assessment of HIV-specific CD4 T cell populations throughout the course of HIV disease.
Labeling with MHC class II tetramers would be suitable for addressing the question of how HIV-specific CD4 T cell populations change over extended periods of time. Scriba et al. (22) recently reported that tetramers of HLA-DR1 loaded with two peptides from HIV p24 could be used for direct ex vivo visualization of p24-specific T cell populations. These authors attempted to examine the kinetics of the CD4 T cell response in relationship to viral replication and measured the frequency of p24-tetramer+ CD4 T cells before, during, and after short-term antiretroviral therapy (ART),3 which was administered over a course of 2–5 mo. During this single cycle of treatment administration and withdrawal, no significant change in the frequency of p24-tetramer+ CD4 T cells was observed, but an increased turnover of these cells was noted based on labeling with the activation marker Ki67. These results were limited, however, by the relatively short duration of follow-up.
We have examined the kinetic relationship between HIV replication and the CD4 T cell response to the virus over an extended period of time using tetramers for multiple HLA-DR alleles loaded with a number of different HIV p24 peptides and observed substantial changes in the frequency of tetramer+ CD4 T cells in response to treatment initiation, short-term cessation of antiretroviral therapy, and sustained viremia. HIV p24-tetramer+ CD4 T cells had the phenotypic characteristics of effector rather than central memory T cells, and the kinetics of expansion and contraction of these cell populations in response to viral replication was characteristic of effector T cell pools that contract in size following substantial reduction or clearance of Ag. Persistent HIV viremia resulted in the physical disappearance of the majority of these cells.
Subjects and Methods
Subjects
Thirteen HIV-positive individuals were recruited to this study at the Massachusetts General Hospital and the Brigham and Women’s Hospital. Freshly isolated lymphocytes were analyzed from 10 patients (see Table II). Frozen cells from four of these patients and an additional three patients were examined at multiple time points to define the kinetics of the CD4 T cell response to the virus over extended periods of time. The appropriate institutional review boards approved the study and all participants gave informed consent. HIV plasma viral loads were measured by RT-PCR (Amplicor; Roche Molecular Systems); the detection limit for this assay was 50 RNA copies per ml. HLA-DR typing was performed by SSP-PCR using a combination of primers that permitted precise definition of DRB1 alleles (23, 24). Six HIV-negative healthy donors were included as control subjects.
Table II.
Analysis of HIV p24-specific CD4 T cells with HLA-DR tetramers in patients
| Subject | Years since Diagnosis | HIV RNA (copies/ml) | CD4 Count (cells/μl) | HLA-DR Allele | Control Tetramer
|
HIV p24 Tetramers
|
||
|---|---|---|---|---|---|---|---|---|
| Peptide | Tetramer+ cells/106 CD4 | Peptides | Tetramer+ cells/106 CD4 | |||||
| Non-progressors | ||||||||
| LT02 | 13 | 1,780 | 626 | DRB1*1501 | PLP | 14 | p24–1 | 25 |
| p24–3 | 14 | |||||||
| DRB5*0101 | Ann II | 16 | p24–4 | 275 | ||||
| LT11 | 14 | ND | ND | DRB1*0401 | Ann II | 26 | p24–1 | 140 |
| p24–2 | 407 | |||||||
| p24–4 | 735 | |||||||
| Low viral load | ||||||||
| AC120 | 1.15 | 2,620 | 659 | DRB1*0401 | Ann II | 3 | p24–4 | 436 |
| 1.41 | 2,020 | 921 | DRB1*0401 | Ann II | 15 | p24–1 | 67 | |
| p24–3 | 58 | |||||||
| p24–4 | 119 | |||||||
| 1.71 | 1,480 | 981 | DRB1*0401 | Ann II | 11 | p24–4 | 51 | |
| ART | ||||||||
| AC62 | 3.47 | <50 | 1098 | DRB1*0401 | Ann II | 11 | p24–2 | 31 |
| p24–4 | 60 | |||||||
| AC29 | 5.71 | 53 | 1111 | DRB1*0401 | Ann II | 9 | p24–2 | 108 |
| p24–4 | 85 | |||||||
| AC03 | 6.72 | 149 | 549 | DRB1*1501 | p24–3 | 31 | ||
| DRB1*0401 | Ann II | 16 | p24–3 | 28 | ||||
| p24–4 | 102 | |||||||
| DRB5*0101 | Ann II | 9 | p24–4 | 120 | ||||
| Off therapy | ||||||||
| AC15 | 5.84 | 41,100 | 448 | DRB1*1501 | p24–3 | 35 | ||
| DRB5*0101 | Ann II | 8.7 | p24–4 | 199 | ||||
| 6.14 | 52,600 | 408 | DRB5*0101 | Ann II | 12 | p24–4 | 125 | |
| 6.53 | 75,700 | 437 | DRB5*0101 | Ann II | 13 | p24–4 | 142 | |
| AC20 | 5.98 | 21,000 | 733 | DRB1*0101 | CLIP | 5 | p24–1 | 9 |
| p24–3 | 3 | |||||||
| p24–4 | 121 | |||||||
| AC06 | 6.51 | >750,000 | 211 | DRB5*0101 | Ann II | 3 | p24–4 | 27 |
| AC05 | 6.93 | 19,600 | 336 | DRB1*1501 | p24–1 | 51 | ||
| p24–3 | 7 | |||||||
| DRB5*0101 | Ann II | 16 | p24–4 | 109 | ||||
| 7.14 | 32,600 | 312 | DRB5*0101 | Ann II | 19 | p24–4 | 25 | |
Analysis of HIV p24-specific CD4 T cells with HLA-DR tetramers in patients with different clinical characteristics and HLA-DR alleles. Freshly isolated CD4 T cells from 10 patients were labeled with control tetramers (proteolipid protein (PLP), annexin II (Ann II) and CLIP self-peptides as controls) or HIV tetramers with four different peptides from the p24 protein. Tetramers correspond to the HLA-DR alleles and peptides listed in Table I. The frequency of tetramer+ T cells per 106 CD4 T cells is indicated, as well as clinical characteristics of these patients (years since diagnosis, viral load, and CD4 T cell count). Labeling with HIV p24 tetramers was considered positive when the T cell population was >3-fold larger compared to control tetramers, and these numbers are underlined.
Tetramer production
Peptide competition assays were used to identify peptides from the HIV p24 protein suitable for the loading of four HLA-DR molecules as previously described (25). Briefly, overlapping HIV gag p24 peptides (20-mer, 10 aa overlaps) were used as competitors for binding of a biotinylated myelin basic protein peptide (residues 85–99) to the DRA, DRB1*1501 molecule (abbreviated as DRB1*1501) or a biotinylated influenza hemagglutinin peptide (residues 306 –318) to DRB1*0101, DRB1*0401 and DRB5*0101 molecules. The HLA-DR molecules represented precursors with a covalently linked CLIP peptide, which was released by thrombin cleavage of the linker. Following an overnight incubation of the HLA-DR molecules with biotinylated indicator peptides and unlabeled competitor peptides at 37°C, HLA-DR molecules were captured on mAb L243-coated 96-well plates, and the HLA-DR-bound biotinylated peptide was quantitated with europium-labeled streptavidin using a DELFIA 1234 fluorometer (Wallac). Tetramers were made from the HLA-DR/CLIP precursors as previously described using the peptides listed in Table I. For tetramer formation, biotinylated HLA-DR/peptide complexes were incubated with R-PE-labeled streptavidin for >1 h at 4°C.
Table I.
Peptides used for the generation of HLA-DR tetramersa
| HLA-DR Allele | Peptide Residues | Abbreviation | Sequence | IC50 (μM) | Loading Efficiency (%) |
|---|---|---|---|---|---|
| DRB1*0101 | CLIP (87–101) | CLIP | PVSKMRMATPLLMQA | ND | ND |
| p24 (33– 46) | p24 –1 | SPEVIPMFSALSEG | 2.3 | 80 | |
| p24 (138 –150) | p24 –3 | LNKIVRMYSPTSI | 1.9 | 53 | |
| p24 (166 –179) | p24 – 4 | DRFYKTLRAEQASQ | 4.5 | 100 | |
| DRB1*0401 | Annexin II (208 –223) | Ann II | DVPKWISIMTERSVPH | 0.58 | 95 |
| p24 (33– 46) | p24 –1 | SPEVIPMFSALSEG | 0.7 | 87 | |
| p24 (113–127) | p24 –2 | EQIGWMTNNPPIPVG | 5 | 78 | |
| p24 (138 –150) | p24 –3 | LNKIVRMYSPTSI | 3 | 90 | |
| p24 (166 –179) | p24 – 4 | DRFYKTLRAEQASQ | 2 | 99 | |
| DRB1*1501 | PLP (90 –109) | PLP | FYTTGAVRQIFGDYKTTICG | ND | 77 |
| p24 (33– 46) | p24 –1 | SPEVIPMFSALSEG | 0.68 | 63 | |
| p24 (138 –150) | p24 –3 | LNKIVRMYSPTSI | 0.22 | 59 | |
| DRB5*0101 | Annexin II (208 –223) | Ann II | DVPKWISIMTERSVPH | 1.8 | 27 |
| p24 (166 –179) | p24 – 4 | DRFYKTLRAEQASQ | 2 | 79 |
A total of 10 tetramers were generated with four distinct HIV p24 peptides and four different HLA-DR molecules. The peptides were identified as ligands for the respective HLA-DR molecule in a competition assay, and the IC50 values from these experiments are listed. HLA-DR molecules with bound peptides from self-proteins were used as controls, the invariant chain-derived CLIP peptide, as well as peptides from annexin II (Ann II) and proteolipid protein (PLP). HIV, annexin, and proteolipid protein peptides carried an N-terminal affinity tag (DNP), and HLA-DR molecules loaded with these peptides were affinity purified using an anti-DNP column. The loading efficiency represents the fraction of HLA-DR that bound to the anti-DNP column.
Tetramer staining
Staining of lymphocytes from fresh blood samples was done by isolating CD4 T cells using the RosetteSep CD4+ T cell enrichment mixture (Stem-Cell Technologies) according to the manufacturer’s instructions. This deletion mixture contains bivalent Abs to CD8, CD16, CD19, CD36, and CD56 that lead to binding of cells expressing these markers to glycophorin A on RBC. Blood samples were incubated with the Ab mixture for 20 min at room temperature (RT) and CD4 T cells were then separated from RBC on a density gradient. The cells were washed with PBS/2%FCS and erythrocytes were lysed using a RBC lysis buffer (Sigma-Aldrich). The cells were then suspended in R-10 medium (RPMI 1640 with 10% human serum, 2 mM L-glutamine, 50 IU/ml penicillin, 50 μg/ml streptomycin, and 10 mM HEPES) and tetramers were added to a final concentration of 10 μg/ml. Staining was done at RT for 50 min and Abs CD4-allophycocyanin-Cy7, CD45RA-allophycocyanin, CCR7-PECy7, as well as exclusion markers CD8-FITC, CD14-FITC, CD16-FITC, CD19-FITC, CD56-FITC, and BD Via-Probe (7-aminoactinomycin D (7AAD)) were then added for 20 min at RT (all BD Immunocytometry Products or BD Pharmingen). Cells were washed and fixed in PBS containing 1% formaldehyde and 1 μg/ml actinomycin D.
HIV-specific T cells were examined in cryopreserved samples by enrichment of tetramer-labeled cells with magnetic microbeads directed against the PE fluorophore bound to the streptavidin component of tetramers. Cryopreserved PBMCs were stained in R-10 medium for 40 min at RT with tetramers at a concentration of 16 μg/ml, and CCR7-allophycocyanin (R&D Systems) was added during the last 10 min of incubation. CD4-Alexa 405 (Caltag Laboratories), CD45RA-FITC, and exclusion Abs (CD8-PerCP, CD14-PerCP, and CD19-PerCP) were then added and the reaction was incubated for 20 min on ice. Washed cells were incubated with anti-PE microbeads (Miltenyi Biotec) and BD Via-Probe for 15 min at 4°C. Cells were then washed with buffer containing 1 μg/ml actinomycin D (Calbiochem), which was present in all subsequent steps; 90% of the cells were used for magnetic enrichment of tetramer+ cells on a MS column (Miltenyi Biotec) according to the manufacturer’s instructions. The other 10% were used to determine the total CD4 T cell count before enrichment. All cells were fixed in PBS containing 1 μg/ml actinomycin D and 1% formaldehyde. For a detailed phenotypic analysis, combinations of CD27-FITC plus CD28-allophycocyanin (BD Pharmingen) or CD57-FITC (Caltag Laboratories) plus IL-7R-allophycocyanin (R&D Systems) were used in the staining reaction. The number of total PBMCs used for each staining reaction varied from 5 to 10 × 106 depending on the availability of frozen samples for the particular time point. PBMCs from healthy donors that had the relevant HLA-DR alleles were also tested as controls with HIV p24 tetramers.
Flow cytometric analysis was done using the FACSCalibur or the LSR2 flow cytometers (BD Immunocytometry Systems). FCS files were then exported and analyzed using FlowJo software (Tree Star). The lymphocyte gate was set based on forward and side scatter. Cells were gated on the CD4+CD8−CD14−CD19−7AAD− population and then plotted as tetramer vs CD4, CD45RA, CCR7, CD27, CD28, CD57, or IL-7R. The number of tetramer+ T cells from tetramer-enriched samples was calculated per 106 CD4 T cells based on the total number of CD4 T cells in the pre-enrichment sample.
Cytokine secretion assays
Fresh or frozen PBMCs were incubated with 10 μg/ml HIV p24-4 or control myelin basic protein (88 –102) peptide for 4 h at 37°C and a CD28 Ab (BD Biosciences) in RPMI 1640 supplemented with 5% human serum, 2 mM L-glutamine, 50 U/ml penicillin, 50 μg/ml streptomycin, and 10 mM HEPES. Cell surface detection of IFN-γ or IL-2 secretion was done using the IFN-γ or IL-2 Secretion Assay kits (Miltenyi Biotec) according to the manufacturer’s protocol. Briefly, cells were washed and labeled for 5 min with either IL-2 or IFN-γ capture reagent and then incubated for 45 min at 37°C for the secretion and capture phase. Cells were washed and stained with CD4-Alexa 405, CD69-allophycocyanin-CY7, CD45RA-FITC, CCR7-allophycocyanin, exclusion markers (CD8-PerCP, CD14-PerCP, CD19-PerCP), and IL-2-PE or IFN-γ-PE detection Abs for 40 min on ice. Washed cells were incubated with anti-PE microbeads and BD Via-Probe for 15 min at 4°C. Cells were then split, 40% for direct analysis by flow cytometry and 60% for magnetic enrichment on a MS column. After BD Via-Probe addition, all subsequent steps were done in the presence of 1 μg/ml actinomycin D in all buffers. Cells were fixed before flow cytometric analysis.
Combined IFN-γ and tetramer labeling of cells was done as described above, except that the cells were first incubated with 13 μg/ml HIV p24-4 tetramer or control tetramer for 40 min at RT and incubated with p24-4 or control peptide. Since the tetramers were labeled with PE, IFN-γ-allophycocyanin, and CD69-PE-CY7 Abs were used. For data analysis, cells were gated based on forward scatter and side scatter and CD4+CD8−CD14−CD19−Via-Probe− populations were analyzed for CD69 vs cytokine labeling. For combined IFN-γ and tetramer-labeling experiments, CD69+ cells were analyzed for IFN-γ vs tetramer fluorescence.
Statistical analysis
The reduction in the frequency of HIV-tetramer+ T cells following initiation of ART during acute infection was examined using the two-sided one-sample t test. Five patients for whom tetramer-based analysis could be performed at a sufficient number of time points during this stage of the disease were included in the analysis.
Results
Identification of HIV p24-specific CD4 T cells in patients with different clinical characteristics and MHC class II alleles
Analysis of CD4 T cell populations with MHC class II tetramers in humans requires the utilization of multiple allelic forms when a significant fraction of the patient population needs to be analyzed. Functional studies can be used to provide information on major T cell epitopes, but such experiments do not precisely define the MHC class II restriction elements because most patients are heterozygous at the MHC locus. To precisely define which HIV p24 peptides would be suitable for the generation of MHC class II tetramers, we performed systematic peptide-binding studies with an overlapping set of peptides covering the entire HIV p24 sequence and four HLA-DR molecules that we had previously expressed (DRB1*0101, DRB1*0401, DRB1*1501, and DRB5*0101) (25). These peptide-binding experiments demonstrated that three HIV p24 peptides (residues 31–52, 131–152, and 161–182) bound to all four HLA-DR molecules studied, indicating that these peptides are promiscuous binders and may thus represent T cell epitopes in a significant fraction of HIV-infected subjects. For DRB1*0401, 4 of a total of 23 p24 peptides were identified as ligands; 3 of these peptides represented the promiscuous binders described above while the fourth peptide (residues 111–132) bound only to DRB1*0401. All four epitopes were used for the generation of tetramers so that a complete analysis of HIV p24-specific, DRB1*0401-restricted T cells could be performed. For the generation of tetramers, the length of these HIV p24 peptides was reduced from 20 to 13–15 aa based on the known HLA-DR-binding motifs. A total of 10 different HLA-DR tetramers were generated with these HIV peptides using an approach in which the CLIP peptide that was covalently linked to the HLA-DRβ chain was released following linker cleavage and then replaced with the peptide of interest (Table I) (25). Homogenous HLA-DR/p24 peptide complexes were isolated by affinity chromatography using an epitope tag (DNP) attached to the N terminus of these peptides. This purification step also permitted determination of the loading efficiency, which ranged from 53% to close to 100% for the different peptides. In addition, a number of different control HLA-DR/peptide complexes were generated using self-peptides (Table I).
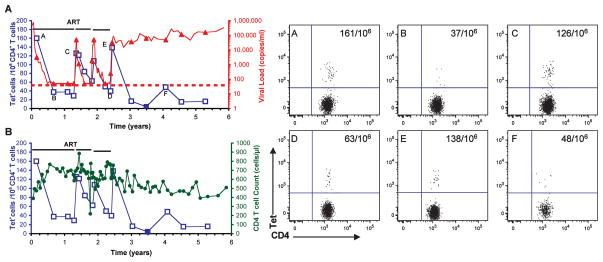
We initially focused our efforts on the analysis of CD4 T cells that were freshly isolated from patients representing different clinical groups (Fig. 1 and Table II). Two of these patients were long-term nonprogressors (LT01 and LT11) with spontaneous control of viremia for 13 and 14 years, respectively, as well as a patient who spontaneously maintained a low viral load for more than 1 year after acute HIV infection (AC120). We also examined three patients receiving ART as well as four patients who were currently off therapy and as a consequence had high viral loads and low CD4 counts. All HLA-DR alleles for which tetramers were available were represented in this patient group: one patient was heterozygous for the DRB1*1501 and the DRB1*0401 haplotypes, four patients carried the DRB1*1501 haplotype, four the DRB1*0401, and one the DRB1*0101 haplotype. HIV p24-specific CD4 T cells could be detected with the HIV tetramers for every p24 peptide identified in the binding studies. We systematically evaluated conditions that reduced background staining and found that enrichment of CD4 T cells from PBMCs, exclusion of irrelevant cell populations (with FITC-labeled Abs to CD8, CD14, CD16, CD19, and CD56), and exclusion of apoptotic cells (by labeling with 7AAD) were useful. The control tetramers showed a low level of background staining, generally <1:50,000 CD4 T cells. We considered cell populations that were 3-fold larger than the background as positive, and most populations identified with the HIV p24 tetramers were >10-fold larger than background.
FIGURE 1.
Ex vivo analysis of HIV p24-tetramer+ CD4 T cells in different patient groups. Representative examples are shown for a long-term nonprogressor (A, patient LT11), a patient off therapy (B, patient AC20), and a patient under ART (C and D, patient AC03). Freshly isolated PBMCs were enriched for CD4 T cells and labeled with control or HIV p24 tetramers (Tet), Abs to CD4, CCR7, and CD45RA, and exclusion markers CD8, CD14, CD16, CD19, CD56, and 7AAD. (CCR7 and CD45RA Abs were omitted for the staining done on LT11.) FACS plots show CD4 vs tetramer staining for the CD4+CD8−CD14−CD16−CD19−CD56−, and 7AAD− population, with the frequency of tetramer+ CD4 cells (per 106 CD4 T cells) indicated in the right-hand corner. The tetramers comprised three different DR alleles, DRB1*0401, DRB5*0101, and DRB1*0101, loaded with four different HIV p24 peptides (p24-1, p24-2, p24-3, or p24-4) or control peptides (CLIP for DRB1*0101, annexin II for DRB5*0101 and DRB1*0401). CD4 T cells specific for the p24-4 peptide were identified in patients with different DR alleles using tetramers of DRB1*0401 (A and D), DRB1*0101 (B), and DRB5*0101 (C) loaded with this HIV peptide. HIV p24-positive CD4 T cells were detected in patients from all three groups and at the highest frequency in the long-term nonprogressor.
HIV p24-tetramer+ cells could be identified in all patients studied even though they belonged to different clinical groups and carried different HLA-DR alleles. Among patients, the frequency of HIV p24-tetramer+ cells spanned a wide range, from 1 per 1,360 cells to 1 per 37,037 CD4 T cells. The highest frequency was observed in the two long-term nonprogressors (LT02 and LT11) and in patient AC120 who had spontaneously maintained a low viral load for more than 1 year after acute HIV infection. In patients with a high frequency of HIV p24-specific T cells, T cells directed against multiple epitopes were identified with these tetramers. The HIV p24-4 peptide emerged as the dominant epitope for the p24 Ag in the context of three different DR molecules (DRB1*0101, DRB1*0401, DRB5*0101) and in every patient the HIV p24-4 tetramer identified the largest population of tetramer+ cells. These results confirm that the promiscuous p24-4 peptide represents a major epitope in patients with different clinical characteristics and MHC class II alleles, consistent with other studies (22, 26, 27).
The dynamics of the CD4 T cell response in relationship to viral replication
Little is known about the kinetics of the CD4 T cell response to HIV throughout the course of the disease, in part because such studies require highly reproducible measurements of CD4 T cell frequency and function. Functional assays are prone to substantial variability across time points, but tetramer-based analysis can be highly reproducible because it only involves the physical interaction of the tetramer with the TCR on the relevant T cell populations. Such analyses require samples collected over extended periods of time so that the CD4 T cell response during acute infection, in response to ART, and during disease progression can be analyzed, and can thus only be performed in a practical manner with frozen T cells. Sensitive techniques for the detection of tetramer+ T cells are required so that these cells can be studied at stages of the disease when CD4 T cell responses decline. We used a method for the magnetic enrichment of tetramer-labeled CD4 T cells which permits investigation of CD4 T cell populations present at very low frequencies (1/100,000) because large numbers of input cells can be analyzed and background staining is substantially reduced (25). A control experiment in which cells from subject AC20 were analyzed from the same time point, both fresh and after freezing, yielded very similar frequencies (92 HIV p24-4-tetramer+ cells per 106 CD4 T cells for the cryopreserved sample, compared with 88 per 106 CD4 T cells for the fresh sample). Scriba et al. (22) recently demonstrated linear recoveries of tetramer+ T cells with this enrichment technique by spiking T cells from a T cell clone into PBMCs from an unrelated donor. Linear recoveries were observed when the frequency of T cells from the clone was varied over more than three orders of magnitude. Frozen PBMCs were labeled with HIV p24 tetramers, Abs to CD4, CCR7, and CD45RA, as well as exclusion markers (Abs to CD8, CD14, and CD19 as well as 7AAD). Tetramer-labeled cells were then enriched with anti-PE magnetic microbeads directed against the fluorophore attached to streptavidin. Labeling with control tetramers demonstrated very low levels of background, ranging from 0 to 2 control tetramer+ cells per sample. The frequency of cells labeled with control tetramers thus ranged from 0 to 1.65 per 106 CD4 T cells; frequencies >10 per 106 were considered to be positive for labeling with HIV tetramers (data not shown). We also conducted control experiments with T cells from six HIV-negative subjects with the appropriate MHC class II alleles and observed low levels of background labeling with the HIV tetramers (0 –3 tetramer+ cells per 106 CD4 T cells).
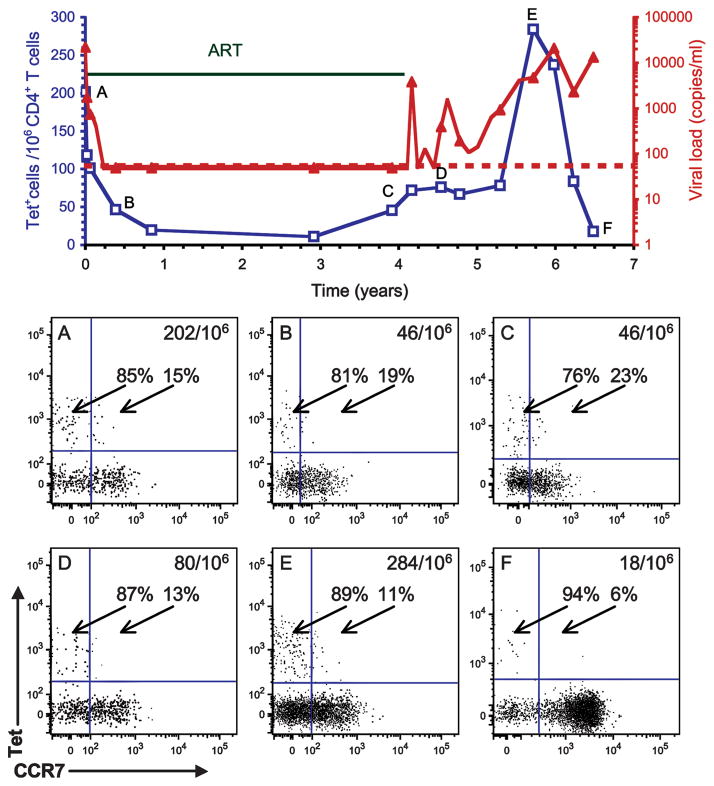
We selected seven patients with the appropriate MHC class II alleles for these studies from whom frozen cells were available for an observation period of several years, starting at the acute stage of infection, and performed detailed kinetic analyses of HIV p24-tetramer+ cells in relationship to viral load and in response to therapy (Figs. 2–4). Analysis of T cells from patient AC26 demonstrated a close relationship between the level of viral replication and the kinetics of the CD4 T cell response to the virus (Fig. 2A, examples of FACS plots are shown for six key time points over the course of observation). Before initiation of ART, this patient had a very high viral load (>750,000 copies/ml) during the acute stage of infection. At this time there was a high frequency of HIV p24-4-specific T cells in the peripheral blood (Fig. 2, time point A). Initiation of ART led to a sharp decline in the viral load, which was mirrored by a substantial reduction in the frequency of tetramer+ CD4 T cells (Fig. 2, time point B). Fifteen months following diagnosis when the virus was undetectable, the patient elected to discontinue therapy. This led to a sharp rise in the viral load, which was mirrored by a substantial expansion of the pool of HIV p24-tetramer+ T cells. ART was reinitiated, and the reduction in viral load again resulted in a drop in the frequency of HIV p24-tetramer+ T cells, a pattern that repeated itself with subsequent interruptions in therapy (Fig. 2, time points C–E). Two and one-half years following diagnosis, the patient discontinued antiviral therapy, resulting in sustained high-level viremia. After an initial expansion of p24-tetramer+ T cells, this population quickly diminished in size with continued high-level virus production (Fig. 2, time point F). These results show that the frequency of p24-tetramer+ cells was high during acute infection and that the frequency of these cells dropped substantially following initiation of ART. Short bursts of viral replication resulted in a transient increase in the number of p24-tetramer+ T cells, but sustained high-level viremia led to the physical disappearance of the majority of HIV p24-tetramer+ cells.
FIGURE 2.
Longitudinal analysis starting at acute infection demonstrates the relationship between viral load and the CD4 T cell response to HIV p24 (A). Frozen PBMCs from patient AC26 were stained with the DRB1*0401 p24-4 tetramer (Tet) and tetramer+ cells were enriched with anti-PE magnetic microbeads directed against the fluorophore linked to streptavidin. The graph shows the viral load (red line, copies of viral RNA per ml) and the frequency of HIV p24-4-tetramer+ CD4 T cells (blue line, tetramer+ cells per 106 CD4 T cells). Filled blue squares indicate the detection of <10 tetramer+ cells per 106 CD4 T cells. Horizontal green bars indicate time periods during which the patient received ART; staggering of this line indicates a change in the drug regimen. The limit of detection for the viral load was 50 copies/ml, indicated by the red dotted line. Examples of FACS plots are shown for six time points (time points A–F). During acute infection, HIV p24-4-tetramer+ T cells were present at a high frequency (time point A) and suppression of viral replication led to a substantial reduction in the frequency of these cells (time point B). Interruptions in ART resulted in spikes of viral replication and expansion of p24-4 T cells (time points C–E), but sustained high levels of virus production led to the physical loss of the majority of these cells (time points beyond 3 years after diagnosis). B, Total CD4 T cell counts (green line, cells per μl) and frequency of HIV p24-4-tetramer+ CD4 T cells (blue line, tetramer+ cells per 106 CD4 T cells) were plotted, demonstrating preferential loss of HIV-specific CD4 T cells.
FIGURE 4.
Effector memory phenotype of HIV p24-specific CD4 T cells during acute infection, periods of undetectable viral replication as well as periods of high-level viremia. Frozen PBMCs from patient AC20 covering a 6-year time span were labeled with DRB1*0101 p24-4 tetramer (Tet), Abs to CD4, CCR7, and CD45RA, and exclusion markers (CD8, CD14, CD19, 7AAD). Tetramer+ cells were enriched with anti-PE microbeads and CD4+ CD8−CD14−CD19−7AAD− cells were plotted for tetramer vs CCR7 staining. The percentage of tetramer+ cells that were CCR7− or CCR7+ is indicated on the FACS plots (A–F) that cover this time span. At all time points during this 6.6-year period, the majority of tetramer+ cells had a CCR7− phenotype.
We also observed important differences between the kinetics of HIV-tetramer+ CD4 T cell populations and the total CD4 T cell pool. First, total numbers of CD4 T cells increased following initiation of ART during acute infection, whereas the frequency of HIV-tetramer+ CD4 T cells declined. Second, numbers of total CD4 T cells remained quite stable during brief treatment interruptions while substantial changes in the frequency of HIV-tetramer+ CD4 T cells were observed. Third, following cessation of therapy, the frequency of HIV-tetramer+ CD4 T cells declined more rapidly and more substantially than total CD4 counts. These results demonstrate a preferential loss of HIV-positive CD4 T cells during periods of sustained viral replication.
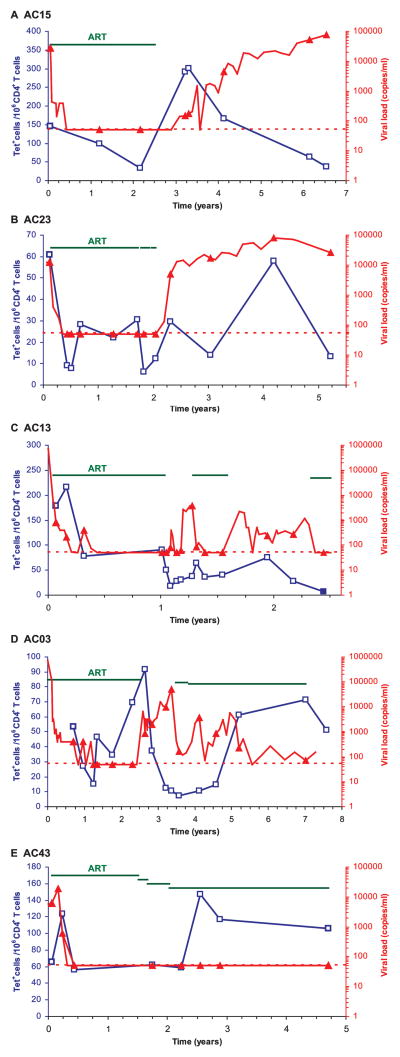
The same general patterns were observed in the other six patients for whom a detailed kinetic analysis was performed, even though in each patient the response had individual features. When treatment was initiated during acute infection, such as in patients AC23, AC13 (Fig. 3, B and C), and AC20 (Fig. 4), there was a substantial decline in the frequency of p24-tetramer+ cells following suppression of viral replication. The reduction in the frequency of HIV-tetramer+ T cells following initiation of therapy during acute infection was statistically significant by the two-sided one-sample t test ( p = 0.0027) for five patients for whom tetramer-based analysis could be performed at a sufficient number of time points during this stage of the disease. Transient increases in virus production led to expansion of HIV p24-tetramer+ cells, e.g., in patient AC20 (Fig. 4). However, in a number of patients, continued high-level viremia resulted in the physical loss of the majority of tetramer+ T cells, as for example seen in patients AC15 and AC23 (Fig. 3, A and B) and in patient AC20 (Fig. 4). In patient AC13 (Fig. 3C), progressively declining levels of HIV-tetramer+ CD4 T cells were observed following cessation of ART, which resulted in intermediate levels of virus production. In one patient (AC43), an increase in the frequency of tetramer+ T cells was observed following switching of medications, even though the virus was undetectable at the rather widely spaced time points at which it was measured. We cannot exclude the possibility that there was a transient increase in virus production that was not captured in this analysis.
FIGURE 3.
The in vivo dynamics of HIV p24-specific CD4 T cells over extended periods of time. Frozen PBMCs from five patients treated during acute infection were labeled with DRB5*0101 p24-4 (patients AC15 and AC03, A and D, respectively), DRB1*0401 p24-4 tetramers (Tet; patients AC23 and AC13, B and C, respectively) or DRB1*0401 p24-1 tetramer (patient AC43, E) and tetramer-labeled cells were enriched with anti-PE magnetic beads. The frequency of tetramer+ cells (blue line) and viral loads (red line) were plotted as in Fig. 2; different scales were used for the frequency of tetramer+ T cells in individual patients because of differences in the frequency of these cells. Filled blue squares indicate detection of <10 tetramer+ cells per 106 CD4 T cells. Staggering of the green line indicates a short interruption of treatment without change of drugs in patient AC03 (D) or change in the drug regimen for patient AC43 (E). Transient increases in virus production resulted in an increased frequency of HIV p24-specific CD4 T cells, while sustained suppression of viral replication resulted in a reduction of the frequency of HIV-tetramer+ cells. Sustained viremia led to substantial loss of these cells.
T cell proliferation assays had been performed on freshly isolated PBMCs from several of these patients at a number of time points and confirmed the well-established pattern of a vigorous proliferative response to the HIV p24 Ag during acute infection which declined over time. Because these analyses had been performed on freshly isolated cells by different investigators over a number of years, there was considerable variability in these measurements (including the proliferative response to the mitogen PHA used as a positive control) so that close correlations to tetramer labeling could not be made. In patients AC20 and AC26, a proliferative response to HIV p24 was absent at the end of the observation period, even though tetramer+ T cells could still be detected, albeit at a substantially lower frequency than at earlier time points.
HIV p24-tetramer+ T cells identified during acute infection had a phenotype characteristic of effector T cells. The majority of tetramer+ cells were CCR7− and CD45RA− in all patients for whom a kinetic analysis was performed, consistent with other studies (22, 28 –31). A surprising finding was that tetramer+ T cells maintained a CCR7−CD45RA− phenotype even when viral replication was not detectable based on RT-PCR analysis of plasma. As shown in Fig. 4, the majority of tetramer+ CD4 cells were CCR7− even during the 3.7-year period in which the virus was undetectable in plasma by RT-PCR. This phenotype was also maintained after the patient had stopped therapy and high-level virus production resumed, as shown for time points D–F in Fig. 4.
The phenotypic properties of these T cells were further defined by analysis of tetramer+ T cells in patient AC20 on day 8 and 4.8 years following initial diagnosis (Fig. 5A). The phenotype of those cells was remarkably similar at both time points. Memory T cells have been classified based on CD28 and CD27 expression as early memory T cells (CD28+CD27+), intermediate memory cells (CD28+CD27−), and late memory cells (both CD27−CD28−) (32, 33). Virtually all tetramer+ T cells were CD28+, some of which were CD27+ or CD27−. These cells were IL-7R− or IL-7Rlow, again consistent with an effector T cell phenotype (34). p24-tetramer+ T cells were negative for CD57, which has been described as a marker of replicative senescence, primarily for CD8 T cells (35). The vast majority of tetramer+ T cells in patient AC15 were CD45RA−CCR7−, indicating a continued effector memory T cell phenotype (Fig. 5B). Virtually all tetramer+ T cells were CD28−, but the majority of these cells had acquired a CD27+ phenotype. These studies are consistent with the conclusion that the majority of p24-tetramer+ T cells represent an effector memory rather than a central memory T cell population.
FIGURE 5.
Detailed phenotypic analysis of HIV p24-tetramer+ CD4 T cells. A, Cells from patient AC20 (time point 4.8 years) were analyzed by five-color FACS analysis using the DRB1*0101 p24-4 tetramer (Tet) and either Abs to CD27 and CD28 or Abs to the IL-7R and CD57. The majority of tetramer+ cells were IL-7Rlow or IL-7R−, and the majority of cells were also CD57−. B, Cells from patient AC15 (time point 6.5 years) were analyzed by five-color FACS analysis, which included the DRB5*0101 p24-4 tetramer and either Abs to CD27 and CD28 or Abs to CD45RA and CCR7. The vast majority of tetramer+ CD4 T cells in this patient were CD28+, CD45RA−, and CCR7−. The majority of tetramer+ cells were also negative for CD27 or CD27low. These cells therefore have the characteristics of an effector memory T cell population.
Functional properties of HIV p24-specific CD4 T cells
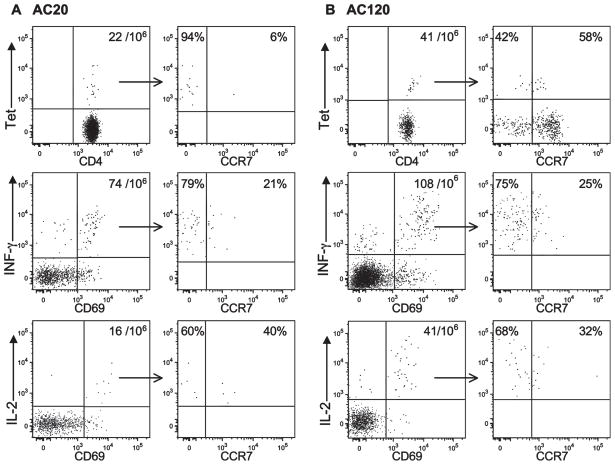
In an effort to further characterize the functional properties of these T cells, we performed parallel tetramer and surface cytokine capture analyses. For surface cytokine capture, PBMCs from patients AC20 and AC120 were stimulated for 4 h with the HIV p24-4 peptide. Following surface cytokine capture and labeling of cell surface-bound cytokine with an anti-PE Ab, cytokine-producing cells were enriched with anti-PE magnetic beads. Cytokine-positive cells that had up-regulated the CD69 activation marker were then analyzed in terms of CCR7 expression. The majority of IFN-γ+ and CD69+ CD4 T cells were negative for CCR7 expression in patient AC20 (79%). Only relatively small numbers of IL-2+ cells were present compared with IFN-γ + CD4 T cells (16 per 106, compared with 74 per 106, respectively) (Fig. 6A). Among IL-2-producing cells, CCR7− cells were again in the majority but a population of IL-2+CCR7+ cells was observed. The frequency of both IFN-γ and IL-2+ CD4 T cells was higher in patient AC120 who had spontaneously controlled viral replication without therapy for more than 1 year (Fig. 6B).
FIGURE 6.
Physical and functional characterization of HIV p24-specific T cells based on tetramer (Tet) labeling as well as IFN-γ and IL-2 surface capture. Freshly isolated PBMCs from patients AC20 (A) and AC120 (B) were labeled with either the DRB1*0101 p24-4 or DRB1*0401 p24-4 tetramer, respectively, and tetramer+ CD4 T cells that had been enriched with anti-PE magnetic beads were analyzed with respect to CCR7 expression (upper panels). In parallel, separate aliquots of cells were stimulated with p24-4 peptide for 4 h, followed by cytokine surface capture assays for IFN-γ (middle panels) or IL-2 (lower panels). IFN-γ + and IL-2+ cells were enriched using anti-PE beads. CD69+ and cytokine+ cells were plotted for cytokine vs CCR7 fluorescence. The percentages of CD69+/cytokine+ cells that were CCR7− or CCR7+ are indicated. The majority of p24-4-specific CD4 T cells secreted IFN-γ and were CCR7−; IL-2-producing cells were observed at a lower frequency. Patient AC120 spontaneously maintained a low viral load (time point 1.9 years), and patient AC20 had a high viral load following discontinuation of therapy (time point 6.5 years).
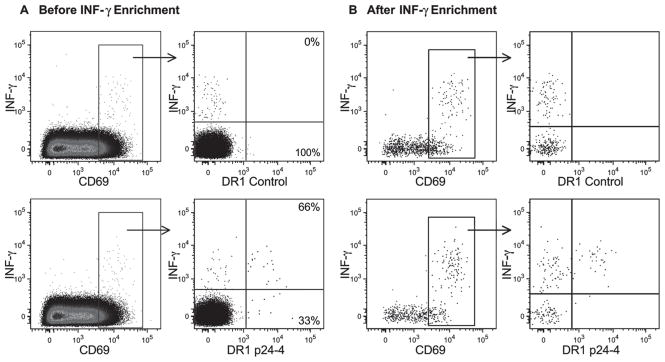
We next combined tetramer labeling with surface IFN-γ capture to determine the fraction of tetramer+ T cells producing this cytokine. PBMCs from patient AC20 were first labeled with a control tetramer or the HIV p24-4 tetramer for 40 min at RT and then cultured for 4 h in the presence of the p24-4 peptide and a CD28 Ab. Cells were gated on CD4+ T cells negative for the exclusion markers and the CD69+IFN-γ+ population was plotted for IFN-γ vs tetramer labeling (Fig. 7). With the DRB1*0101-CLIP control tetramer, no tetramer+ cells that were also IFN-γ+ were identified (Fig. 7, upper panels). However, a discrete population of cells that were both IFN-γ+ and tetramer+ was identified with the specific DRB1*0101 p24-4 tetramer. The majority of DRB1*0101 p24-4-tetramer+ cells were also labeled with the IFN-γ Ab (66%), indicating that the majority of tetramer+ cells produced this cytokine. Enrichment of IFN-γ-labeled cells with anti-allophycocyanin magnetic microbeads (Fig. 7B) confirmed the specificity of labeling. This experiment demonstrated that the majority of tetramer-labeled cells are functional and that these cells produce IFN-γ, a finding that is consistent with the effector phenotype of these cells. Cells labeled with IFN-γ but not the p24-4 tetramer were also identified and these cells may have recognized the promiscuous p24-4 peptide in the context of another HLA-DR molecule in this patient.
FIGURE 7.
The majority of HIV p24-4-tetramer+ CD4 T cells produce IFN-γ. Frozen PBMCs from patient AC20 were incubated with DRB1*0101 CLIP control tetramer or DRB1*0101 p24-4 tetramer for 40 min at RT and then cultured for 4 h at 37°C in the presence of p24-4 peptide and anti-CD28. In the cytokine capture assay, cells were labeled with an allophycocyanin-conjugated IFN-γ Ab and IFN-γ-producing cells were then enriched with magnetic anti-allophycocyanin beads. Cells were gated on the CD4+CD8−CD14−CD19−7AAD− population, and data are shown before (A) and after (B) enrichment of IFN-γ-producing T cells. CD69+ cells were plotted for IFN-γ vs tetramer labeling and the percentage of tetramer+ cells that were IFN-γ+ or IFN-γ− was evaluated. None of the cells labeled with the DRB1*0101 CLIP control tetramer were IFN-γ+ (upper panel) and 66% of the DRB1*0101 p24-4-tetramer+ cells produced IFN-γ. The magnetic enrichment (B) confirmed the specificity of labeling. IFN-γ+ cells that were negative for the DRB1*0101 p24-4 tetramer presumably recognized this HIV peptide in the context of another MHC class II molecule.
Discussion
These data demonstrate a close relationship between the magnitude and duration of HIV replication and the dynamics of the CD4 T cell pool to HIV p24. During acute infection, the frequency of HIV p24-tetramer+ CD4 T cells was relatively high, but declined rapidly following suppression of viral replication. Short-term cessation of ART and resulting viremia triggered expansion of the pool of circulating HIV p24-tetramer+ CD4 T cells, which contracted following reinitiation of therapy with kinetics characteristic of an effector T cell population. Because tetramer labeling permits assessment of T cell frequency independent of a particular T cell function, we could directly demonstrate that the majority of HIV p24-specific CD4 T cells are lost as a consequence of sustained viremia, even though smaller numbers of these cells remained detectable.
Throughout the course of infection and even in the setting of successful suppression of HIV viremia, these cells had all of the characteristics of an effector rather than a central memory CD4 T cell population (CCR7−CD45RA−, predominance of IFN-γ over IL-2-producing cells) (36). Our data thus resolve a long-standing issue in the field in which proliferation assays had demonstrated weak or absent proliferative responses during the chronic stages of infection (12, 13), while intracellular cytokine analysis had demonstrated the presence of CD4 T cells that produce IFN-γ (21). We conclude that the population of HIV p24-specific CD4 T cells is initially responsive to changes in the levels of viral Ags, but that the majority of these cells are lost in a setting of chronic viremia.
The kinetics of expansion and contraction of the HIV-specific CD4 T cell pool are consistent with the behavior of effector CD8 and CD4 T cells that have been defined in animal models of viral infection. In mice infected with the lymphocytic choriomeningitis virus (LCMV), a dramatic expansion of virus-specific CD8 T cells is observed which peaks at day 8 following infection, such that the majority of CD8 T cells in the spleen are specific for LCMV (~50 –70%). Following clearance of the virus, this virus-specific CD8 T cell pool rapidly contracts due to apoptosis of ~90 –95% of these effector cells, which express low levels of Bcl-2. The remaining virus-specific CD8 T cells form a memory T cell pool that is maintained at a constant size over extended periods of time (34, 37). The expansion of LCMV-specific CD4 T cells is less pronounced compared with CD8 T cells, and in contrast to the stable CD8 T cell population, the frequency of CD4 T cells continues to decline with a half-life of ~400 days, a finding that correlates with a lower level of Bcl-2 expression in memory CD4 compared with CD8 T cells (38). The less pronounced expansion and continued contraction of the virus-specific CD4 T cell pool account for the fact that the LCMV-specific memory CD4 T cell population is substantially smaller than the corresponding CD8 T cell population. The kinetics with which the HIV-specific CD4 T cell pool contracts following initiation of ART during acute infection resembles those observed for effector CD4 T cells in the murine LCMV model. The conclusion that the kinetic behavior of HIV-specific CD4 T cells is characteristic of effector T cells is supported by the phenotypic and functional studies performed on this T cell population. Specifically, the majority of HIV p24-tetramer+ CD4 T cells displayed a CD45RA−CCR7−CD28+CD27− effector memory phenotype.
Our kinetic data are also consistent with the observation that the HIV-specific CD4 and CD8 T cell responses (15, 39) decline following initiation of ART and demonstrate that the reduced levels of proliferation or cytokine production by CD4 T cells observed in other studies are due to contraction in the size of the pool of circulating HIV p24-specific CD4 T cells. This contraction of the pool of HIV p24-tetramer+ T cells was not observed by Scriba et al. (22), potentially because their study only assessed the kinetics of HIV-tetramer+ T cell populations during one short cycle of treatment that was stopped after 2–5 mo. Furthermore, treatment was initiated during acute infection when the frequency of these cells was high, and the duration of treatment was not sufficient for definition of the kinetic patterns that we observed by long-term analysis over a number of years. A substantial reduction in the frequency of circulating HIV-specific CD8 T cells has also been observed following initiation of ART based on MHC class I tetramer labeling (39), and the pools of both CD4 and CD8 T cells directed against the virus therefore contract when viral replication is suppressed.
During the acute stage of the infection characterized by very high viral loads, HIV p24-tetramer+ CD4 T cells had a phenotype characteristic of effector T cells. Surprisingly, this phenotype (CCR7−CD45RA−) was maintained over extended periods of time during which virus production was undetectable by RT-PCR as a result of ART (several years in some patients). Even treatment of acute infection did not result in the emergence of a significant number of cells with a central memory phenotype. There are two potential explanations for this finding: these T cells have acquired a differentiation state that does not permit conversion to a central memory CD4 T cell phenotype, or the effector state is maintained by continued, low level production of viral Ags. Studies in a LCMV TCR-transgenic mouse model have demonstrated that virus-specific CD4 T cells lose the ability to produce IL-2 in chronically infected mice, and that transfer of such T cells into uninfected mice does not restore their ability to produce IL-2 or to proliferate (40). Younes et al. (29) demonstrated a functional distinction between HIV-specific CD4 T cell populations from viremic vs aviremic patients for whom treatment had been initiated early during infection: IL-2-producing CCR7+ CD4 T cells were present only in aviremic, but not viremic patients, while the frequency of CCR7−IFN-γ+ CD4 T cells was higher in viremic patients. Taken together, these findings suggest an early defect in the generation of IL-2-producing central memory CD4 T cells, which may be related to the level and/or duration of virus production before the initiation of therapy.
Short-term treatment interruptions result in transient increases in the frequency of HIV p24-tetramer+ T cells, but the pool of these cells again contracts following reinitiation of therapy. Multiple short-term cycles of treatment interruption thus do not substantially expand the pool of these cells, and there is also no evidence for a desirable change in their functional properties. This feature of the antiviral CD4 T cell response needs to be taken into account in the design of immunotherapeutic approaches. The finding that the majority of HIV p24-tetramer+ CD4 T cells are lost with prolonged viremia underscores the importance of suppressing HIV viremia whenever possible. The availability of MHC class II tetramers to multiple MHC class II alleles and with multiple HIV p24 peptides should prove useful for the investigation of CD4 T cell populations following therapeutic interventions, in particular when combined with single-cell analysis of T cell function.
Acknowledgments
We thank Dr. Donna Neuberg for performing statistical analysis of the data and Daryld Strick for processing and cryopreservation of PBMCs from study subjects.
Footnotes
This work was supported by grants from the National Institutes of Health (PO1 AI45757 to K.W.W, RO1 AI40873 to E.S.R. and K.W.W., and KO1 DK068383 to N.S.) and a grant from the Schweizerische Stiftung fuer medizinisch-biologische Stipendien (to D.E.K.).
Abbreviations used in this paper: ART, antiretroviral therapy; RT, room temperature; 7AAD, 7-aminoactinomycin D; LCMV, lymphocytic choriomeningitis virus.
Disclosures
The authors have no financial conflict of interest.
References
- 1.McCune JM. The dynamics of CD4+ T-cell depletion in HIV disease. Nature. 2001;410:974–979. doi: 10.1038/35073648. [DOI] [PubMed] [Google Scholar]
- 2.McMichael AJ, Rowland-Jones SL. Cellular immune responses to HIV. Nature. 2001;410:980–987. doi: 10.1038/35073658. [DOI] [PubMed] [Google Scholar]
- 3.Altman JD, Moss PA, Goulder PJ, Barouch DH, McHeyzer-Williams MG, Bell JI, McMichael AJ, Davis MM. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. doi: 10.1126/science.274.5284.94. [DOI] [PubMed] [Google Scholar]
- 4.Bourgeois C, Rocha B, Tanchot C. A role for CD40 expression on CD8+ T cells in the generation of CD8+ T cell memory. Science. 2002;297:2060–2063. doi: 10.1126/science.1072615. [DOI] [PubMed] [Google Scholar]
- 5.Grakoui A, Shoukry NH, Woollard DJ, Han JH, Hanson HL, Ghrayeb J, Murthy KK, Rice CM, Walker CM. HCV persistence and immune evasion in the absence of memory T cell help. Science. 2003;302:659–662. doi: 10.1126/science.1088774. [DOI] [PubMed] [Google Scholar]
- 6.Janssen EM, Lemmens EE, Wolfe T, Christen U, von Herrath MG, Schoenberger SP. CD4+ T cells are required for secondary expansion and memory in CD8+ T lymphocytes. Nature. 2003;421:852–856. doi: 10.1038/nature01441. [DOI] [PubMed] [Google Scholar]
- 7.Kaech SM, Ahmed R. Immunology: CD8 T cells remember with a little help. Science. 2003;300:263–265. doi: 10.1126/science.1084511. [DOI] [PubMed] [Google Scholar]
- 8.Seder RA, Ahmed R. Similarities and differences in CD4+ and CD8+ effector and memory T cell generation. Nat Immunol. 2003;4:835–842. doi: 10.1038/ni969. [DOI] [PubMed] [Google Scholar]
- 9.Shedlock DJ, Shen H. Requirement for CD4 T cell help in generating functional CD8 T cell memory. Science. 2003;300:337–339. doi: 10.1126/science.1082305. [DOI] [PubMed] [Google Scholar]
- 10.Sun JC, Bevan MJ. Defective CD8 T cell memory following acute infection without CD4 T cell help. Science. 2003;300:339–342. doi: 10.1126/science.1083317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bachmann MF, Hunziker L, Zinkernagel RM, Storni T, Kopf M. Maintenance of memory CTL responses by T helper cells and CD40-CD40 ligand: antibodies provide the key. Eur J Immunol. 2004;34:317–326. doi: 10.1002/eji.200324717. [DOI] [PubMed] [Google Scholar]
- 12.Pontesilli O, Carlesimo M, Varani AR, Ferrara R, Guerra EC, Bernardi ML, Ricci G, Mazzone AM, D’Offizi G, Aiuti F. HIV-specific lymphoproliferative responses in asymptomatic HIV-infected individuals. Clin Exp Immunol. 1995;100:419–424. doi: 10.1111/j.1365-2249.1995.tb03716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenberg ES, Billingsley JM, Caliendo AM, Boswell SL, Sax PE, Kalams SA, Walker BD. Vigorous HIV-1-specific CD4+ T cell responses associated with control of viremia. Science. 1997;278:1447–1450. doi: 10.1126/science.278.5342.1447. [DOI] [PubMed] [Google Scholar]
- 14.Pontesilli O, Carotenuto P, Kerkhof-Garde SR, Roos MT, Keet IP, Coutinho RA, Goudsmit J, Miedema F. Lymphoproliferative response to HIV type 1 p24 in long-term survivors of HIV type 1 infection is predictive of persistent AIDS-free infection. AIDS Res Hum Retroviruses. 1999;15:973–981. doi: 10.1089/088922299310485. [DOI] [PubMed] [Google Scholar]
- 15.Oxenius A, Price DA, Easterbrook PJ, O’Callaghan CA, Kelleher AD, Whelan JA, Sontag G, Sewell AK, Phillips RE. Early highly active antiretroviral therapy for acute HIV-1 infection preserves immune function of CD8+ and CD4+ T lymphocytes. Proc Natl Acad Sci USA. 2000;97:3382–3387. doi: 10.1073/pnas.97.7.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson JD, Imami N, Watkins A, Gill J, Hay P, Gazzard B, Westby M, Gotch FM. Loss of CD4+ T cell proliferative ability but not loss of human immunodeficiency virus type 1 specificity equates with progression to disease. J Infect Dis. 2000;182:792–798. doi: 10.1086/315764. [DOI] [PubMed] [Google Scholar]
- 17.McNeil AC, Shupert WL, Iyasere CA, Hallahan CW, Mican JA, Davey RT, Jr, Connors M. High-level HIV-1 viremia suppresses viral antigen-specific CD4+ T cell proliferation. Proc Natl Acad Sci USA. 2001;98:13878–13883. doi: 10.1073/pnas.251539598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lichterfeld M, Kaufmann DE, Yu XG, Mui SK, Addo MM, Johnston MN, Cohen D, Robbins GK, Pae E, Alter G, et al. Loss of HIV-1-specific CD8+ T cell proliferation after acute HIV-1 infection and restoration by vaccine-induced HIV-1-specific CD4+ T cells. J Exp Med. 2004;200:701–712. doi: 10.1084/jem.20041270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Douek DC, Brenchley JM, Betts MR, Ambrozak DR, Hill BJ, Okamoto Y, Casazza JP, Kuruppu J, Kunstman K, Wolinsky S, et al. HIV preferentially infects HIV-specific CD4+ T cells. Nature. 2002;417:95–98. doi: 10.1038/417095a. [DOI] [PubMed] [Google Scholar]
- 20.Yue FY, Kovacs CM, Dimayuga RC, Gu XX, Parks P, Kaul R, Ostrowski MA. Preferential apoptosis of HIV-1-specific CD4+ T cells. J Immunol. 2005;174:2196–2204. doi: 10.4049/jimmunol.174.4.2196. [DOI] [PubMed] [Google Scholar]
- 21.Pitcher CJ, Quittner C, Peterson DM, Connors M, Koup RA, Maino VC, Picker LJ. HIV-1-specific CD4+ T cells are detectable in most individuals with active HIV-1 infection, but decline with prolonged viral suppression. Nat Med. 1999;5:518–525. doi: 10.1038/8400. [DOI] [PubMed] [Google Scholar]
- 22.Scriba TJ, Zhang HT, Brown HL, Oxenius A, Tamm N, Fidler S, Fox J, Weber JN, Klenerman P, Day CL, Lucas M, Phillips RE. HIV-1-specific CD4+ T lymphocyte turnover and activation increase upon viral rebound. J Clin Invest. 2005;115:443–450. doi: 10.1172/JCI23084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olerup O, Zetterquist H. HLA-DRB1*01 subtyping by allele-specific PCR amplification: a sensitive, specific and rapid technique. Tissue Antigens. 1991;37:197–204. doi: 10.1111/j.1399-0039.1991.tb01872.x. [DOI] [PubMed] [Google Scholar]
- 24.Zetterquist H, Olerup O. Identification of the HLA-DRB1*04, -DRB1*07, and -DRB1*09 alleles by PCR amplification with sequence-specific primers (PCR-SSP) in 2 hours. Hum Immunol. 1992;34:64–74. doi: 10.1016/0198-8859(92)90086-3. [DOI] [PubMed] [Google Scholar]
- 25.Day CL, Seth NP, Lucas M, Appel H, Gauthier L, Lauer GM, Robbins GK, Szczepiorkowski ZM, Casson DR, Chung RT, et al. Ex vivo analysis of human memory CD4 T cells specific for hepatitis C virus using MHC class II tetramers. J Clin Invest. 2003;112:831–842. doi: 10.1172/JCI18509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malhotra U, Holte S, Dutta S, Berrey MM, Delpit E, Koelle DM, Sette A, Corey L, McElrath MJ. Role for HLA class II molecules in HIV-1 suppression and cellular immunity following antiretroviral treatment. J Clin Invest. 2001;107:505–517. doi: 10.1172/JCI11275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaufmann DE, Bailey PM, Sidney J, Wagner B, Norris PJ, Johnston MN, Cosimi LA, Addo MM, Lichterfeld M, Altfeld M, et al. Comprehensive analysis of human immunodeficiency virus type 1-specific CD4 responses reveals marked immunodominance of gag and nef and the presence of broadly recognized peptides. J Virol. 2004;78:4463–4477. doi: 10.1128/JVI.78.9.4463-4477.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harari A, Rizzardi GP, Ellefsen K, Ciuffreda D, Champagne P, Bart PA, Kaufmann D, Telenti A, Sahli R, Tambussi G, et al. Analysis of HIV-1-and CMV-specific memory CD4 T-cell responses during primary and chronic infection. Blood. 2002;100:1381–1387. doi: 10.1182/blood-2001-11-0080. [DOI] [PubMed] [Google Scholar]
- 29.Younes SA, Yassine-Diab B, Dumont AR, Boulassel MR, Grossman Z, Routy JP, Sekaly RP. HIV-1 viremia prevents the establishment of interleukin 2-producing HIV-specific memory CD4+ T cells endowed with proliferative capacity. J Exp Med. 2003;198:1909–1922. doi: 10.1084/jem.20031598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harari A, Petitpierre S, Vallelian F, Pantaleo G. Skewed representation of functionally distinct populations of virus-specific CD4 T cells in HIV-1-infected subjects with progressive disease: changes after antiretroviral therapy. Blood. 2004;103:966–972. doi: 10.1182/blood-2003-04-1203. [DOI] [PubMed] [Google Scholar]
- 31.Palmer BE, Boritz E, Wilson CC. Effects of sustained HIV-1 plasma viremia on HIV-1 Gag-specific CD4+ T cell maturation and function. J Immunol. 2004;172:3337–3347. doi: 10.4049/jimmunol.172.5.3337. [DOI] [PubMed] [Google Scholar]
- 32.Appay V, Dunbar PR, Callan M, Klenerman P, Gillespie GM, Papagno L, Ogg GS, King A, Lechner F, Spina CA, et al. Memory CD8+ T cells vary in differentiation phenotype in different persistent virus infections. Nat Med. 2002;8:379–385. doi: 10.1038/nm0402-379. [DOI] [PubMed] [Google Scholar]
- 33.Yue FY, Kovacs CM, Dimayuga RC, Parks P, Ostrowski MA. HIV-1-specific memory CD4+ T cells are phenotypically less mature than cyto-megalovirus-specific memory CD4+ T cells. J Immunol. 2004;172:2476–2486. doi: 10.4049/jimmunol.172.4.2476. [DOI] [PubMed] [Google Scholar]
- 34.Kaech SM, Tan JT, Wherry EJ, Konieczny BT, Surh CD, Ahmed R. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat Immunol. 2003;4:1191–1198. doi: 10.1038/ni1009. [DOI] [PubMed] [Google Scholar]
- 35.Brenchley JM, Karandikar NJ, Betts MR, Ambrozak DR, Hill BJ, Crotty LE, Casazza JP, Kuruppu J, Migueles SA, Connors M, et al. Expression of CD57 defines replicative senescence and antigen-induced apoptotic death of CD8+ T cells. Blood. 2003;101:2711–2720. doi: 10.1182/blood-2002-07-2103. [DOI] [PubMed] [Google Scholar]
- 36.Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 37.Murali-Krishna K, Altman JD, Suresh M, Sourdive DJ, Zajac AJ, Miller JD, Slansky J, Ahmed R. Counting antigen-specific CD8 T cells: a reevaluation of bystander activation during viral infection. Immunity. 1998;8:177–187. doi: 10.1016/s1074-7613(00)80470-7. [DOI] [PubMed] [Google Scholar]
- 38.Homann D, Teyton L, Oldstone MB. Differential regulation of antiviral T-cell immunity results in stable CD8+ but declining CD4+ T-cell memory. Nat Med. 2001;7:913–919. doi: 10.1038/90950. [DOI] [PubMed] [Google Scholar]
- 39.Ogg GS, Jin X, Bonhoeffer S, Moss P, Nowak MA, Monard S, Segal JP, Cao Y, Rowland-Jones SL, Hurley A, et al. Decay kinetics of human immunodeficiency virus-specific effector cytotoxic T lymphocytes after combination antiretroviral therapy. J Virol. 1999;73:797–800. doi: 10.1128/jvi.73.1.797-800.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oxenius A, Zinkernagel RM, Hengartner H. Comparison of activation versus induction of unresponsiveness of virus-specific CD4+ and CD8+ T cells upon acute versus persistent viral infection. Immunity. 1998;9:449–457. doi: 10.1016/s1074-7613(00)80628-7. [DOI] [PubMed] [Google Scholar]