Abstract
Small mammals from the Democratic Republic (DR) of the Congo and Tanzania were tested to determine the prevalence and genetic diversity of Bartonella species. The presence of Bartonella DNA was assessed in spleen samples of the animals by rpoB- and gltA-polymerase chain reactions (PCRs). By rpoB-PCR, Bartonella was detected in 8 of 59 animals of DR Congo and in 16 of 39 Tanzanian animals. By gltA-PCR, Bartonella was detected in 5 and 15 animals of DR Congo and Tanzania, respectively. The gene sequences from Arvicanthis neumanni were closely related to Bartonella elizabethae. The genotypes from Lophuromys spp. and from Praomys delectorum were close to Bartonella tribocorum. Five genogroups were not genetically related to any known Bartonella species. These results suggest the need to conduct further studies to establish the zoonotic risks linked with those Bartonella species and, in particular, to verify whether these agents might be responsible for human cases of febrile illness of unknown etiology in Africa.
Introduction
Bartonella species are zoonotic and vector-borne bacteria associated with an increasing array of emerging infections in humans and animals.1–3 These bacteria are responsible for a wide range of clinical manifestations, including trench fever, cat-scratch disease, and endocarditis in immunocompetent patients, and bacillary angiomatosis and peliosis hepatitis in immunocompromised patients.4 Bartonellae typically parasitize the erythrocytes of mammalian hosts, resulting in long-lasting infections. Several new Bartonella species have been isolated recently from a wide range of wild mammals, including rodents,5–15 lagomorphs,16,17 carnivores,1,18 and ruminants.1,19 The close association between rodents and humans throughout the world, especially in rural environments and in the overcrowded metropololis of sub-Saharan Africa, makes the study of rodent-borne Bartonella essential to determine the extent to which rodents may serve as a source of human infections.20
Bartonella species associated with small mammals have been detected in Asia, Australia, North America, and Europe.11,15,20–27 Recent studies showed that bartonellae were widely distributed among rodents in South Africa14 and among fleas in Democratic Republic of the Congo (DR Congo).28 However, no study was conducted in small mammals of DR Congo or Tanzania. The aims of this study were 1) to investigate the prevalence of Bartonella infections in small mammal populations of selected areas of these two countries; 2) to evaluate the genetic diversity of Bartonella communities by analyzing partial sequences of gltA and rpoB genes; and 3) to compare Bartonella genotypes obtained from small animals in DR Congo and Tanzania with genotypes identified in Africa and other regions of the world.
Materials and Methods
Mammal sampling.
Small mammals were sampled either in crop fields or fallow land during March–April 2007 in the Rethy village of the Ituri district (N2.09176-E30.88982), DR Congo at elevations ranging from 1,960 to 2,120 m above sea level. In Mbulu district, northern Tanzania, rodents were trapped during February–March 2007 in two villages, namely Arri and Tumati, which are located in the Division of Dongobesh (S040 04-E0350 22) at altitudes ranging from 1,930 to 2,250 m a.s.l. Trapping was conducted in the forest near the hamlet of Mongahay in Tumati and in crop fields of Arri. Details of the captured animals are given in Tables 1 and 2.
Table 1.
Prevalence of Bartonella in small mammals of Democratic Republic of Congo
| Mammal species | Common name | Habitat | Area/district | No. positive/studied (% positive) | Total no. of positive/studied (% positive) |
|---|---|---|---|---|---|
| Arvicanthis neumanni | Neumann's grass rat | Crop field | Kpandruma | 0/1 (0) | 2/5 (40) |
| – | Rethy | 2/3 (66.7) | |||
| – | Zaa | 0/1 (0) | |||
| Crocidura sp. | White-toothed shrew | Swamp area | Djalusene | 0/1 (0) | 0/8 (0) |
| – | Kpandruma | 0/1 (0) | |||
| – | Rethy | 0/3 (0) | |||
| – | Zaa | 0/3 (0) | |||
| Lophuromys rita | Yellow-spotted brush-furred rat | Crop field | Djalusene | 0/1 (0) | 1/4 (25) |
| – | Rethy | 1/3 (33.3) | |||
| Mastomys coucha | Multimammate rat | Crop field | Kpandruma | 0/4 (0) | 0/10 (0) |
| House | Zaa | 0/6 (0) | |||
| Mus minutoides | African pygmy mouse | Bushes | Djalusene | 0/1 (0) | 4/6 (66.7) |
| Bushes | Rethy | 3/4 (75) | |||
| Swamp area | Rethy | 1/1 (100) | |||
| Otomys sp. | African vlei rat | Crop filed | Rethy | 0/1 (0) | 0/1 (0) |
| Rattus rattus | Black rat | House, domestic environment | Djalusene | 0/1 (0) | 1/25 (4) |
| Kpandruma | 0/5 (0) | ||||
| Rethy | 0/2 (0) | ||||
| Zaa | 1/17 (5.9) | ||||
| Total (7 species) | 8/59 (13.6) |
Table 2.
Prevalence of Bartonella in rodents of Tanzania
| Rodent species | Common name | Habitat | Area/district | No. of positive/studied (% positive) | Total no. of positive/studied (% positive) |
|---|---|---|---|---|---|
| Grammomys sp. | African thicket rat | Crop field and fallow land | Ari | 1/1 (100) | 1/1 (100) |
| Lophuromys sp. (dudui-related) | Brush-furred rat | Crop field and fallow land | Tumati | 3/6 (50) | 9/18 (50) |
| Natural forest | Ari | 0/1 (0) | |||
| – | Tumati | 6/11 (54.5) | |||
| Mus minutoides | African pigmy mouse | Crop field and fallow land | Ari | 0/2 (0) | 0/11 (0) |
| – | Tumati | 0/9 (0) | |||
| Praomys delectorum | African soft-furred rat | Crop field and fallow land | Tumati | 2/3 (66.7) | 6/9 (66.7) |
| Natural forest | – | 4/6 (66.7) | |||
| Total (4 species) | 16/39 (41) |
Small mammals were captured mainly using Sherman traps (model LFA, 3 × 3, 5 × 9 in.; Sherman Traps Inc., Tallahassee, FL) baited with peanut butter mixed with maize flour. Occasionally, Tomahawk collapsible traps (model 202, Tomahawk Live Traps Co., WI), and locally made box-traps were also used. Rodents were trapped in various habitats including primary altitude natural forest, fallow land (shrubs and bushes), and crop fields. In Mbulu District, rodents were trapped in two principal habitats: 1) the natural rain forest with dense undergrowth and tall trees and 2) the crop field in the fringes of the forest. Trapping of the small mammals in these habitats was carried out for three consecutive nights with 100 Sherman traps per habitat per night. Each captured animal was transferred to the laboratory in a tissue bag and euthanized with ether. Tissue samples were taken from spleen and stored in 90% ethanol.
Each mammal was initially identified to genus level in the field. Before sample collection, the gender and species of animals were recorded. Species identification of the animals was confirmed in the laboratories of the University of Antwerp and the Royal Institute of Natural Science in Brussels (Belgium) by combining craniometrical measurements and mitochondrial DNA cytochrome-b sequencing.
Molecular screening for bartonellae DNA.
Genomic DNA was isolated from spleen samples using tissue protocol of the QIAamp DNA Mini Kit (Qiagen, Valencia, CA) according to manufacturer's instructions and stored at –20°C. The quantity of DNA was measured using NanoDrop 2000 (Thermo Scientific, Wilmington, DE). Extracted DNA was used in all polymerase chain reaction (PCR) assays. Primers used were 1400F and 2300R29 for an 825-bp specific fragment of rpoB gene.29 For the specific fragments of the gltA, we used two combinations of the primers: CS140f-BhCS.1137n30,31 and CS443f-BhCS.1137n30 for 327 bp. The PCR was performed with 50-μL mixtures containing 20 ng of the DNA, 5 × Green GoTaq reaction buffer (10 μL), 200 μM of each dNTP, 1.25 U Taq DNA polymerase (Promega, Madison, WI), and 1.0 μM of each primer. Each PCR was conducted in a PTC-200 Peltier Thermal Cycler (MJ Research, Waltham, MA). The PCR was incubated at 94°C for 2 min to denature DNA and the thermal cycle reaction programmed for 38 cycles of 30 s at 94°C, 30 s at 52°C (for rpoB) and 48°C (for gltA), and 2 min at 72°C, with a 7-min final extension step at 72°C. The PCR products were subjected to electrophoresis on a 1.5% agarose gel and stained with ethidium bromide. Amplicons of the expected size were identified by size comparison to the positive control. The PCR products were purified using the QIAquick PCR Purification Kit (Qiagen, Germantown, MD). Primers 1400F, 2028F, 1596R, 1873R, and 2300R for rpoB29 and CS140f, CS443f, and BhCS.1137n for gltA30 were used for DNA sequencing. Sequencing reactions were carried out with a PTC-200 Peltier Thermal Cycler, using Dye Terminator Cycle Sequencing with the Quick Start kit (Beckman Coulter, Fullerton, CA) using the following program: initial denaturing step for 1 min at 96°C, and 96°C for 10 s, 50°C for 5 s, 60°C for 4 min, and each step was repeated for 25 cycles. The DNA sequences were analyzed using Lasergene version 8 sequence analysis software (DNASTAR, Madison, WI). The SeqMan program (DNASTAR) was used to obtain consensus sequences for the amplified regions of the target genes. The DNA sequences of this study were deposited in GenBank (Table 3).
Table 3.
Bartonella rpoB and gltA genotypes found in the small mammals of Democratic Republic of Congo and Tanzania
| Mammal species | Animal ID | Country | rpoB genotyping | gltA genotyping | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| GenBank accession nos. | Genotype | Group/% similarity within group | Closest Bartonella spp./% similarity | GenBank accession nos. | Genotype | Group/% similarity within group | Closest Bartonella spp./% similarity | |||
| Arvicanthis neumanni | An615drc | DR Congo | FJ851128 | 1-r | A-r | B. elizabethae/98.1 | FJ851106 | 1-g | A-g/96–98.5 | B. elizabethae/98.4 |
| Praomys delectorum | Pd5696t | Tanzania | FJ851133 | 2-r | B-r | B. elizabethae/93.8 | FJ851110 | 3-g | A-g | B. elizabethae/95.8 |
| Lophuromys sp. (dudui-related) | Ld482t | Tanzania | FJ851146 | 3-r | C-r/96–99.6 | B. tribocorum/93.9 | FJ851122 | 5-g | C-g/96.4–99.7 | B. elizabethae/96 |
| Lophuromys sp. (dudui-related) | Ld481t | Tanzania | FJ851144 | 4-r | C-r | B. tribocorum/93.8 | FJ851121 | 6-g | C-g | B. elizabethae/96 |
| Lophuromys rita | Lr601drc | DR Congo | FJ851123 | 5-r | C-r | B. tribocorum/94.1 | FJ851103 | 16-g | G-g | B. birtlesii/91.1 |
| Lophuromys sp. (dudui-related) | Ld5743t | Tanzania | FJ851132 | 6-r | C-r | B. tribocorum/93.6 | FJ851109 | 7-g | C-g | B. elizabethae/96.3 |
| Lophuromys sp. (dudui-related) | Ld5707t | Tanzania | FJ851136 | 6-r | C-r | B. tribocorum/93.6 | FJ851113 | 7-g | C-g | B. elizabethae/96.3 |
| Lophuromys sp. (dudui-related) | Ld5706t | Tanzania | FJ851140 | 6-r | C-r | B. tribocorum/93.6 | FJ851117 | 7-g | C-g | B. elizabethae/96.3 |
| Lophuromys sp. (dudui-related) | Ld5742t | Tanzania | FJ851134 | 7-r | C-r | B. tribocorum/93.6 | FJ851111 | 15-g | G-g | B. birtlesii/92.6 |
| Mus minutoides | Mm625drc | DR Congo | FJ851125 | 8-r | C-r | B. tribocorum/94.3 | FJ851104 | 4-g | B-g | B. elizabethae/93.6 |
| Mus minutoides | Mm627drc | DR Congo | FJ851126 | 8-r | C-r | B. tribocorum/94.3 | FJ851105 | 4-g | B-g | B. elizabethae/93.6 |
| Mus minutoides | Mm628drc | DR Congo | FJ851129 | 8-r | C-r | B. tribocorum/94.3 | FJ851107 | 4-g | B-g | B. elizabethae/93.6 |
| Mus minutoides | Mm604drc | DR Congo | FJ851124 | 8-r | C-r | B. tribocorum/94.3 | nd | |||
| Lophuromys sp. (dudui-related) | Ld480t | Tanzania | FJ851143 | 9-r | C-r | B. tribocorum/94.8 | FJ851120 | 8-g | D-g | B. queenslandensis/95.4 |
| Grammomys sp. | Gs5686t | Tanzania | FJ851135 | 10-r | D-r | B. grahamii/94.1 | FJ851112 | 9-g | E-g | B. queenslandensis/95.7 |
| Arvicanthis neumanni | An616drc | DR Congo | FJ851127 | 11-r | E-r | B. grahamii/92.8 | nd | |||
| Praomys delectorum | Pd5700t | Tanzania | FJ851138 | 12-r | F-r/97.9–99.9 | B. grahamii/92.5 | FJ851115 | 2-g | A-g | B. elizabethae/97.1 |
| Praomys delectorum | Pd5695t | Tanzania | FJ851137 | 13-r | F-r | B. grahamii/92.4 | FJ851114 | 10-g | F-g/96.3–99.7 | B. tribocorum/97.9 |
| Praomys delectorum | Pd5708t | Tanzania | FJ851139 | 14-r | F-r | B. grahamii/92.2 | FJ851116 | 11-g | F-g | B. tribocorum/97.5 |
| Praomys delectorum | Pd5728t | Tanzania | FJ851142 | 14-r | F-r | B. grahamii/92.2 | FJ851119 | 12-g | F-g | B. tribocorum/96.6 |
| Rattus rattus | Rr641drc | DR Congo | FJ851130 | 15-r | F-r | B. grahamii/92.6 | nd | |||
| Praomys delectorum | Pd5692t | Tanzania | FJ851131 | 16-r | F-r | B. grahamii/92.4 | FJ851108 | 13-g | F-g | B. tribocorum/96.3 |
| Lophuromys sp. (dudui-related) | Ld479t | Tanzania | FJ851145 | 17-r | F-r | B. grahamii/92.4 | nd | |||
| Lophuromys sp. (dudui-related) | Ld5693t | Tanzania | FJ851141 | 18-r | G-r | B. birtlesii/91.2 | FJ851118 | 14-g | G-g/96.0–98.2 | B. birtlesii/92.9 |
nd = not detected.
Phylogenetic analysis.
Analysis of DNA sequences and phylogenetic relationships were done using MEGA4.32 The DNA sequences of this study and the known Bartonella species retrieved from the GenBank were aligned using the Clustal X.33 Phylogenetic trees were drawn separately based on the rpoB (825 bp) and gltA (327 bp) gene fragments, using the neighbor-joining method34 with the Kimura 2-parameter distance model35 in MEGA4.32 The stability of inferred phylogenies was assessed by bootstrap analysis of 1,000 randomly generated sample trees.36
Results
Animal collection.
A total of 98 small mammals of 10 genera were used for this study (Tables 1 and 2). Fifty-nine animals, including 51 rodents of six genera, Arvicanthis, Lophuromys, Mastomys, Mus, Otomys, and Rattus, all belong to Muridae and 8 white-toothed shrews of the genus Crocidura of Soricidae, were sampled in DR Congo (Table 1). Thirty-nine rats of 4 genera (Muridae), Grammomys, Lophuromys, Mus, and Praomys were trapped in Tanzania (Table 2). All field procedures involving animal sample collection were conducted under protocols approved by the University of Antwerp.
Prevalence of Bartonella in DR Congo and Tanzania.
By rpoB-PCR, Bartonella DNA was detected in 8 of 59 (13.6%) rodents sampled in DR Congo: in 2 Arvicanthis neumanni, 1 Lophuromys rita, 4 Mus minutoides, and 1 Rattus rattus (Table 1). The highest prevalence was observed in M. minutoides, 4 of 6 (67.0%). The lowest rate of Bartonella infection was found in R. rattus, 1 of 25 (4.0%). By gltA-PCR, Bartonella DNA was detected in 5 (8.5%) animals collected in DR Congo. The gltA-PCR was negative in one of A. neumanni and M. minutoides and in R. rattus animals, which were positive by the rpoB-PCR. No evidence of infection was found in 19 animals: Crocidura sp. (N = 8), Mastomys coucha (N = 10), and Otomys sp. (N = 1).
In Tanzanian rodents, 1 Grammomys sp. (100%), 9 Lophuromys sp. (50%), and 6 Praomys delectorum (66.7%) were infected by Bartonella (Table 2). The rpoB sequences were obtained in 16 out of a total 39 animals (41.0%). By gltA-PCR, Bartonella DNA was detected in 15 (38.5%) animals of Tanzania. The gltA-PCR was negative in one of Lophuromys sp., in which Bartonella DNA was detected by the rpoB gene. No evidence of infection was found in 9 M. minutoides.
Genetic diversity of Bartonella DNA sequences.
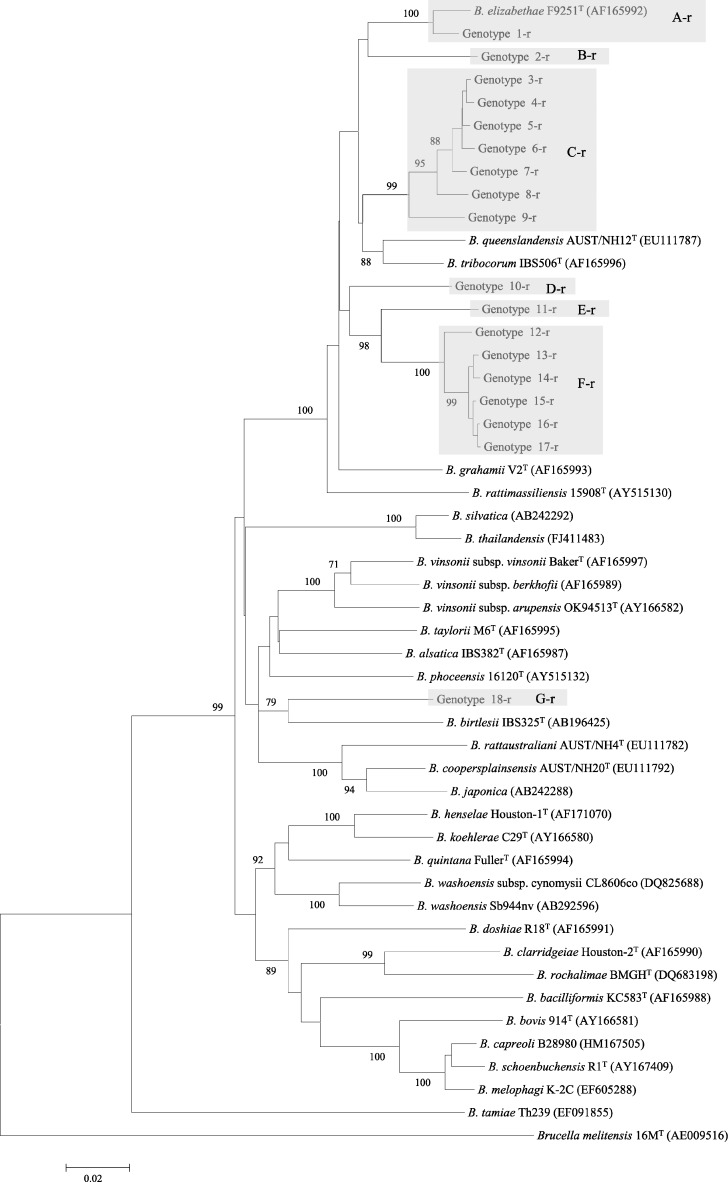
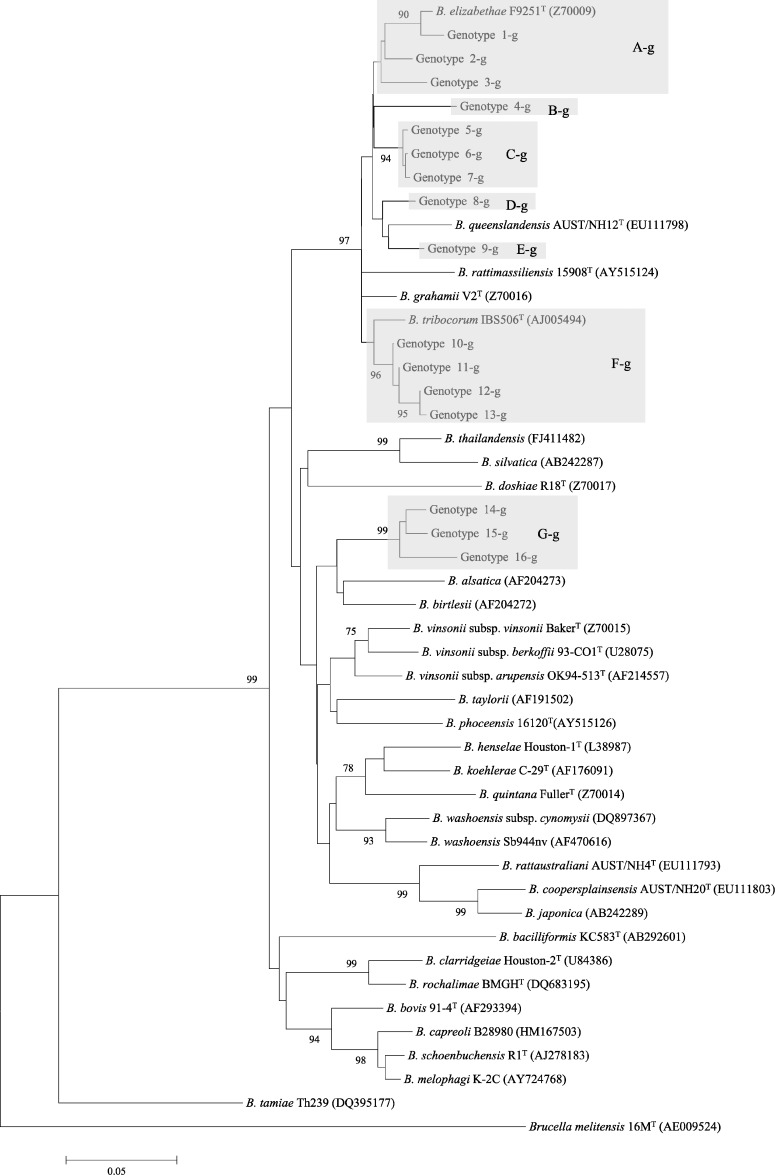
In this study, rpoB (825 bp) and gltA (327 bp) sequences were obtained from 24 and 20 animals, respectively (Table 3). Comparison of the 24 rpoB gene sequences revealed 18 genotypes (Table 3). The sequence homology ranging from 88.2% to 99.9% was found among rpoB genotypes. Among 20 gltA sequences, we found 16 genotypes with 87.4–99.7% homology (Table 3). All genotypes were further categorized into seven groups, A-r to G-r for rpoB and A-g to G-g for gltA, based on sequence similarities and a clustered pattern in phylogeny (Figures 1 and 2; Table 3). In this study, some Bartonella groups appear to follow a specific pattern that reflects host specificity. This specific pattern was clearly observed especially in the gltA phylogeny (Figure 2). For example, the gltA genotypes of groups B-g, C-g, F-g, and G-g identified in Mus, Lophuromys, Praomys, and Lophuromys animals, respectively, were clustered together in different branches in the phylogeny (Figure 2). In this study, several groups were close to two well-known rat-associated bartonellae: rpoB groups A-r, B-r, and gltA groups A-g, B-g, C-g, for B. elizabethae; rpoB group C-r and gltA group F-g for Bartonella tribocorum. However, only the genotype 1-r of rpoB and 1-g of gltA were concordant in both phylogenies and very close to Bartonella elizabethae with respective similarities 98.1% and 98.4%, which were higher than the cut-off values 95.4% and 96% for rpoB and gltA genes, respectively, according to La Scola and others.37 Similarly, rpoB 18-r of G-r group was concordant with gltA 14-g of G-g group and clustered with Bartonella birtlesii in both phylogenies. In contrast, other genotypes were not concordant with both phylogenies. For example, the genotypes of rpoB groups C-g to F-g were close to B. tribocorum and Bartonella grahamii; however, counterparts of the gltA groups from the same animals were close to B. elizabethae, B. birtlesii, Bartonella queenslandensis, and B. tribocorum. Overall, several genotypes found in the animals were not closely related to any described Bartonella species and were mostly belonging to unknown species, as per cut-off values determined by La Scola and others.37
Figure 1.
Phylogenetic classification of Bartonella genotypes based on rpoB gene sequences. Only the bootstrap values above 70% obtained are given. The GenBank accession numbers for reference sequences are given in parentheses. The rpoB gene sequences of Brucella melitensis 16MT were included as an out-group.
Figure 2.
Phylogenetic classification of Bartonella genotypes based on gltA gene sequences. Only the bootstrap values above 70% obtained are given. The GenBank accession numbers for reference sequences are given in parentheses. The gltA gene sequences of Brucella melitensis 16MT were included as an out-group.
Discussion
Our study reports the first detection and prevalence of Bartonella species in rodent populations from DR Congo and Tanzania. To our knowledge, it is also the first report of detection of Bartonella spp. in several rodent species: A. neumanni, L. rita, and M. minutoides, Grammomys sp., Lophuromys sp., and P. delectorum. The overall prevalence of Bartonella in six species (A. neumanni, L. rita, M. minutoides from DR Congo; Grammomys sp., Lophuromys sp., and P. delectorum from Tanzania) of the 11 examined rodent species was remarkably high (25–100%), although it was 4% in R. rattus. However, the number of animals trapped for this study was very low for four rodent species, which could explain our failure to identify Bartonella in these animals (Tables 1 and 2).
The prevalence and diversity of Bartonella species was reported previously in 10 species of small mammals collected in Free State province of South Africa.14 In this study, 9 Mastomys natalensis (N = 15), 1 Otomys irroratus (N = 2) were positive, and the only R. rattus examined was not infected by Bartonella.14 On the contrary, in our study, 10 and 1 specimen of related species, Mastomys coucha and Otomys sp., respectively, were found to be negative and 1 R. rattus was positive for Bartonella. A total of 24 out of 98 animals were infected by Bartonella species in our study. This prevalence was comparable to previous reports from other countries; ranging from 9% to 44% in Asia, 17% to 64% in Europe, 42% in North America, 44% in South Africa, and 29% in Australia.5,8,9,11,13–15,20,22,24,38–41 Such high rates of Bartonella prevalence could be significant with respect to the risks of humans becoming infected with these agents. The commensal mammals harboring pathogenic microorganisms are often found in biotopes where they can come into close contact with humans who might therefore be at some risk of exposure. In this study, considerably high levels of heterogeneities were found among rpoB and gltA gene sequences. Moreover, several rpoB genotypes were not concordant with gltA genotypes sequenced in the same animal, and hence, a similar branching pattern was not observed between two phylogenetic trees. This genotypic heterogeneity might be caused by the environmental conditions, host animal and its specificity, ectoparasites, etc., in the respective geographical locations.
In this study, gltA genotypes of genogroups B-g, C-g, and F-g obtained, respectively, from M. minutoides, Lophuromys sp., and P. delectorum were very close to each other in their respective groups and clustered together as separate branches (Figure 2). This finding suggests the host specificity between the genotypes in these genogroups and the host. Moreover, this host specificity was also observed in the rpoB genotyping (Figure 1). According to Ellis and others,21 Bartonella associated with hosts native to the Old World are phylogenetically distinct from those associated with a host-specific native to the New World. Several genotypes of this study were completely novel and had no evolutionary relationships with other known Bartonella, which supports the hypothesis of Ellis and others. However, some genotypes from Congolese and Tanzanian small mammals in the phylogenetic trees clustered with well-known rodent-associated species of bartonellae, including B. elizabethae, B. tribocorum, and B. queenslandensis (Figures 1 and 2). In this context, further studies should be conducted on a large collection of rodents and small animals from Africa to determine the evolutionary, genetic, and pathogenic relationships between African and other isolates.
The results suggest the need to conduct further studies to verify whether these agents might be responsible for human cases of febrile illness of unknown etiology in these countries. These preliminary data will allow us to design further studies on the comprehensive survey of the risks associated with exposure to rodent-associated Bartonella in these countries and other regions in Africa. In this context, future studies will be concentrated on isolation of bartonellae from African small animals.
ACKNOWLEDGMENTS
We thank Ying Bai and Hidenori Kabeya at CDC for their help.
Footnotes
Financial support: This research was supported in part by an appointment to the Emerging Infectious Diseases (EID) Fellowship Program administered by the Association of Public Health Laboratories (APHL) and funded by the Centers for Disease and Control and Prevention (CDC). We wish to acknowledge the financial support from the Belgium funds for Scientific Research (FNRS and FRIA), the University of Antwerp (Belgium), the Sokoine University of Agriculture, Morogoro, Tanzania, and the logistical support from the Pest Management Centre that enabled the scientific expedition to the Rift Valley District of Mbulu in northern Tanzania.
Disclosure: Dr. Gundi was an EID Fellow during 2008–2009. He is working in Bartonella Laboratory, Division of Vector-Borne Infectious Diseases, Centers for Disease Control and Prevention, Fort Collins, Colorado. His research interests include tropical and emerging infectious diseases.
Authors' addresses: Vijay A. K. B. Gundi and Michael Y. Kosoy, Centers for Disease Control and Prevention, Division of Vector-Borne Diseases, Fort Collins, CO, E-mail: mck3@cdc.gov. Rhodes H. Makundi, Sokoine University of Agriculture, Morogoro, Tanzania. Anne Laudisoit, Veterinary and Agrochemical Research Centre, Brussels; University of Antwerp, Antwerp; and University of Liège, Liège, Belgium.
References
- 1.Breitschwerdt EB, Kordick DL. Bartonella infection in animals: carriership, reservoir potential, pathogenicity, and zoonotic potential for human infection. Clin Microbiol Rev. 2000;13:428–438. doi: 10.1128/cmr.13.3.428-438.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chomel BB, Boulouis HJ, Maruyama S, Breitschwerdt EB. Bartonella spp. in pets and effect on human health. Emerg Infect Dis. 2006;12:389–394. doi: 10.3201/eid1203.050931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mogollon-Pasapera E, Otvos L, Jr, Giordano A, Cassone M. Bartonella: emerging pathogen or emerging awareness? Int J Infect Dis. 2009;13:3–8. doi: 10.1016/j.ijid.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 4.Koehler JE. Bartonella infections. Adv Pediatr Infect Dis. 1996;11:1–27. [PubMed] [Google Scholar]
- 5.Kosoy MY, Regnery RL, Tzianabos T, Marston EL, Jones DC, Green D, Maupin GO, Olson JG, Childs JE. Distribution, diversity, and host specificity of Bartonella in rodents from the southeastern United States. Am J Trop Med Hyg. 1997;57:578–588. doi: 10.4269/ajtmh.1997.57.578. [DOI] [PubMed] [Google Scholar]
- 6.Heller R, Riegel P, Hansmann Y, Delacour G, Bermond D, Dehio C, Lamarque F, Monteil H, Chomel B, Piemont Y. Bartonella tribocorum sp. nov., a new Bartonella species isolated from the blood of wild rats. Int J Syst Bacteriol. 1998;48:1333–1339. doi: 10.1099/00207713-48-4-1333. [DOI] [PubMed] [Google Scholar]
- 7.Hofmeister EK, Kolbert CP, Abdulkarim AS, Magera JM, Hopkins MK, Uhl JR, Ambyaye A, Telford SR, 3rd, Cockerill FR, 3rd, Persing DH. Cosegregation of a novel Bartonella species with Borrelia burgdorferi and Babesia microti in Peromyscus leucopus. J Infect Dis. 1998;177:409–416. doi: 10.1086/514201. [DOI] [PubMed] [Google Scholar]
- 8.Birtles RJ, Hazel SM, Bennett M, Bown K, Raoult D, Begon M. Longitudinal monitoring of the dynamics of infections due to Bartonella species in UK woodland rodents. Epidemiol Infect. 2001;126:323–329. doi: 10.1017/s095026880100526x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holmberg M, Mills JN, McGill S, Benjamin G, Ellis BA. Bartonella infection in sylvatic small mammals of central Sweden. Epidemiol Infect. 2003;130:149–157. doi: 10.1017/s0950268802008075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tea A, Alexiou-Daniel S, Papoutsi A, Papa A, Antoniadis A. Bartonella species isolated from rodents, Greece. Emerg Infect Dis. 2004;10:963–964. doi: 10.3201/eid1005.030430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gundi VA, Davoust B, Khamis A, Boni M, Raoult D, La Scola B. Isolation of Bartonella rattimassiliensis sp. nov. and Bartonella phoceensis sp. nov. from European Rattus norvegicus. J Clin Microbiol. 2004;42:3816–3818. doi: 10.1128/JCM.42.8.3816-3818.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jardine C, Appleyard G, Kosoy MY, McColl D, Chirino-Trejo M, Wobeser G, Leighton FA. Rodent-associated Bartonella in Saskatchewan, Canada. Vector Borne Zoonotic Dis. 2005;5:402–409. doi: 10.1089/vbz.2005.5.402. [DOI] [PubMed] [Google Scholar]
- 13.Knap N, Duh D, Birtles R, Trilar T, Petrovec M, Avsic-Zupanc T. Molecular detection of Bartonella species infecting rodents in Slovenia. FEMS Immunol Med Microbiol. 2007;50:45–50. doi: 10.1111/j.1574-695X.2007.00226.x. [DOI] [PubMed] [Google Scholar]
- 14.Pretorius AM, Beati L, Birtles RJ. Diversity of bartonellae associated with small mammals inhabiting Free State province, South Africa. Int J Syst Evol Microbiol. 2004;54:1959–1967. doi: 10.1099/ijs.0.03033-0. [DOI] [PubMed] [Google Scholar]
- 15.Gundi VA, Taylor C, Raoult D, La Scola B. Bartonella rattaustraliani sp. nov., Bartonella queenslandensis sp. nov. and Bartonella coopersplainsensis sp. nov., identified in Australian rats. Int J Syst Evol Microbiol. 2009;59:2956–2961. doi: 10.1099/ijs.0.002865-0. [DOI] [PubMed] [Google Scholar]
- 16.Gundi VA, Bourry O, Davous B, Raoult D, La Scola B. Bartonella clarridgeiae and B. henselae in dogs, Gabon. Emerg Infect Dis. 2004;10:2261–2262. doi: 10.3201/eid1012.040359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heller R, Kubina M, Mariet P, Riegel P, Delacour G, Dehio C, Lamarque F, Kasten R, Boulouis HJ, Monteil H, Chomel B, Piemont Y. Bartonella alsatica sp. nov., a new Bartonella species isolated from the blood of wild rabbits. Int J Syst Bacteriol. 1999;49:283–288. doi: 10.1099/00207713-49-1-283. [DOI] [PubMed] [Google Scholar]
- 18.Kordick DL, Swaminathan B, Greene CE, Wilson KH, Whitney AM, O'Connor S, Hollis DG, Matar GM, Steigerwalt AG, Malcolm GB, Hayes PS, Hadfield TL, Breitschwerdt EB, Brenner DJ. Bartonella vinsonii subsp. berkhoffii subsp. nov., isolated from dogs; Bartonella vinsonii subsp. vinsonii; and emended description of Bartonella vinsonii. Int J Syst Bacteriol. 1996;46:704–709. doi: 10.1099/00207713-46-3-704. [DOI] [PubMed] [Google Scholar]
- 19.Chang CC, Chomel BB, Kasten RW, Heller RM, Ueno H, Yamamoto K, Bleich VC, Pierce BM, Gonzales BJ, Swift PK, Boyce WM, Jang SS, Boulouis HJ, Piemont Y, Rossolini GM, Riccio ML, Cornaglia G, Pagani L, Lagatolla C, Selan L, Fontana R. Bartonella spp. isolated from wild and domestic ruminants in North America. Emerg Infect Dis. 2000;6:306–311. doi: 10.3201/eid0603.000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castle KT, Kosoy M, Lerdthusnee K, Phelan L, Bai Y, Gage KL, Leepitakrat W, Monkanna T, Khlaimanee N, Chandranoi K, Jones JW, Coleman RE. Prevalence and diversity of Bartonella in rodents of northern Thailand: a comparison with Bartonella in rodents from southern China. Am J Trop Med Hyg. 2004;70:429–433. [PubMed] [Google Scholar]
- 21.Ellis BA, Regnery RL, Beati L, Bacellar F, Rood M, Glass GG, Marston E, Ksiazek TG, Jones D, Childs JE. Rats of the genus Rattus are reservoir hosts for pathogenic Bartonella species: an Old World origin for a New World disease? J Infect Dis. 1999;180:220–224. doi: 10.1086/314824. [DOI] [PubMed] [Google Scholar]
- 22.Ying B, Kosoy MY, Maupin GO, Tsuchiya KR, Gage KL. Genetic and ecologic characteristics of Bartonella communities in rodents in southern China. Am J Trop Med Hyg. 2002;66:622–627. doi: 10.4269/ajtmh.2002.66.622. [DOI] [PubMed] [Google Scholar]
- 23.Fournier PE, Robson J, Zeaiter Z, McDougall R, Byrne S, Raoult D. Improved culture from lymph nodes of patients with cat scratch disease and genotypic characterization of Bartonella henselae isolates in Australia. J Clin Microbiol. 2002;40:3620–3624. doi: 10.1128/JCM.40.10.3620-3624.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Engbaek K, Lawson PA. Identification of Bartonella species in rodents, shrews and cats in Denmark: detection of two B. henselae variants, one in cats and the other in the long-tailed field mouse. APMIS. 2004;112:336–341. doi: 10.1111/j.1600-0463.2004.apm1120603.x. [DOI] [PubMed] [Google Scholar]
- 25.De Sousa R, Edouard-Fournier P, Santos-Silva M, Amaro F, Bacellar F, Raoult D. Molecular detection of Rickettsia felis, Rickettsia typhi and two genotypes closely related to Bartonella elizabethae. Am J Trop Med Hyg. 2006;75:727–731. [PubMed] [Google Scholar]
- 26.Marie JL, Fournier PE, Rolain JM, Briolant S, Davoust B, Raoult D. Molecular detection of Bartonella quintana, B. Elizabethae, B. koehlerae, B. doshiae, B. taylorii, and Rickettsia felis in rodent fleas collected in Kabul, Afghanistan. Am J Trop Med Hyg. 2006;74:436–439. [PubMed] [Google Scholar]
- 27.Gundi VA, Kosoy MY, Myint KS, Shrestha SK, Shrestha MP, Pavlin JA, Gibbons RV. Prevalence and genetic diversity of Bartonella species detected in different tissues of small mammals in Nepal. Appl Environ Microbiol. 2010;76:8247–8254. doi: 10.1128/AEM.01180-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sackal C, Laudisoit A, Kosoy M, Massung R, Eremeeva ME, Karpathy SE, Van Wyk K, Gabitzsch E, Zeidner NS. Bartonella spp. and Rickettsia felis in fleas, Democratic Republic of Congo. Emerg Infect Dis. 2008;14:1972–1974. doi: 10.3201/eid1412.080610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Renesto P, Gouvernet J, Drancourt M, Roux V, Raoult D. Use of rpoB gene analysis for detection and identification of Bartonella species. J Clin Microbiol. 2001;39:430–437. doi: 10.1128/JCM.39.2.430-437.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Birtles RJ, Raoult D. Comparison of partial citrate synthase gene (gltA) sequences for phylogenetic analysis of Bartonella species. Int J Syst Bacteriol. 1996;46:891–897. doi: 10.1099/00207713-46-4-891. [DOI] [PubMed] [Google Scholar]
- 31.Norman AF, Regnery R, Jameson P, Greene C, Krause DC. Differentiation of Bartonella-like isolates at the species level by PCR-restriction fragment length polymorphism in the citrate synthase gene. J Clin Microbiol. 1995;33:1797–1803. doi: 10.1128/jcm.33.7.1797-1803.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 33.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 35.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 36.Felsenstein J. Estimating effective population size from samples of sequences: a bootstrap Monte Carlo integration method. Genet Res. 1992;60:209–220. doi: 10.1017/s0016672300030962. [DOI] [PubMed] [Google Scholar]
- 37.La Scola B, Zeaiter Z, Khamis A, Raoult D. Gene-sequence-based criteria for species definition in bacteriology: the Bartonella paradigm. Trends Microbiol. 2003;11:318–321. doi: 10.1016/s0966-842x(03)00143-4. [DOI] [PubMed] [Google Scholar]
- 38.Inoue K, Maruyama S, Kabeya H, Yamada N, Ohashi N, Sato Y, Yukawa M, Masuzawa T, Kawamori F, Kadosaka T, Takada N, Fujita H, Kawabata H. Prevalence and genetic diversity of Bartonella species isolated from wild rodents in Japan. Appl Environ Microbiol. 2008;74:5086–5092. doi: 10.1128/AEM.00071-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bai Y, Montgomery SP, Sheff KW, Chowdhury MA, Breiman RF, Kabeya H, Kosoy MY. Bartonella strains in small mammals from Dhaka, Bangladesh, related to Bartonella in America and Europe. Am J Trop Med Hyg. 2007;77:567–570. [PubMed] [Google Scholar]
- 40.Marquez FJ, Rodriguez-Liebana JJ, Pachon-Ibanez ME, Docobo-Perez F, Hidalgo-Fontiveros A, Bernabeu-Wittel M, Muniain MA, Pachon J. Molecular screening of Bartonella species in rodents from south western Spain. Vector Borne Zoonotic Dis. 2008;8:695–700. doi: 10.1089/vbz.2007.0257. [DOI] [PubMed] [Google Scholar]
- 41.Welc-Faleciak R, Paziewska A, Bajer A, Behnke JM, Sinski E. Bartonella spp. infection in rodents from different habitats in the Mazury Lake District, Northeast Poland. Vector Borne Zoonotic Dis. 2008;8:467–474. doi: 10.1089/vbz.2007.0217. [DOI] [PubMed] [Google Scholar]