Abstract
Amorphous aggregation is a major problem for protein biopharmaceuticals, and aggregate formation in a drug formulation can have serious health implications for the patient. In many cases, an immunogenic response is generated from the administration of a drug product containing aggregated protein. This becomes especially significant when the patient requires long-term or repeated administration of the drug, because the likelihood of a severe immune response increases. While the prevention of protein aggregation is critically important for the future of protein pharmaceuticals, the process is still poorly understood. The lack of understanding regarding non-fibrillar aggregation is largely due to the fact that assembly is difficult to study. In particular the role that various structural features (i.e. α-helix, β-structure, disulfide bonds) play in the aggregation process varies with the amino acid sequence and is dependent upon tertiary structure and solution conditions. Well-structured proteins do not readily aggregate in solution, whereas partially unfolded proteins tend to aggregate rapidly and often become insoluble. Here, we present a unique and simple system for studying amorphous protein aggregation. We have previously reported the isolation of the basic leucine zipper (bZIP) domain of activating transcription factor 5 (ATF5), a protein notable for its potential as a pharmaceutical target for treatment of glioblastoma multiforme. This domain consists of a single α-helix and possesses a single cysteine residue. It is only partially structured and displays marginal stability in solution under physiological conditions. We have modulated solution conditions that affect backbone solubility and the oxidation state of the thiol to successfully investigate the role that α-helical structure and disulfide bond formation play in protein stability. Our data indicate that covalent cross-linking helps to retain ATF5’s helicity, which inhibits the formation of large aggregates. These studies have led to the identification of stabilizing conditions for ATF5, which will enable further study of the protein as a pharmaceutical target. Moreover, this work has general implications for analyzing stability of helical proteins in vitro and the specific atomic-level interactions in ATF5 that contribute to instability and self-association.
Keywords: ATF5, ATFx, bZIP, disulfide bond, helix, aggregation, stability
Introduction
Amorphous aggregation is a common phenomenon and a major obstacle to handling proteins in vitro. The aggregation of proteins isolated for use as pharmaceutical products poses a serious concern, as aggregates can stimulate the immune system, which may have devastating affects on the patient.1–3 Although the study of protein aggregation has wide-reaching implications, the process by which aggregation occurs is still difficult to predict. The study of amyloids has provided understanding about the structure and formation of fibrillar aggregates, but a lack of data regarding amorphous structure has prevented verifiable application to amorphous aggregation. In particular, the role that disulfide bonds play in this process is poorly understood, because it varies among different systems. Reports indicate in some instances that disulfide bond formation promotes protein aggregation, yet in others it dampens this process.4–8 These effects appear to be largely protein specific and dependent on features of tertiary structure. Alpha-helices can also play an important role in aggregation. In most instances, retention of α-helical structure has been linked to improved protein stability; 9, 10 however, several recent reports indicate that α-helical structure can facilitate aggregate formation.11–13
The lack of understanding surrounding non-fibrillar aggregation is largely due to the fact that the assembled species lacks a regular repeating pattern, making it is difficult to study. Well-structured and natively disordered proteins do not readily aggregate in solution, whereas partially unfolded proteins tend to aggregate rapidly, suggesting structured features facilitate aggregation. Here we present a unique and simple system, which is amenable to investigating the mechanism of protein aggregation involving a helix to coil transition. We have previously reported the isolation of the bZIP domain of activating transcription factor 5 (ATF5), a protein notable for its potential as a pharmaceutical target for treatment of glioblastoma multiforme.14 This domain consists of a single α-helix and possesses a single cysteine residue. It is only partially structured (~25% helix) and displays marginal stability in solution under physiological conditions. We have tested solution conditions that modulate helical propensity and the oxidation state of the thiol, to successfully investigate the role that α-helical structure and disulfide bond formation play in protein stability. These studies have led to the identification of stabilizing conditions for ATF5, which will enable further study of the protein as a pharmaceutical target and provided a model system for further examination of residue-specific contributions to the formation of amorphous aggregates.
Materials and Methods
Protein Expression and Purification
The cDNA of ATF5 was obtained through ATCC (MGC-842) and the bZIP domain was PCR amplified and inserted into a Novagen (San Diego, CA) pET-42b vector as described previously.14 Site-directed mutagenesis was performed to create the C240A point mutation in the bZIP region using the following primers: 5’GAGGCCCTGGAGGGCGAGGCCCAGGGGCTGGAGGCACGG (forward primer), 5’-CCGTGCCTCCAGCCCCTGGGCCTCGCCCTCCAGGGCCTC (reverse primer). Plasmids were transformed into Novagen BL21(DE3) E. Coli for expression. Purification was done using a refolding procedure described previously.14 Protein concentrations were determined using a standard Bradford assay. ATF5 purification and isolation was confirmed using SDS-PAGE and LC/MS.14 These methods were also used to confirm intermolecular disulfide bond formation for wild-type ATF5.14 Isotopic labeling for two-dimensional NMR was accomplished with the substitution of 15N-ammonium chloride. The 15N-ammonium chloride (>99% N-15) was obtained from Spectra Stable Isotopes (Columbia, MD).
Circular Dichroism
Circular dichroism (CD) spectra were acquired with a Jasco-810 polarimeter (Tokyo, Japan) equipped with a Peltier-type temperature controller and a six-position sample holder. Samples were prepared at a final protein concentration of 100 µM in 20 mM MES buffer at pH 6.0 in the absence or presence of dithiothreitol (DTT) or trifluoroethanol (TFE). Scans were performed at 4°C from 260 to 190 nm in a 0.1-cm path length cell. A scanning speed of 50 nm/min and a response time of 8 sec were used. The selected bandwidth was 5 nm and the data pitch was 1 nm. Scans were run in duplicate for each sample and two samples were prepared and analyzed for each set of solution conditions. Using the Jasco Spectra Manager v1.18.00 software the measured values were converted to molar ellipticity. The resulting average and standard deviations were plotted for each condition using Plot. Percent helicity was calculated using the mean residue ellipticity observed at 222 nm as previously described.14, 15
To evaluate protein stability the absorbance at 222 nm was monitored at a function of temperature. The temperature was adjusted from 4 to 85°C at a ramping rate of 15°C/hour and the absorbance was monitored every 0.5°C. The results were converted to molar ellipticity using the instrument software. Two samples were prepared and analyzed for each set of solution conditions. The resulting average and standard deviations were plotted using Plot. Midpoints of thermal transition were calculated using Microcal Origin sigmoidal fit graphing tools. None of the solution conditions used produced signals above background at any temperature and therefore, no buffer subtraction manipulations were performed.
Static Light Scattering
Static light scattering (SLS) data were acquired with a Photon Technology International (PTI) spectrafluorometer (Lawrenceville, NJ) equipped with a Peltier-type temperature controller and a four-position sample holder. Samples were prepared at a final protein concentration of 100 µM in 20 mM MES buffer at pH 6.0 in the absence or presence of dithiothreitol (DTT) or trifluoroethanol (TFE). The intensity of scattered light was measured as a function of temperature and detected at an angle of 90° to the light source by a photomultiplier tube. An arc lamp white light source was used in all cases. The excitation wavelength was set at 275 nm. The emission wavelength range was set from 250 nm to 350 nm. Spectra were obtained following a 5 min equilibration period at each temperature. Data were collected every 2.5°C between the temperature range of 4 to 81.5°C. The background was subtracted from each data point based on a blank containing the corresponding buffer solution. Two samples were prepared and analyzed for each set of solution conditions. The resulting average and standard deviations were plotted using Plot.
Dynamic light scattering
The average hydrodynamic diameter was monitored as a function of temperature using a Brookhaven (Holtsville, NY) BLS-9000 DLS instrument equipped with a 50 mW HeNe diode laser operating at 532 nm and a Brookhaven BI-200SM goniometer. Samples were prepared in 20 mM MES buffer pH 6.0 at a final concentration of 100 µM. Five consecutive 30 sec data collection intervals were taken every 2.5°C over the range of 10 to 90 °C. Light scattering was monitored 90° to the incident light and autocorrelation functions were generated using a Brookhaven BI-9000AT digital autocorrelator. The hydrodynamic diameter was calculated from the translational diffusion coefficient using the Stokes-Einstein equation.16
Nuclear Magnetic Resonance
Two-dimensional 1H-15N heteronuclear single quantum coherence (HSQC) spectra were recorded at 25°C using a Bruker AVANCE 800 MHz spectrometer equipped with a triple-resonance CRYO-probe with pulse field gradients. Samples were prepared at a concentration of 0.2 mM in 50 mM sodium phosphate, 100mM NaCl pH 6.5, containing 5% D2O. Water suppression was accomplished using flip-back pulses. Data were acquired in 128 scans with 2048 points in 1H and 128* increments in 15N. 1H chemical shifts were referenced with respect to an external DSS standard in D2O.17 Indirect referencing relative to 1H was determined for 15N, assuming a ratio of 15N/1H = 0.101329118. Data were processed using NMRPipe and Sparky software.18, 19
Results
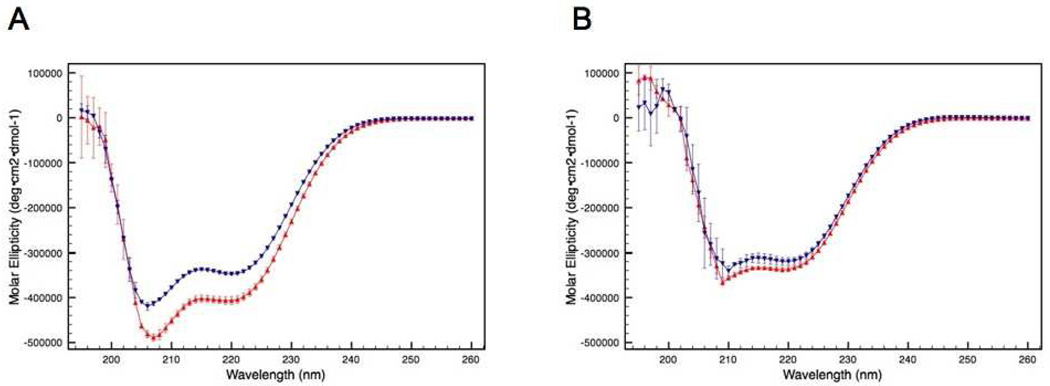
CD spectroscopy was used to investigate the influence of a single intermolecular disulfide bond (C240–C240) on the structure of the bZIP domain of ATF5. The spectra reveal absorption characteristics consistent with the presence of α-helical structure for both the disulfide-bound dimer and the C240A monomer (Figure 1A). This is depicted by the double absorption minima pattern detected at 222 nm and 208 nm.20 The ratio of 222 to 208 nm is 0.84 for wild type and 0.85 for C240A, which is consistent with α-helical but not coiled-coil structure.21, 22 An increase in helical content is observed for the dimer in comparison to the monomer. This difference can be quantified for each species using the mean residue ellipticy at 222 nm.14, 15 The percent helicity calculated for the dimer under these conditions is 23.4% ± 0.3%, whereas the percent helicity of the C240A monomer is 19.0% ± 1.0%.
Figure 1.
CD absorption spectra of the disulfide-linked ATF5 dimer (red upright triangles) and the C240A monomer (blue downturned triangles) in the absence (A) or presence (B) of 10 mM DTT.
Upon the addition of reducing agent to wild-type (WT) ATF5 (10 mM DTT), a decrease in absorption signal that corresponds to α-helix is observed (Figure 1B). The absorption intensity for the reduced WT more closely resembles that of the C240A monomer under reducing conditions with a calculated percent helicity of 17.9% (± 0.2%) and 16.7% (± 0.1%), respectively. This indicates the intermolecular disulfide bond is responsible for the increased helicity originally observed for the dimeric species. It is important to note that the decrease in absorption intensity observed at 208 nm relative to 222 nm in the presence of DTT is due to substantially increased interference caused by DTT on the detection at shorter wavelengths. For this reason, absorption measurements taken at wavelengths below 210 nm in the presence of DTT should be considered less accurate. As such, the ratio of 222 nm to 208 nm was not assessed because the interpretation of this value is also unreliable for determining α-helical vs coiled-coil content.
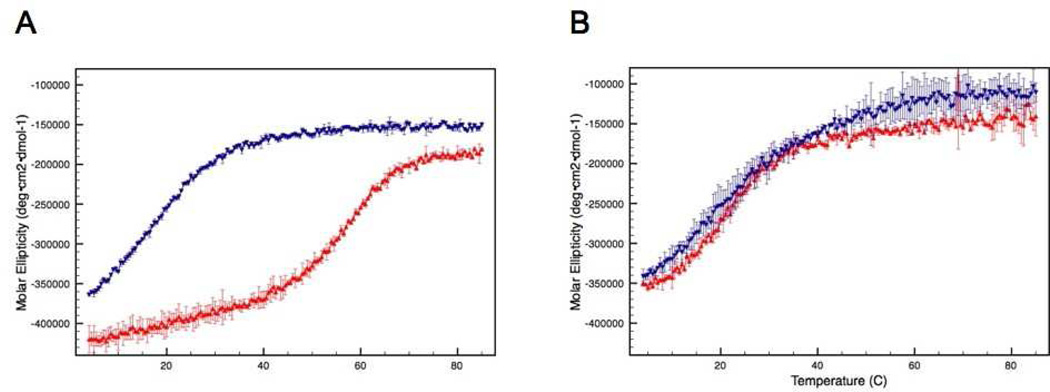
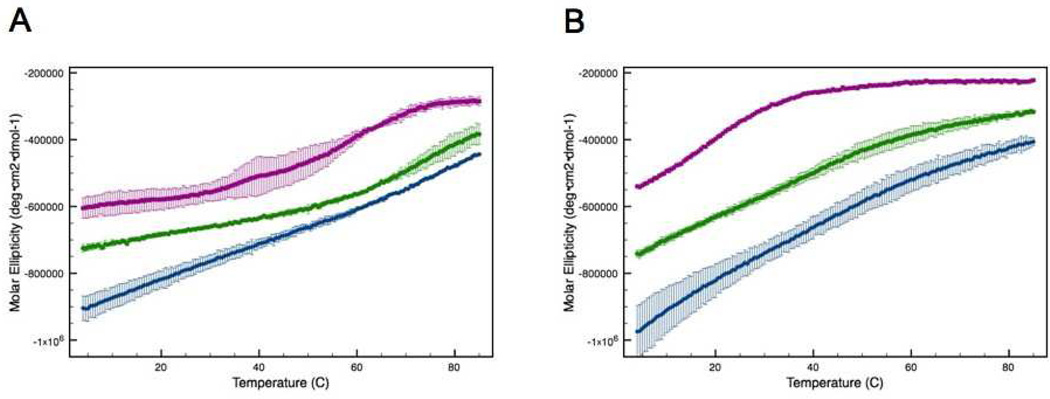
The impact of the intermolecular disulfide bond on thermal stability was evaluated by monitoring the α-helix CD absorption signal at 222 nm as a function of temperature. A decrease in absorption signal at 222 nm indicates a loss of secondary structure, which reflects protein unfolding. The disulfide-bound dimer is more stable than the C240A monomer. The calculated melting temperature (Tm) of the dimer is 55.5°C, whereas it is 15.9°C for the monomer (Figure 2A). In the presence of reducing agent (10 mM DTT) the Tm for the C240A mutant remains relatively unchanged (15.1°C), whereas the melting curve for the dimer shifts dramatically to more closely resemble that of the monomer (Figure 2B). The calculated transition temperature for the dimer under these conditions is 19.9°C. Notably, unfolding of all forms of ATF5 was found to be largely irreversible due to precipitation of aggregated species (data not shown).
Figure 2.
CD absorption at 222 nm of the disulfide-bound ATF5 dimer (red upright triangles) and C240A monomer (blue downturned triangles) in the absence (A) or presence (B) of 10 mM DTT as a function of temperature.
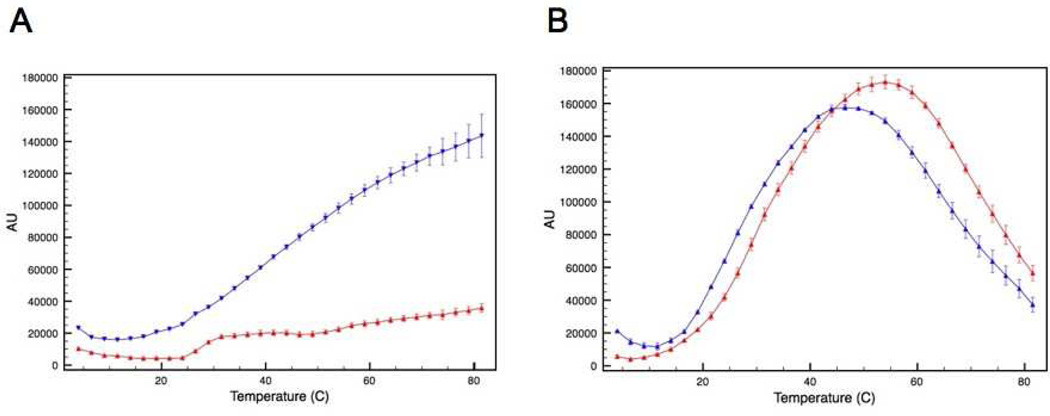
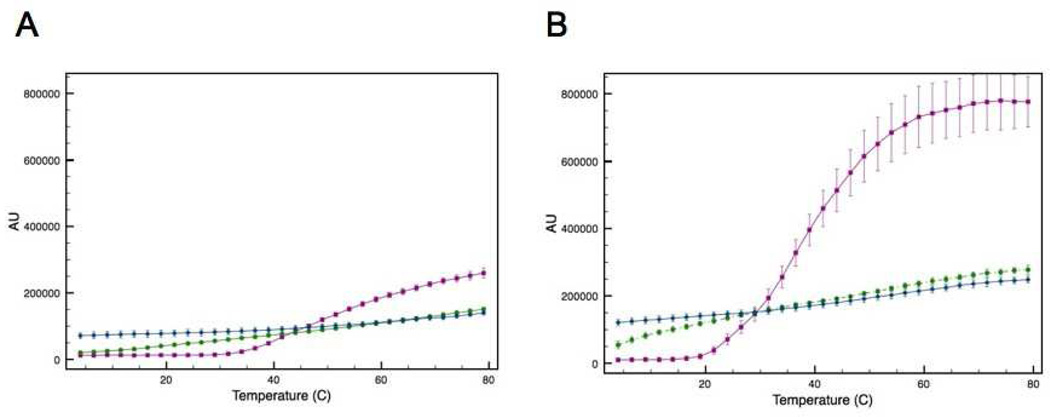
The effect this intermolecular disulfide bond has on protein aggregation was then evaluated using a static light scattering assay. The intensity of scattered light was measured as a function of temperature for the two forms of ATF5. In this assay, an increase in the intensity of scattered light indicates increased aggregate formation. The data show that the onset of aggregation is more rapid, and the extent of aggregation is greater for the C240A mutant compared to the WT under non-reducing conditions (Figure 3A). The onset of aggregation is more gradual and the extent lessened for the disulfide-bound dimer (Figure 3A). Upon reduction of the disulfide bond via the addition of reducing agent (10 mM DTT), the scattering profile of the dimer more closely parallels that of the C240A monomer (Figure 3B). The decrease in signal seen at higher temperatures in Figure 3B indicates the formation of very large aggregates that have settled out of solution, resulting in diminished absorbance intensity. This was verified using dynamic light scattering analysis, which indicated that the average hydrodynamic diameter of protein in solution was much greater in the presence of DTT (See Supporting Information; Table S1). Clearly, reduction of the disulfide bond decreases thermal stability, as the two proteins now behave similarly, but it is also apparent that the inclusion of DTT in solution has an additional adverse effect on protein stability, possibly due to the more hydrophobic character or decreased dielectric constant of this co-solvent.
Figure 3.
SLS analysis of the disulfide bound ATF5 dimer (red upright triangles) and C240A monomer (blue downturned triangles) in the absence (A) or presence (B) of 10 mM DTT as a function of temperature.
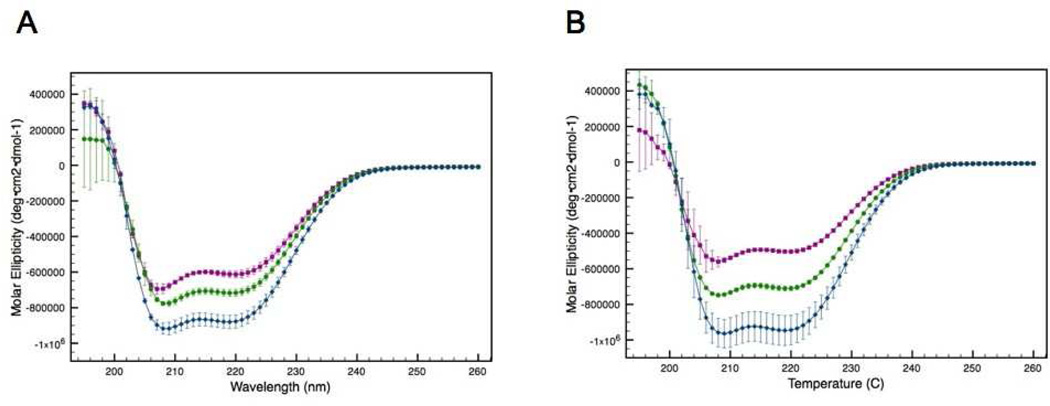
The effect that α-helical structure has on protein stability was evaluated using the addition of the co-solvent trifluoroethanol (TFE) to induce helical structure. The use of TFE for the purpose of inducing protein structure is common and well cited.23, 24 TFE is added to peptides to enhance α-helicity, because the solubility of the backbone changes. Low percentages of TFE typically increase helicity, but at high concentrations (often >40%) β-sheets may form in some peptides.23 The impact of TFE on the structure of ATF5 was assessed using CD spectroscopy. The helical content of both the disulfide-bound dimer and the C240A monomer increased in a dose-dependent fashion upon the addition of TFE within the experimental range used (Figure 4). The increase in helicity observed for a given amount of TFE added was greater for the C240A mutant than for the dimer, but at 10% TFE and above both forms displayed equivalent amounts of helical structure. The percent helicity was quantified as described above for both forms of ATF5. The dimer displayed a percent helicity of 25.5% (± 0.6%), 30.8% (± 0.6%), and 39.3% (± 1.0%) in the presence of no TFE, 10% TFE and 20% TFE, respectively. The comparative values for the C240A monomer under the same conditions are 19.5% (± 0.3%), 30.5% (± 0.3%), and 42.0% (± 2.2%), respectively.
Figure 4.
CD absorption spectra of the disulfide-linked ATF5 dimer (A) and the C240A monomer (B) in the presence of increasing amounts of TFE: purple squares (no TFE), green circles (10% [v/v] TFE) and blue diamonds (20%[v/v] TFE).
The thermal stability of both forms of ATF5 in the presence of increasing concentration of TFE was investigated by monitoring the α-helical CD absorption signal at 222 nm as a function of temperature. The melting profiles of both forms changed in the presence of TFE (Figure 5). The data show that increasing the helicity of either form of ATF5 improves thermal stability, because greater α-helical structure is retained at all temperatures in the presence of TFE. A standard quantitative analysis comparing Tm values is not possible for these data, because the melting profiles in the presence of 10% and 20% TFE could not be fit to a sigmoidal curve. The melting temperature calculated for the WT form of ATF5 in the absence of TFE is 56.0°C. At that same temperature in the presence of 10% and 20% TFE, there is greater helical structure present. The molar ellipticity values for the WT dimer at 56°C in the presence of 0%, 10% and 20% TFE are −426,189, −581,870, and −630946, respectively. This equates to approximately 9% more α-helix at 10% TFE than at 0% TFE, and 3% more helix at 20% TFE than at 10%. The melting temperature calculated for the C240A monomer in the absence of TFE is 17.2°C.
Figure 5.
CD absorption at 222 nm of the disulfide-linked ATF5 dimer (A) and the C240A monomer (B) as a function of temperature in the presence of increasing amounts of TFE: purple squares (no TFE), green circles (10%[v/v] TFE) and blue diamonds (20%[v/v] TFE).
The molar ellipticity values for the C240A mutant at 17°C in the presence of 0%, 10% and 20% TFE are −428,822, −653,246 and −846,000, respectively, equating to 12% more helix at 10% TFE than 0% TFE, and 11% more helix at 20% TFE than 10% TFE.
Analysis of static light scattering data was used to evaluate the effect of helicity on protein aggregation. Increasing the α-helical structure of both the cross-linked WT dimer and the C240A monomer prevents aggregation (Figure 6). The maximum intensity of scattered light detected for the WT form is 259,404, 150,731 and 139,743 in the presence of 0%, 10% and 20% TFE, respectively. Notably, a greater difference in the extent of aggregation is observed between 0% and 10% TFE than between 10% and 20% TFE. This same trend is observed for the C240A monomer, where the maximum intensity of scattered light detected in the presence of 0%, 10% and 20% is 779,386, 277,567, and 248,188, respectively. The greatest impact on aggregation in all cases is observed for the C240A mutant in the presence of 10% TFE, where the inclusion of the co-solvent dramatically reduces the onset and extent of aggregation. The impact on aggregation for the WT form at the same concentration of 10% TFE is much less dramatic, even though the percent helicity induced is roughly the same as that of the C240A monomer (approximately 30%). At low temperatures, the addition of TFE leads to a small amount of increased light scattering for both the WT and C240A. There is more scattering at 20% TFE compared to 10% TFE, suggesting that an alternative mode of association may be responsible for aggregation in the TFE-containing samples.
Figure 6.
SLS analysis of the disulfide-linked ATF5 dimer (A) and the C240A monomer (B) in the presence of increasing amounts of TFE: (purple squares) no TFE, (green circles) 10% TFE and (blue diamonds) 20% TFE.
NMR experiments were performed to evaluate general differences in structure between the WT and C240A forms of ATF5 at higher resolution. The 2D 1H-15N HSQC experiment used here selectively detects all NH pairs in the protein and correlates each 1H to the directly attached 15N atom. The resultant spectrum is considered a “fingerprint” of the protein, because the chemical shifts depicted reflect the conformationally-weighted average of the environment experienced by the nuclei, which is defined by the overall protein fold. Consequently, the 1H-15N HSQC experiment can provide a quick indication of protein structure. A well-behaved globular protein will display good signal dispersion, typically ranging from 6 to 12 ppm on the 1H-axis and 100–140 ppm on the 15N-axis. This dispersion results from the presence of stable, structural features that confine the individual nuclei to unique chemical environments. More disordered proteins typically have peaks clustered more closely around random coil values (~8.3 ppm in 1H). This is because, on average, the nuclei experience more similar chemical environments in solution than in the structured protein. The HSQC spectra of proteins composed of only a single helix, like the bZIP of ATF5, are difficult to interpret without full assignments and additional data, because in a standard alpha helix, all the amides are in similar environments, which can result in overlapping NMR signals. Typically, when bZIP proteins functionally dimerize they interact to form a two-stranded coiled-coil. Higher-order protein structure, such as coiled-coil, would place the NH nuclei in more distinct chemical environments, resulting in greater dispersion of the NMR signal, as has been demonstrated experimentally for a number of bZIP proteins examined structurally using solution NMR.25–29
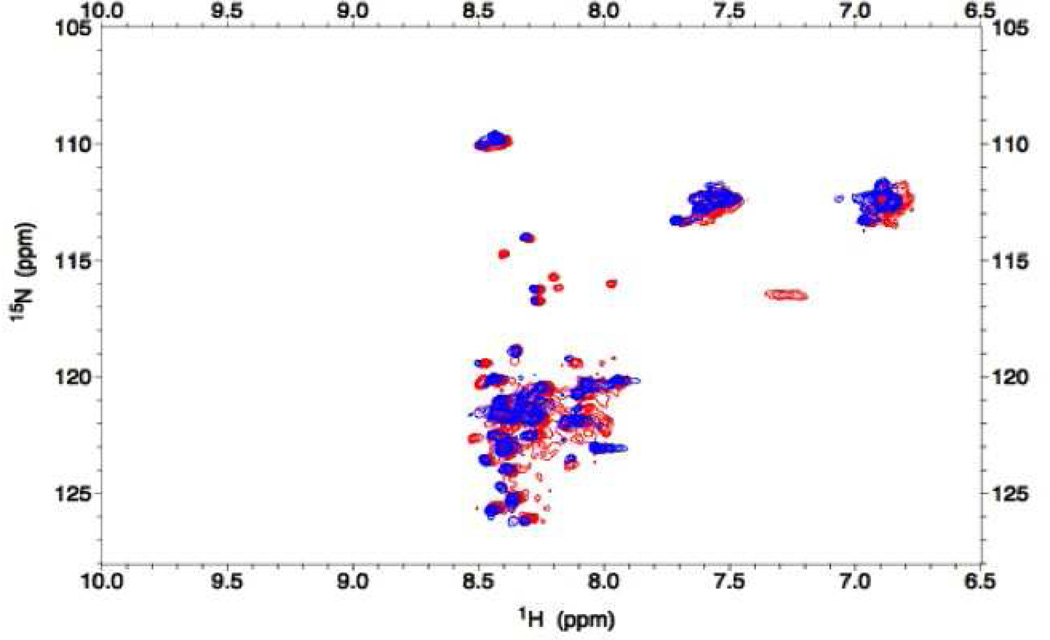
An overlay of the 2D HSQC spectrum of the C240A mutant onto the WT spectrum shows that the chemical shifts for both forms are largely clustered around 8.3 ppm in 1H and only a few differences in resonance positions between the spectra are apparent (Figure 7). This indicates the average structure of the two forms is quite similar and both proteins are conformationally labile. The limited chemical shift dispersion observed for both forms of ATF5 represents α-helix and/or random coil structure. The lack of dispersion in the NMR spectra further indicates coiled-coil structure is not developed, even in the obligate wild-type dimer. This is consistent with the CD spectra in which the ratio of 222 to 208 nm indicate α-helix is present but coiled-coil structure is not. The unique red peaks in the WT dimer likely correspond to residues adjacent to C240 involved in the helical region that are stabilized by disulfide bond formation. The NH signal generated by the side chains of Asn and Gln residues cluster in a different chemical shift range and account for the signal observed between 6.8 and 7.6 ppm on the 1H -axis. There is little evidence for the existence of a coiled-coil structure for either form of ATF5, even at the high concentrations required for these studies. These data suggest that despite the presence of 23% helical content in the covalent dimer and its resistance to aggregation when the helix is maintained, ATF5 does not develop well-defined tertiary structure. Interestingly, the addition of 20% TFE alters the spectrum to only a small extent (data not shown), suggesting TFE may temporarily stabilize residues already prone to helix formation rather than propagating helicity to other portions of the sequence.
Figure 7.
2D 1H-15N HSQC spectrum of 0.2 mM 15N-labeled ATF5 WT dimer (red peaks) and C240A monomer (blue peaks) in 20 mM MES, pH 6.5.
Discussion
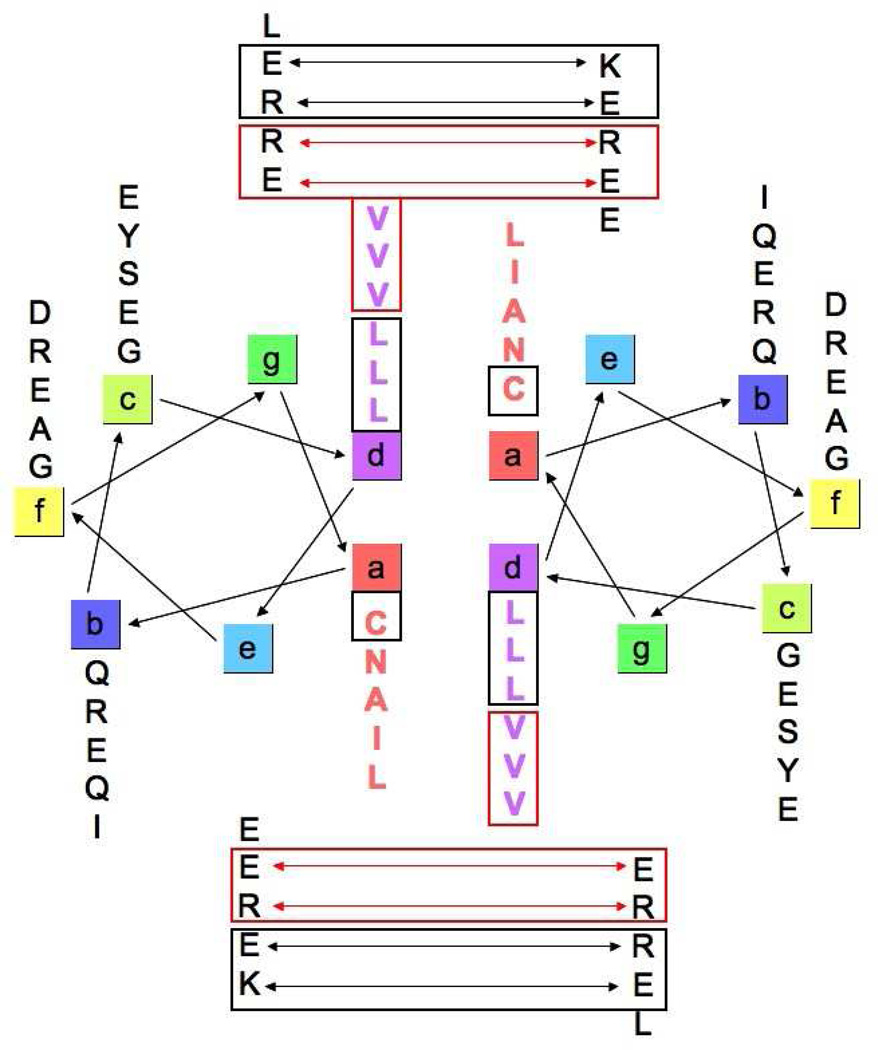
ATF5 belongs to the basic leucine zipper (bZIP) family of transcription factors. These proteins function as dimers that bind DNA and regulate transcription. The bZIP domain, present in all family members, contains a repeating pattern of leucine in every seventh position, which facilitates dimerization through formation of a parallel two-stranded coiled-coil.30 Favorable hydrophobic interactions between residues in the a and d positions of the leucine zipper heptad in conjunction with attractive electrostatic interactions between residues in the e and g positions typically drive coiled-coil formation in bZIP proteins.30 The basic regions of each monomer then bind DNA in what has been described as a “scissors-grip” model of binding.31 bZIP proteins can either homodimerize or heterodimerize with other bZIP family members. Previous findings suggest that ATF5 forms a bZIP homodimer (Figure 8).32 We have identified some favorable interactions that exist in our helical wheel diagram of the ATF5 homodimer that could assist in formation of a coiled-coil. For instance, the leucine residues in the d position (boxed in black) have a high propensity to adopt a helical conformation and are known to pack favorably in two-stranded coiled-coils.33–35 We have shown that disulfide bond formation at the cysteine residue in the a position (boxed in black) retains helical structure, which could be a stabilizing force for a coiled-coil. Additionally, there are a few favorable electrostatic interactions that exist between residues in the e and g positions (depicted by black double-headed arrows and boxed) that could help stabilize a coiled-coil homodimer. However, there are also several unfavorable interactions present here that are not found in other bZIP coiled-coils. The valine residues in the d position (outlined in red) have a much lower propensity to adopt helical structure.33 β-branched amino acids, like valine, are also not favored in the packing arrangement of two-stranded coiled-coils.34, 35 A series of unfavorable electrostatic interactions exist between residues in the e and g positions (delineated by red double-headed arrows and boxed) that would further prevent formation of a stable coiled-coil. We have collected structural data on the bZIP domain of ATF5 at high concentrations using CD and solution NMR,14 and our data indicate that the ATF5 protein does not form a stable coiled-coil in solution under physiological pH, ionic strength and reducing conditions.14 The absence of valine residues and repulsive electrostatic interactions in the ATF4 and other bZIP coiled-coil structures implies that these features are responsible for the prevention of a stable coiled-coil for the ATF5 homodimer.
Figure 8.
Helical wheel diagram of an ATF5 homodimer depicting standard coiled-coil interactions. Favorable interactions that would stabilize formation of a coiled-coil are delineated in black. Repulsive interactions that would be destabilizing are shown in red. The diagram reveals the lack of consensus among homotypic interactions to support coiled-coil stabilization.
ATF5 shares 74% sequence homology with another bZIP protein family member, ATF4 (Figure 9). A crystal structure has been solved for ATF4, in which the protein participates in formation of a bZIP heterodimer with CCAAT enhancer binding protein β (C/EBPβ).36 In this structure, ATF4 is shown to exist as a very straight α-helix, while C/EBPβ curves to wrap around ATF4 and create the coiled-coil. This is a unique feature of ATF4, as most bZIP proteins display more flexibility, with two bZIP monomers curving equally to wrap around each other.37–41 The structure suggests that the rigidity of the ATF4 α-helix disfavors homodimerization, i.e. coiled-coil formation, and therefore must dimerize with a more flexible partner. The lack of stable homodimer formation under reducing conditions suggests ATF5 may heterodimerize in vivo and that it is prone to aggregation in vitro because its hydrophobic side chains are exposed and unable to form native interactions without the alignment imposed by covalent attachment.
Figure 9.
Sequence alignment of the bZIP domain of ATF4 and ATF5. The basic residues comprising the basic DNA binding domain are highlighted in blue. The leucine and valine residue located in the d-position of the leucine zipper heptad are highlighted in red. The unusual cysteine residue poised to form an intermolecular disulfide bond is underlined. The region of ATF5 most likely to form α-helical structure is boxed.
Alternatively, it has been suggested that intermolecular disulfide bond formation might facilitate formation of an ATF4 or ATF5 homodimer.36 A unique cysteine residue is located at the center of the bZIP region of ATF4.36 This analogous cysteine is positioned such that it would be at the interface of a coiled-coil dimer. ATF4 and ATF5 are the only bZIP proteins that contain a cysteine residue in this position, suggesting a functional role for this residue in vivo. We have collected NMR data in the presence and absence of this intermolecular disulfide bond for ATF5. The notable lack of dispersion in 1H signal in the spectra of both the reduced monomer and oxidized dimer compared to spectra of other coiled-coil proteins indicates disulfide bond formation at this position does not confer formation of a stable coiled-coil (Figures 7 and 8). This disulfide bond does, however, play a role in retention of α-helical structure and physical stability of the protein to resist aggregation. If homodimerization has a biological function, the protein is likely to be redox regulated, as has been reported for other transcription factors.42
A naturally occurring mutant of apolipoprotein A-I (ApoA-I) known as the Milano variant (ApoA-IM) was found to affect the stability of this protein in a parallel manner to that observed for ATF5. The Milano mutation introduces a non-native Cys residue that results in the formation of a disulfide-bonded homodimer. Cys substitution significantly increases both the percent helical content and the stability of this protein at high concentration.43 Cross-linking stabilizes a conformational change in ApoA-IM that increases the helical content to match that of the native dimerized form, which is present at higher concentrations of protein. At lower protein concentration this mutation and other substitutions at the same site leads to diminished helicity and stability due to disruption of a salt bridge.44 Addition of TFE to monomeric ApoA-I also increases its helical content to match that of the Milano form and concomitantly improves stability of the native protein against aggregation. ApoA-I is prone to form amyloid aggregates in vivo, whereas the disulfide-bound Milano form resists aggregation.45 Interestingly, mutation at the same or adjacent positions in the protein that do not confer covalent attachment render ApoA-I more susceptible to aggregation.45 Despite that ApoA-I and ATF5 are unrelated in sequence or function, these two systems display parallel behaviors with respect to the influence of helicity and disulfide cross-linked dimerization on stability and aggregation. Further investigation of ATF5 and comparison to ApoA-I may provide insights to better understand the basis of how helical structure and dynamics influence stability and aggregate formation.
The bZIP domain of ATF5 contains several valine residues instead of leucine residues at the C-terminal end of this region. While valine residues are commonly located in the a-position of the leucine zipper heptad, they rarely occupy the d-position. ATF5 is the only bZIP protein to possess three consecutive valines in this domain. ATF4 encodes only a single valine residue in the last d-position of its bZIP domain. It is not known what affect these β-branched amino acids would have on the binding partner selectively of ATF5 in vivo, but they seem to prevent coiled-coil formation and are likely to be a key component of the protein’s instability in vitro.
Our data show that the bZIP domain of ATF5 is only partially structured in solution, with its percent helicity increasing from 19.0% to 23.4% in the presence of the intermolecular disulfide bond. Using the DISEMBLTM server we predicted the intrinsic disorder of this domain (data not shown).46, 47 The N-terminal basic domain (residues 203–238) and the end of the C-terminal leucine zipper domain (residues 275–282) are the most likely regions to be disordered. Additionally, a number of studies have shown that the helical propensity of leucine is much greater than valine.33 This suggests that the central portion of the leucine zipper of ATF5 is most likely to be helical. There are seventeen residues between the cysteine at position 240 and the valine at position 257, which make up the first half of the leucine zipper domain. If this region displayed 100% helicity and the rest of the protein was disordered, the predicted percent helicity would be 22.8%. This matches closely with the experimental values obtained for ATF5. An intermolecular disulfide bond positioned at cysteine 240 would likely help to stabilize the helical structure in this region by constraining the two chains and increasing intermolecular contact between them.
Computational methods for predicting protein aggregation have been developed, and we used several to examine ATF5. TANGO is a statistical mechanical model used to identify nucleation sites for aggregation based on the observations that aggregates often contain increased β-structure and the core regions of an aggregate are completely buried, such that nucleating sequences will have their hydrogen bonding potential largely satisfied.48 This program predicts that the region most likely to initiate aggregation in ATF5 lies between residues 260 and 276. This corresponds to the valine-containing portion of the leucine zipper, which is largely hydrophobic (Figure 9). AGGRESCAN is another sequence-based tool, which relies upon the aggregation-propensity for each of the individual amino acids to identify sequences likely to aggregate.49 An evaluation of the bZIP domain of ATF5 identified the region contained between residues 265 and 273 as most likely to aggregate, which again, falls within the valine-containing portion of the leucine zipper. Based upon this information, it is likely that the structural features demonstrated to impart improved stability to ATF5 function protect the less-structured and aggregation-prone valine-containing portion of the protein from intermolecular association. A third program was used to evaluate the propensity of protein molecules to self-associate. Prediction of amyloid structure aggregation (PASTA) is based on the alignment observed in most cross-β structures.50 PASTA predicted that it is unlikely for ATF5 to form cross-β fibrillar aggregates, which is consistent with our experimental observations of amorphous aggregates. The PASTA results, however, indicated that the antiparallel alignment of ATF5 in solution is energetically preferred over parallel alignment. This small preference could explain why aggregates form more rapidly between monomers. Covalent dimerization may not so much alter the structure of the monomer as constrain the Val-containing sequences in close proximity and provide an adjacent hydrophobic surface with which to interact.51
We have shown that the retention of α-helical structure, either through formation of an intermolecular disulfide bond or via the addition of TFE, prevents rapid, thermally-induced protein aggregation. Based on the prediction results derived from first principles, it is most likely that the disordered regions of ATF5 play a role in initiating the process of aggregation rather than the uncoiled helical sequence. As electrostatic repulsion will dominate and prevent association between the basic regions, we propose that the hydrophobic residues present in the valine-containing portion of the protein are involved in initiating protein aggregation. Reduction of the disulfide bond at C240 would result in greater conformational freedom and exposure of hydrophobic residues in this region, which would increase protein aggregation. This explains why aggregation proceeds so much more quickly for the reduced WT and C240A monomer. The addition of TFE likely stabilizes helicity of the Leu-containing region, providing enhanced rigidity and reduced dynamic motion in the Val-containing region that would diminish solvent exposure and prevent self-association. Based upon our experimental evidence along with the results acquired using the computational tools identified above, we have created a model that depicts the aggregation mechanism of ATF5 (Figure 10). In using such a simple system to investigate protein aggregation, we have clear evidence to support the notion that increasing the structural integrity of native α-helices improves protein physical stability. This conclusion has implications for handling other helical proteins in vitro and provides a basis for further investigating amorphous aggregation at higher resolution.
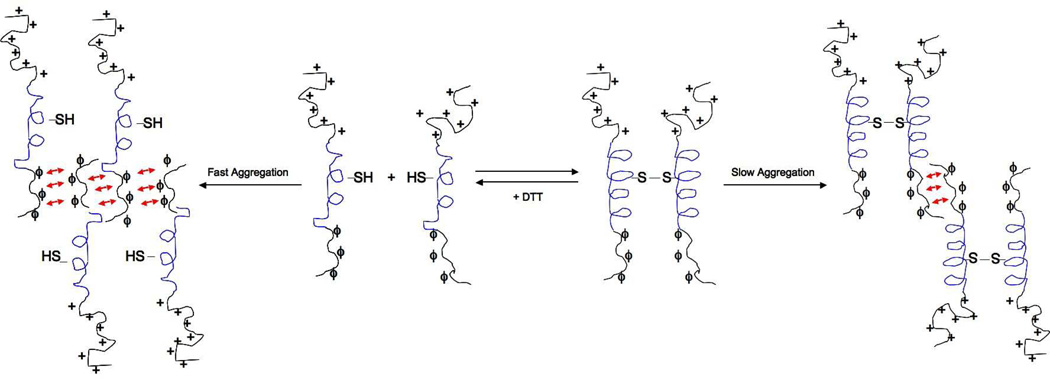
Fig 10.
Aggregation model of ATF5. Slow aggregation of the cross-linked ATF5 dimer is facilitated by antiparallel association of the disordered valine-containing region (ϕ). Reduction of the disulfide bond creates the monomeric species, which displays decreased helicity and participates in fast aggregation through antiparallel alignment of the same valine-containing region. The N-terminal basic domain is illustrated by the addition of symbols indicating positive charge (+). The helical portion of the leucine zipper is depicted in blue. Note: A limited set of arrangements is shown here, but others are equally possible.
Supplementary Material
Table 1.
Summary of the results from the CD and SLS experiments for the WT and C240A forms of ATF5 in the absence and presence of the 10 mM DTT.
| Percent Helicity (%) |
Melting Temperature (°C) |
Maximum Intensity of Scattered Light (AU) |
|
|---|---|---|---|
| WT, no DTT | 23.4 ± 0.3 | 55.5 | 35,815 ± 2,658 |
| WT + DTT | 17.9 ± 0.2 | 19.9 | 173,132 ± 4,151 |
| C240A, no DTT | 19.0 ± 1.0 | 15.9 | 143,560 ± 13,475 |
| C240A + DTT | 16.7 ± 0.1 | 15.1 | 175,415 ± 1,149 |
Table 2.
Summary of the results from the CD and SLS experiments for the WT and C240A forms of ATF5 in the presence of increasing amounts of the co-solvent TFE. A melting temperature could not be calculated for the protein in the presence of 10% and 20% TFE, because the data could not be fit to a sigmoidal curve. NA= not applicable.
| Percent Helicity (%) |
Melting Temperature (°C) |
Maximum Intensity of Scattered Light (AU) |
|
|---|---|---|---|
| WT, no TFE | 25.5 ± 0.6 | 56.0 | 259,404 ± 15,061 |
| WT, 10% TFE | 30.8 ± 0.6 | NA | 150,731 ± 6,096 |
| WT, 20% TFE | 39.3 ± 1.0 | NA | 139,743 ± 6,404 |
| C240A, no TFE | 19.5 ± 0.3 | 17.2 | 779,386 ± 87,881 |
| C240A, 10% TFE | 30.5 ± 0.3 | NA | 277,567 ± 13,598 |
| C240A, 20% TFE | 42.0 ± 2.2 | NA | 248,188 ± 9,972 |
Acknowledgements
The authors thank Dr. Russ Middaugh and Brooke Barrett for their technical assistance and discussion of the data and manuscript. This publication was made possible by NIH Grant Number P20 RR-17708 from the National Center for Research Resources and the Kansas University Center for Research. Additional support was provided by the Madison and Lila Self Graduate Fellowship for N. Ciaccio.
Footnotes
Supporting Information
A table containing average hydrodynamic diameter values for the WT and C240A forms of the protein at temperatures ranging from 10°C to 80°C is included as supplemental data (Table S1). This information is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Hermeling S, Schellekens H, Maas C, Gebbink MF, Crommelin DJ, Jiskoot W. Antibody response to aggregated human interferon alpha2b in wild-type and transgenic immune tolerant mice depends on type and level of aggregation. J Pharm Sci. 2006;95(5):1084–1096. doi: 10.1002/jps.20599. [DOI] [PubMed] [Google Scholar]
- 2.Purohit VS, Middaugh CR, Balasubramanian SV. Influence of aggregation on immunogenicity of recombinant human Factor VIII in hemophilia A mice. J Pharm Sci. 2006;95(2):358–371. doi: 10.1002/jps.20529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schellekens H. How to predict and prevent the immunogenicity of therapeutic proteins. Biotechnol Annu Rev. 2008;14:191–202. doi: 10.1016/S1387-2656(08)00007-0. [DOI] [PubMed] [Google Scholar]
- 4.Pecher P, Arnold U. The effect of additional disulfide bonds on the stability and folding of ribonuclease A. Biophys Chem. 2008 doi: 10.1016/j.bpc.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 5.Kumar S, Ravi VK, Swaminathan R. How do surfactants and DTT affect the size, dynamics, activity and growth of soluble lysozyme aggregates? Biochem J. 2008;415(2):275–288. doi: 10.1042/BJ20071499. [DOI] [PubMed] [Google Scholar]
- 6.Knowles TP, Zahn R. Enhanced stability of human prion proteins with two disulfide bridges. Biophys J. 2006;91(4):1494–1500. doi: 10.1529/biophysj.106.081653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu BY, Chang JY. A 3-disulfide mutant of mouse prion protein expression, oxidative folding, reductive unfolding, conformational stability, aggregation and isomerization. Arch Biochem Biophys. 2007;460(1):75–84. doi: 10.1016/j.abb.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 8.Huang K, Maiti NC, Phillips NB, Carey PR, Weiss MA. Structure-specific effects of protein topology on cross-beta assembly: studies of insulin fibrillation. Biochemistry. 2006;45(34):10278–10293. doi: 10.1021/bi060879g. [DOI] [PubMed] [Google Scholar]
- 9.Dima RI, Thirumalai D. Probing the instabilities in the dynamics of helical fragments from mouse PrPC. Proc Natl Acad Sci U S A. 2004;101(43):15335–15340. doi: 10.1073/pnas.0404235101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu W, Prausnitz JM, Blanch HW. Amyloid fibril formation by peptide LYS (11–36) in aqueous trifluoroethanol. Biomacromolecules. 2004;5(5):1818–1823. doi: 10.1021/bm049841e. [DOI] [PubMed] [Google Scholar]
- 11.Kunjithapatham R, Oliva FY, Doshi U, Perez M, Avila J, Munoz V. Role for the alpha-helix in aberrant protein aggregation. Biochemistry. 2005;44(1):149–156. doi: 10.1021/bi048564t. [DOI] [PubMed] [Google Scholar]
- 12.Morgan GJ, Giannini S, Hounslow AM, Craven CJ, Zerovnik E, Turk V, Waltho JP, Staniforth RA. Exclusion of the native alpha-helix from the amyloid fibrils of a mixed alpha/beta protein. J Mol Biol. 2008;375(2):487–498. doi: 10.1016/j.jmb.2007.10.033. [DOI] [PubMed] [Google Scholar]
- 13.Watzlawik J, Skora L, Frense D, Griesinger C, Zweckstetter M, Schulz-Schaeffer WJ, Kramer ML. Prion protein helix1 promotes aggregation but is not converted into beta-sheet. J Biol Chem. 2006;281(40):30242–30250. doi: 10.1074/jbc.m605141200. [DOI] [PubMed] [Google Scholar]
- 14.Ciaccio NA, Moreno ML, Bauer RL, Laurence JS. High-yield expression in E. coli and refolding of the bZIP domain of activating transcription factor 5. Protein Expr Purif. 2008;62(2):235–243. doi: 10.1016/j.pep.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weiss MA. Thermal unfolding studies of a leucine zipper domain and its specific DNA complex: implications for scissor's grip recognition. Biochemistry. 1990;29(35):8020–8024. doi: 10.1021/bi00487a004. [DOI] [PubMed] [Google Scholar]
- 16.Priddy TS, Middaugh CR, Carlson GM. Electrostatic changes in phosphorylase kinase induced by its obligatory allosteric activator Ca2+ Protein Sci. 2007;16(3):517–527. doi: 10.1110/ps.062577507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, Sykes BD. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J Biomol NMR. 1995;6(2):135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- 18.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6(3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 19.TD Goddard DGK. In. [Google Scholar]
- 20.Johnson WC. Analyzing protein circular dichroism spectra for accurate secondary structures. Proteins. 1999;35(3):307–312. [PubMed] [Google Scholar]
- 21.Manning MC, Woody RW. Theoretical CD studies of polypeptide helices: examination of important electronic and geometric factors. Biopolymers. 1991;31(5):569–586. doi: 10.1002/bip.360310511. [DOI] [PubMed] [Google Scholar]
- 22.Zhou NE, Kay CM, Hodges RS. The role of interhelical ionic interactions in controlling protein folding and stability. De novo designed synthetic two-stranded alpha-helical coiled-coils. J Mol Biol. 1994;237(4):500–512. doi: 10.1006/jmbi.1994.1250. [DOI] [PubMed] [Google Scholar]
- 23.Buck M. Trifluoroethanol and colleagues: cosolvents come of age. Recent studies with peptides and proteins. Q Rev Biophys. 1998;31(3):297–355. doi: 10.1017/s003358359800345x. [DOI] [PubMed] [Google Scholar]
- 24.Povey JF, Smales CM, Hassard SJ, Howard MJ. Comparison of the effects of 2,2,2-trifluoroethanol on peptide and protein structure and function. J Struct Biol. 2007;157(2):329–338. doi: 10.1016/j.jsb.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 25.Atkinson RA, Saudek V, Huggins JP, Pelton JT. 1H NMR and circular dichroism studies of the N-terminal domain of cyclic GMP dependent protein kinase: a leucine/isoleucine zipper. Biochemistry. 1991;30(39):9387–9395. doi: 10.1021/bi00103a001. [DOI] [PubMed] [Google Scholar]
- 26.Santiago-Rivera ZI, Williams JS, Gorenstein DG, Andrisani OM. Bacterial expression and characterization of the CREB bZip module: circular dichroism and 2D 1H-NMR studies. Protein Sci. 1993;2(9):1461–1471. doi: 10.1002/pro.5560020910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lumb KJ, Carr CM, Kim PS. Subdomain folding of the coiled coil leucine zipper from the bZIP transcriptional activator GCN4. Biochemistry. 1994;33(23):7361–7367. doi: 10.1021/bi00189a042. [DOI] [PubMed] [Google Scholar]
- 28.Ishigaki T, Ohki I, Utsunomiya-Tate N, Tate SI. Chimeric structural stabilities in the coiled-coil structure of the NECK domain in human lectin-like oxidized low-density lipoprotein receptor 1 (LOX-1) J Biochem. 2007;141(6):855–866. doi: 10.1093/jb/mvm093. [DOI] [PubMed] [Google Scholar]
- 29.Nikolaev Y, Pervushin K. NMR spin state exchange spectroscopy reveals equilibrium of two distinct conformations of leucine zipper GCN4 in solution. J Am Chem Soc. 2007;129(20):6461–6469. doi: 10.1021/ja0685295. [DOI] [PubMed] [Google Scholar]
- 30.O'Shea EK, Rutkowski R, Kim PS. Evidence that the leucine zipper is a coiled coil. Science. 1989;243(4890):538–542. doi: 10.1126/science.2911757. [DOI] [PubMed] [Google Scholar]
- 31.Vinson CR, Sigler PB, McKnight SL. Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science. 1989;246(4932):911–916. doi: 10.1126/science.2683088. [DOI] [PubMed] [Google Scholar]
- 32.Peters CS, Liang X, Li S, Kannan S, Peng Y, Taub R, Diamond RH. ATF-7, a novel bZIP protein, interacts with the PRL-1 protein-tyrosine phosphatase. J Biol Chem. 2001;276(17):13718–13726. doi: 10.1074/jbc.M011562200. [DOI] [PubMed] [Google Scholar]
- 33.Krittanai C, Johnson WC., Jr The relative order of helical propensity of amino acids changes with solvent environment. Proteins. 2000;39(2):132–141. doi: 10.1002/(sici)1097-0134(20000501)39:2<132::aid-prot3>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 34.Moitra J, Szilak L, Krylov D, Vinson C. Leucine is the most stabilizing aliphatic amino acid in the d position of a dimeric leucine zipper coiled coil. Biochemistry. 1997;36(41):12567–12573. doi: 10.1021/bi971424h. [DOI] [PubMed] [Google Scholar]
- 35.Harbury PB, Zhang T, Kim PS, Alber T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993;262(5138):1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 36.Podust LM, Krezel AM, Kim Y. Crystal structure of the CCAAT box/enhancer-binding protein beta activating transcription factor-4 basic leucine zipper heterodimer in the absence of DNA. J Biol Chem. 2001;276(1):505–513. doi: 10.1074/jbc.M005594200. [DOI] [PubMed] [Google Scholar]
- 37.Junius FK, O'Donoghue SI, Nilges M, Weiss AS, King GF. High resolution NMR solution structure of the leucine zipper domain of the c-Jun homodimer. J Biol Chem. 1996;271(23):13663–13667. doi: 10.1074/jbc.271.23.13663. [DOI] [PubMed] [Google Scholar]
- 38.Glover JN, Harrison SC. Crystal structure of the heterodimeric bZIP transcription factor c-Fos-c-Jun bound to DNA. Nature. 1995;373(6511):257–261. doi: 10.1038/373257a0. [DOI] [PubMed] [Google Scholar]
- 39.Schumacher MA, Goodman RH, Brennan RG. The structure of a CREB bZIP.somatostatin CRE complex reveals the basis for selective dimerization and divalent cation-enhanced DNA binding. J Biol Chem. 2000;275(45):35242–35247. doi: 10.1074/jbc.M007293200. [DOI] [PubMed] [Google Scholar]
- 40.Miller M, Shuman JD, Sebastian T, Dauter Z, Johnson PF. Structural basis for DNA recognition by the basic region leucine zipper transcription factor CCAAT/enhancer-binding protein alpha. J Biol Chem. 2003;278(17):15178–15184. doi: 10.1074/jbc.M300417200. [DOI] [PubMed] [Google Scholar]
- 41.Ellenberger TE, Brandl CJ, Struhl K, Harrison SC. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein-DNA complex. Cell. 1992;71(7):1223–1237. doi: 10.1016/s0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- 42.Arrigo AP. Gene expression and the thiol redox state. Free Radic Biol Med. 1999;27(9–10):936–944. doi: 10.1016/s0891-5849(99)00175-6. [DOI] [PubMed] [Google Scholar]
- 43.Calabresi L, Vecchio G, Longhi R, Gianazza E, Palm G, Wadensten H, Hammarstrom A, Olsson A, Karlstrom A, Sejlitz T, et al. Molecular characterization of native and recombinant apolipoprotein A-IMilano dimmer. The introduction of an interchain disulfide bridge remarkably alters the physicochemical properties of apolipoprotein A-I. J Biol Chem. 1994;269(51):32168–32174. [PubMed] [Google Scholar]
- 44.Alexander ET, Tanaka M, Kono M, Saito H, Rader DJ, Phillips MC. Structural and functional consequences of the milano mutation (R173C) in human apolipoprotein A-I. J Lipid Res. 2009 doi: 10.1194/jlr.M800578-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Obici L, Franceschini G, Calabresi L, Giorgetti S, Stoppini M, Merlini G, Bellotti V. Structure, function and amyloidogenic propensity of apolipoprotein A-I. Amyloid. 2006;13(4):191–205. doi: 10.1080/13506120600960288. [DOI] [PubMed] [Google Scholar]
- 46.Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, Russell RB. Protein disorder prediction: implications for structural proteomics. Structure. 2003;11:1453–1459. doi: 10.1016/j.str.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 47.Iakoucheva LM, Dunker AK. Order, disorder, and flexibility: prediction from protein sequence. Structure. 2003;11(11):1316–1317. doi: 10.1016/j.str.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 48.Fernandez-Escamilla AM, Rousseau F, Schymkowitz J, Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat Biotechnol. 2004;22(10):1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- 49.Conchillo-Sole O, de Groot NS, Aviles FX, Vendrell J, Daura X, Ventura S. AGGRESCAN: a server for the prediction and evaluation of "hot spots" of aggregation in polypeptides. BMC Bioinformatics. 2007;8:65. doi: 10.1186/1471-2105-8-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trovato A, Seno F, Tosatto SC. The PASTA server for protein aggregation prediction. Protein Eng Des Sel. 2007;20(10):521–523. doi: 10.1093/protein/gzm042. [DOI] [PubMed] [Google Scholar]
- 51.Ofran Y, Rost B. Analysing six types of protein-protein interfaces. J Mol Biol. 2003;325(2):377–387. doi: 10.1016/s0022-2836(02)01223-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.