Abstract
Paraná state presents the fourth highest number of accumulated cases of hantavirus pulmonary syndrome in Brazil. To map the risk areas for hantavirus transmission we carried out a study based on rodent trapping and determined the anti-hantavirus seroprevalence in these animals and in the inhabitants of these localities. Overall seroprevalence in rodents and humans were 2.5% and 2.4%, respectively. Eighty-two percent of the seropositive rodents were genetically analyzed. Phylogenetic analyses revealed that hantaviruses from rodent samples cluster with Araucária (Juquitiba-like) or Jaborá hantavirus genotypes. The Jaborá strain was identified in Akodon serrensis and Akodon montensis, whereas the Araucária strain was detected in Oligoryzomys nigripes, Oxymycterus judex, A. montensis, and Akodon paranaensis, with the latter species being identified for the first time as a natural host. These findings expose the complex relationships between virus and reservoirs in Brazil, which could have an impact on hantavirus transmission dynamics in nature and human epidemiology.
Introduction
Hantaviruses (Bunyaviridae) are emerging rodent-borne and soricomorpha-borne viruses. Two diseases related to hantavirus infection are known, hemorrhagic fever with renal syndrome (HFRS) occurring in Eurasia, and hantavirus pulmonary syndrome (HPS), in the Americas.1,2 New World hantaviruses are hosted by rodents of the Cricetidae family (subfamilies Sigmodontinae, Arvicolinae, and Neotominae), and by Soricomorpha mammals (families Soricidae and Talpidae). Sigmodontine and neotomine rodents are the main hosts of hantaviruses known to cause HPS. The hantavirus genus, includes pathogenic and non-pathogenic viruses, with more than 21 species and 30 genotypes.3–5
Since the first description of HPS in the Americas, hantavirus infections are considered a reportable disease in Brazil. Until October 2011, a total of 1,404 cases were confirmed in the country, with a 39% mortality rate (Secretary of Health Surveillance, Brazilian Health Ministry, 2011). Southern Brazil reports the highest number of hantavirus infection in Brazil (∼40%) and 38% of these cases occur in the state of Paraná.
In previous studies, we reported the complete genetic characterization of the S and M segments from the Araucária (Juquitiba-like) hantavirus involved in HPS cases in Paraná state, South Brazil.6,7 This region borders Paraguay and Argentina in the West and the states of Santa Catarina (South) and São Paulo (North). Its main economic activities are agriculture, reforestation, and ecotourism. Over the last 7 years our group has monitored HPS cases reported in Paraná by performing genetic analyses to determine the viral genotypes associated with these cases and rodent trapping to identify the species involved in hantavirus transmission. Epidemiological studies have revealed substantial differences in antibody prevalence in humans and rodents among the various regions of South Brazil, indicating that some communities are experiencing high frequencies of virus exposures7–9 Since 2008, a collaborative network involving our institute, the State Health Department of Paraná and the Laboratory of Biology and Parasitology of Wild Mammals Reservoirs in Fiocruz, Rio de Janeiro has been responsible for carrying out programmed collections of wild rodents in different ecosystems throughout the state of Paraná and for collecting sera from individuals living in these areas. The aims of this program are to determine the distribution of wild rodents, the prevalence of anti-hantavirus antibodies in rodents and humans, and to identify the virus genotypes circulating in the surveyed areas. This information is then used to delineate necessary control measures.
Material And Methods
A cross-sectional study was conducted from November 2006 through March 2011 to measure the anti-hantavirus IgG antibody prevalence in rodents and human population. In addition, the detected viruses were genetically characterized.
The procedures involving the use of human serum samples and the manipulation of small mammals were reviewed and approved by the Ethical Committee from the Brazilian Ministry of Health (CONEP) under protocol no. 10573 for the human samples, and no. IAP/PR 292/11 for the animal handling.
Study locations.
Expedition sites were defined on the basis of reported cases of HPS and on the environmental diversity of the locations, taking into account vegetation type, economic activity, and population density.
Rodent capture.
A professional staff trained in the capturing and handling of small mammals captured the animals using Tomahawk (40.6 cm × 12.7 cm × 12.7 cm; Tomahawk, WI) and Sherman (7.6 cm × 9.5 cm × 30.5 cm; Tallahassee, FL) live traps set at different sites ranging from wild environments to peridomestic areas (including altered habitats, secondary forests, and rural areas. In addition, in the municipalities with reported human cases, live traps were also set around probable infection sites. Traps were baited with a mixture of peanut butter, banana, oats, and bacon. The animals were identified by morphologic characteristics and further by karyological and genetic analyses. Blood and organs (lung, liver, kidney, and spleen) were collected aseptically. Tissues were frozen in liquid nitrogen and blood was kept at 4°C during transport to the laboratory facilities. All procedures were performed after previously reported standards of biosafety.10
Antibodies assays.
Rodent blood samples were screened for immunoglobulin G (IgG) anti-hantavirus antibodies using an indirect enzymatic immunoassay kit (Hantec, Curitiba, Paraná, Brazil) employing a recombinant Araucária nucleoprotein, according to the manufacture's instructions.8 The rodents found to be seropositive and reverse transcription-polymerase chain reaction (RT-PCR) positive to hantavirus were further confirmed at species level by mitochondrial DNA (cytochrome B) sequencing.11 Serum samples were collected from members of the resident population by peripheral blood collection, after they had signed a consent form to participate in the study. Individuals < 18 years of age were excluded from the study. Human sera samples were collected depending on the availability of health care professionals to perform vein puncture procedure at the same locals of rodent trapping. Sampling of humans and rodents were done at same time. Blood samples were screened for IgG anti-hantavirus antibodies using the Hantec kit and positive samples were confirmed by an immunoblotting assay.
Hantavirus genetic characterization.
Tissues (lung, liver, and/or kidney) from antibody-positive rodents were analyzed by RT-PCR to amplify the partial S segment. The molecular tests were carried out using various sets of primers that have been described previously.6,7,12 The PCR products were purified (High Pure PCR kit, Roche Inc., Mannheim, Germany) and both strands were sequenced by the commercial Macrogen facility (Seoul, Republic of Korea).
Sequence analyses.
Genomic sequences of the partial S segment were aligned with hantavirus sequences retrieved from GenBank. A total of 60 S segment sequences were analyzed, including the main South American hantavirus genotypes, and representative virus genotypes from North American and Eurasia (see figure legends for details). Alignments were constructed using the BioEdit v7.0.9.0 package. For phylogenetic inferences, the best-fit model of evolution and associated parameters were calculated using ModelGenerator software (http://bioinf.may.ie/software/modelgenerator). Phylogenies were constructed using the Bayesian inference method (MrBayes v3.1.2; http://mrbayes.csit.fsu.edu) and the maximum likelihood (ML) method (PhyML v3.0.13 Bayesian analyses were conducted under the general time reversible + gamma + proportion invariant model. Two runs of four chains each (one cold, three heated, temperature 0.20) were run for three million generations; trees were sampled every 100 generations. Convergence was assessed by using the average standard deviation in partition frequency values across independent analyses with a threshold value of 0.01; burn-in was set to 25%. For the ML estimation of phylogeny, the model of evolution and parameters used were as described above, initial trees were calculated using the BioNJ option and the tree searching option was set to NNI. For both analyses, sequences of the Hantaan and Seoul hantaviruses were used as outgroup species. The node supports were calculated using the approximate likelihood ratio test (aLRTs).14
Results
Places of collection.
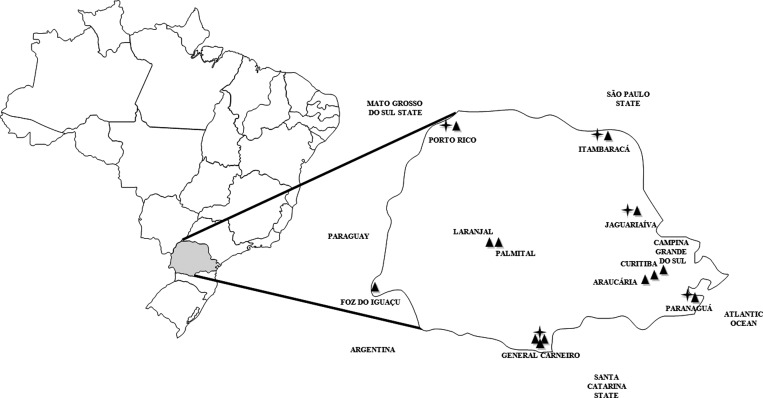
A total of 11 trapping expeditions were carried out until March 2011 with a general trap success near 4.5%. Figure 1 shows the regions studied and those having hantavirus-positive results. Seven hundred forty-eight small mammals were captured during the study; 667 of them were rodents. The genus and/or species of these animals and their distribution by region are shown in Table 1.
Figure 1.
State of Paraná, Brazil. ▲: Places of collections. ✦: Rodents with positive serology for hantavirus.
Table 1.
Small mammals captured in State of Paraná, Southern Brazil, 2006–2009
| Place and date of collection Total of collection | Small mammals | N (%) |
|---|---|---|
| Southern | ||
| General Carneiro (26°25′39″S 51°18′56″W) November, 2006 N = 72 | RODENTIA | |
| Akodon montensis | 31 (44%) | |
| Akodon serrensis | 20 (28%) | |
| Akodon paranaensis | 4 (6%) | |
| Oligoryzomys nigripes | 7 (10%) | |
| Thaptomys nigrita | 3 (4%) | |
| Oxymycterus judex | 2 (2%) | |
| Sooretamys angouya | 2 (2%) | |
| DIDELPHIMORPHIA | ||
| Monodelphis sp. | 3 (4%) | |
| General Carneiro December, 2009 N = 57 | RODENTIA | |
| Akodon serrensis | 30 (53%) | |
| Akodon montensis | 10 (18%) | |
| Akodon paranaensis | 4 (7%) | |
| Oxymycterus judex | 7 (12%) | |
| Thaptomys nigrita | 3 (5%) | |
| Sooretamys angouya | 1 (2%) | |
| DIDELPHIMORPHIA | ||
| Monodelphis sp. | 2 (3%) | |
| General Carneiro March, 2010 N = 85 | RODENTIA | |
| Akodon serrensis | 26 (31%) | |
| Akodon montensis | 24 (28%) | |
| Akodon paranaensis | 6 (7%) | |
| Oxymycterus judex | 18 (21%) | |
| Oligoryzomys nigripes | 2 (2%) | |
| DIDELPHIMORPHIA | ||
| Philander frenatus | 4 (5%) | |
| Monodelphis sp. | 4 (5%) | |
| Lutreolina crassicaudata | 1 (1%) | |
| Northeastern | ||
| Itambaracá (22°58′21″S 50°28′44″W) N = 74 | RODENTIA | |
| Akodon montensis | 14 (19%) | |
| Mus musculus | 25 (34%) | |
| Oligoryzomys nigripes | 11 (15%) | |
| Rattus rattus | 4 (5%) | |
| DIDELPHIMORPHIA | ||
| Didelphis albiventris | 20 (27%) | |
| Jaguariaíva (24°15′04″S 49°42′21″W) N = 54 | RODENTIA | |
| Akodon montensis | 21 (39%) | |
| Calomys tener | 12 (22%) | |
| Oligoryzomys nigripes | 6 (11%) | |
| Euryoryzomys russatus | 8 (15%) | |
| Oxymycterus sp. | 5 (9%) | |
| DIDELPHIMORPHIA | ||
| Didelphis albiventris | 2 (4%) | |
| Northwestern | ||
| Porto Rico (22°46′20″S 53°16′01″W) N = 45 | RODENTIA | |
| Oligoryzomys sp. | 18 (40%) | |
| Akodon sp. | 10 (22%) | |
| Oecomys bicolor | 6 (13%) | |
| Thaptomys nigrita | 5 (11%) | |
| Mus musculus | 3 (7%) | |
| Calomys tener | 2 (5%) | |
| Rattus rattus | 1 (2%) | |
| Western | ||
| Foz do Iguaçu (25°32′52″S 54°35′17″W) N = 115 | RODENTIA | |
| Akodon montensis | 62 (54%) | |
| Mus musculus | 44 (39%) | |
| Oligoryzomys nigripes | 3 (2%) | |
| Thaptomys nigrita | 3 (2%) | |
| DIDELPHIMORPHIA | ||
| Didelphis aurita | 2 (2%) | |
| Didelphis albiventris | 1 (1%) | |
| Central-Western | ||
| Laranjal/Palmital (24°53′12″S 52°28′10″W/24°53′35″S 52°12′10″W N = 25 | RODENTIA | |
| Akodon sp. | 17 (68%) | |
| Mus musculus | 6 (24%) | |
| DIDELPHIMORPHIA | ||
| Monodelphis sp. | 2 (8%) | |
| Eastern | ||
| Paranaguá (Coast) (25°31′12″S 48°30′33″W) N = 71 | RODENTIA | |
| Akodon montensis | 25 (35%) | |
| Oligoryzomys nigripes | 18 (25%) | |
| Thaptomys nigrita | 9 (13%) | |
| Nectomys squamipes | 8 (11%) | |
| Euryoryzomys russatus | 4 (6%) | |
| Rattus rattus | 3 (4%) | |
| Sooretamys angouya | 1 (2%) | |
| DIDELPHIMORPHIA | ||
| Didelphis sp. | 3 (4%) | |
| Curitiba (25°25′40″S 49°16′23″W) N = 27 | RODENTIA | |
| Akodon montensis | 7 (26%) | |
| Akodon paranaensis | 7 (26%) | |
| Oligoryzomys nigripes | 6 (22%) | |
| Mus musculus | 3 (11%) | |
| DIDELPHIMORPHIA | ||
| Monodelphis sp. | 4 (15%) | |
| Araucária (25°35′35″S 49°24′37″W) N = 33 | RODENTIA | |
| Akodon montensis | 12 (37%) | |
| Akodon paranaensis | 5 (15%) | |
| Oligoryzomys nigripes | 9 (27%) | |
| Mus musculus | 7 (21%) | |
| Campina Grande do Sul (25°18′20″S 49°03′19″W) N = 90 | RODENTIA | |
| Akodon sp. | 54 (60%) | |
| Thaptomys nigrita | 16 (18%) | |
| Rattus rattus | 7 (8%) | |
| Oxymycterus sp. | 4 (5%) | |
| Nectomys squamipes | 3 (3%) | |
| Rattus rattus | 1 (1%) | |
| Oligoryzomys flavescens | 1 (1%) | |
| Oryzomys russatus | 1 (1%) | |
| Oryzomys angouya | 1 (1%) | |
| DIDELPHIMORPHIA | ||
| Monodelphis sp. | 2 (2%) | |
Prevalence of anti-hantavirus IgG in rodent and human populations.
A total of 2.5% (17 of 667) of the trapped rodents were anti-hantavirus IgG-positive and 88% (15 of 17) were also found to be hantavirus-positive by RT-PCR. Human seroprevalence analysis was performed for a total of 1,038 sera randomly collected from adults (> 18 years of age) without reported classical HPS signs, in which an overall of 2.4% (25 of 1,038) were anti-hantavirus IgG-positive (Table 2).
Table 2.
Collection locations, numbers of small mammals captured, rodent seroprevalence, positive species, hantavirus genotype, and human seroprevalence, Brazil
| Region of state of Paraná Cities | Total small mammals | Rodent seroprevalence N (% positive) Species (N) | Hantavirus genotype | Human seroprevalence N (% positive) | HPS reported area |
|---|---|---|---|---|---|
| Southern | |||||
| General Carneiro | 214 | 8 (3.7%) | N = 107 | Yes | |
| – | Oxymycterus judex (01) | Araucaria (01) | 8.4% | ||
| Akodon montensis (05) | Araucaria (01) | ||||
| Jaborá (04) | |||||
| Akodon serrensis (01) | Jaborá (01) | ||||
| Palmas | Akodon paranaensis (01) | Araucaria (01) | N = 145 | Yes | |
| – | – | 2.7% | |||
| Northeastern | 1 (1.3%) | N = 164 | No | ||
| Itambaracá | 74 | Oligoryzomys nigripes (01) | Araucária (01) | 0% | |
| Jaguariaíva | 54 | 2 (3.7%) | N = 153 | No | |
| Akodon montensis (01) | Jaborá (01) | 1.3% | |||
| Oligoryzomys nigripes (01) | Araucária (01) | ||||
| Northwestern | |||||
| Porto Rico | 45 | 2 (4.4%) | Not done | Yes | |
| Oligoryzomys nigripes (02) | Araucária (02) | ||||
| Western | – | N = 199 | Yes | ||
| Foz do Iguaçu | 115 | 0 (0%) | 0.5% | ||
| Central-Western | – | N = 270 | Yes | ||
| Palmital/Laranjal | 25 | 0 (0%) | 3.3% | ||
| Eastern | |||||
| Paranaguá (Coast) | 71 | 4 (5.6%) | Not done | Yes | |
| Akodon montensis (01) | – | ||||
| Oligoryzomys nigripes (03) | Araucária (02) | ||||
| Curitiba | 27 | 0 (0%) | Not done | Yes | |
| Araucária | 33 | 0 (0%) | Not done | No | |
| Campina Grande do Sul | 90 | 0 (0%) | Not done | Yes | |
General Carneiro county, located in the southern area of Paraná, is the region with the highest incidence of HPS cases. In this locality, three capture expeditions were conducted at three different times (November 2006, December 2009, and March 2010). Regarding the other regions reported in this study, one capture expedition was performed per locality. The original vegetation of General Carneiro county was Atlantic interior forest, but this has been gradually replaced by pine tree reforestation, which represents the main economic activity. The majority of the HPS patients in this region worked in wood extraction in the secondary growth forests.7 A total of 214 small mammals were captured in three expeditions and their identification is displayed in Table 1. Eight seropositive rodents were detected in General Carneiro County and this locality displayed the highest prevalence of anti-hantavirus antibodies in human population, compared with the other studied areas (8.4%), (Table 2).
The municipalities of Itambaracá and Jaguariaíva, located in the northeastern portion of Paraná, are both areas of savannah and Atlantic forest fragments, where agriculture is the main economic activity. A total of 128 small mammals were captured in Itambaracá and Jaguariaíva during the study (Table 1). Anti-hantavirus antibodies were found in 5% of the captured animals, and no HPS cases have been reported in these areas. No seropositive human samples were found in Itambaracá but 1.3% of the serum samples from Jaguariaíva were positive.
Porto Rico county, located in the northwest part of Paraná, borders with Mato Grosso do Sul. Tourism is its main source of income. Only one rodent capture expedition was carried out in this region, and the seropositivity prevalence for these animals was 4.4% (Table 1).
Foz do Iguaçu (Iguaçu Falls), located in the western part of the state, is a plateau region with a subtropical climate and a subtropical rain forest, and is bordered by Paraguay and Argentina. The main economic activity is tourism, electricity generation, and trade. One hundred fifteen small mammals were captured (Table 1) and all were found to be negative for hantavirus antibodies. One seropositive human serum sample was found in this region (0.5%).
Palmital and Laranjal counties are both located in Midwestern region of Parana. Twenty-three rodents were trapped in this region (Table 1), but none were positive for anti-hantavirus IgG. However, HPS cases have been reported in this locality and the human anti-hantavirus seroprevalence was 3.3%.
The city of Paranaguá is located in eastern Paraná on the Atlantic coast. The main economic activities of the region are related to the Paranaguá port and trade. Seventy-two small mammals were caught here (Table 1), four of them were positive for anti-hantavirus IgG (5.6%). Hantavirus pulmonary syndrome cases have been reported in this locality.
Curitiba (the state capital of Paraná) and the metropolitan region (Araucária and Campina Grande do Sul), located in the eastern part of the state, have a humid subtropical climate. In Curitiba, animals were trapped in peridomestic habitats near the home of a patient who died of HPS. One hundred forty-nine animals were caught (Table 1), however none of them were found to be seropositive for anti-hantavirus antibodies.
Phylogenetic relationships among hantaviruses.
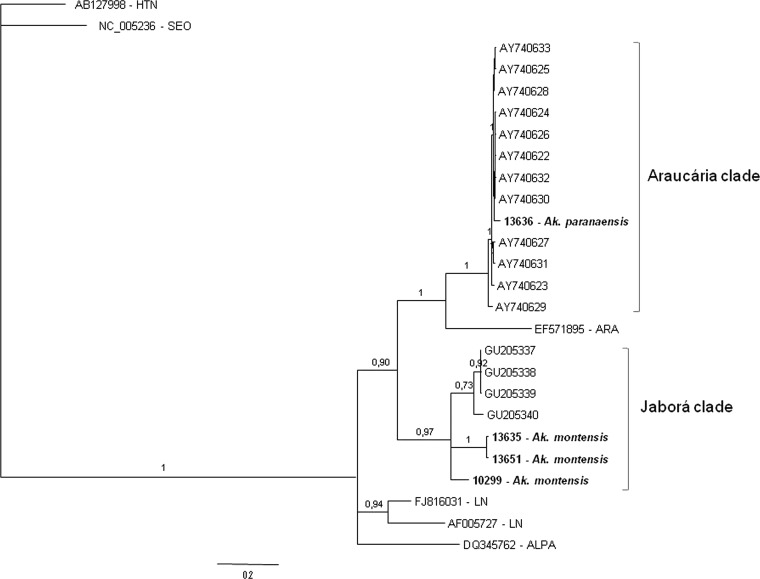
The Bayesian phylogenies of hantavirus partial S segment sequences are shown in Figures 2 and 3. In Figure 2 the phylogenetic tree corresponds to a 631 nt alignment, spanning nucleotides 152–782 of S segment, and in Figure 3 the tree was constructed using a subset of sequences from Figure 2, spanning nucleotides 978–1,250 of S segment (nucleotide positions relative to ARAUV strain AY740633). In both trees, genomic sequences of hantaviruses identified in rodents from different localities of Paraná grouped with high statistical support into the Araucária (ARAUV) and Jaborá (JABV) clades. The majority (7 of 12) of the S sequences belonged to the Araucária clade. They were retrieved from two Oligoryzomys nigripes specimens trapped in Paranaguá (10104 and 10091) and from rodents 10056 (Oxymycterus judex) and 10028 (Akodon montensis) collected in General Carneiro. Two additional sequences were found in O. nigripes specimens captured at Jaguariaiva (10351) and Itambaracá (10217) also belong to this clade (Figure 2). An additional sequence retrieved from a rodent captured in General Carneiro groups within the ARAUV clade (Figure 3) with a posterior probability (pp) of 1. This rodent was identified as A. paranaensis by its morphology, karyotype, and cytochrome B gene sequence.
Figure 2.
Bayesian phylogenetic analysis of partial S segment sequences from rodents captured in different Paraná localities (in boldface). For comparison, a set of representative hantavirus sequences from Brazil, South America, North America, and Eurasia were included in the analysis. Alignment used in the analysis was 631 nt long, spanning nucleotides 152–782 of S segment, respect to ARAUV strain AY740633. Hantaan (HTNV) and Seoul (SEOV) sequences were used as outgroup species. Posterior probabilities (pp) are depicted above the nodes. GenBank accession nos.: 12460: HQ337904; 10217: HQ337905; 10351: HQ337906; 10104: HQ3379047; 13663: JN252310; 13635: JN252313; 13651: JN252311; 10299: JN252312.
Figure 3.
Bayesian phylogenetic analysis of partial S segment sequences from rodents captured in different Paraná localities (in boldface). For comparison, a subset of the sequences shown in Figure 2 (from Brazil, Paraná, and Paraguay) was used in the analysis. Paraná strains are 272 nt long and spanned nucleotides 978–1,250 of S segment, respect to ARAUV strain AY740633. Hantaan (HTNV) and Seoul (SEOV) sequences were used as outgroup species. Posterior probabilities (pp) are depicted above the nodes. GenBank accession nos.: 13636: JN252309.
Three viral genomic S sequences from rodents trapped in General Carneiro in March 2010 and one collected in Jaguariaíva are placed in the Jaborá clade, with significant statistical support (P = 0.99), together with Jaborá strains from Brazil and Paraguay. An additional viral S sequence from an A. serrensis specimen, collected in General Carneiro in December 2009 is placed into Jaborá clade. All these animals were identified as A. montensis and A. serrensis by their morphologies, karyotypes, and cytochrome B gene sequences (Figure 2).
Maximum likelihood analyses (not shown, available upon request) were also performed. Phylogenies obtained under this method displayed the same topology as the Bayesian analysis at the relevant nodes.
Discussion
The HPS has been reported every year in the state of Paraná since 1993, with the highest number of confirmed cases occurring in southern and southwestern regions. The biogeography with its particular vegetation landscape and agricultural practices of these regions are distinct from other regions in Paraná. Nevertheless, there is little information available on the epidemiology, severity of disease, and ecology of the hantavirus reservoirs.
Previously, we had reported a correlation between rodent density and virus transmission, with the description of outbreaks associated to pine tree reforestation area, with bamboo blooming and mast seeding in southern Brazil.6 Subsequently, we demonstrated the presence of the same hantavirus in three different rodent species, and the co-circulation of two different strains in the same rodent species, highlighting the complexity of hantavirus transmission dynamics in nature.7 Similar findings were previously reported for a Juquitiba-like hantavirus harbored by two different rodent species in Uruguay,15 stressing the point that hantavirus/reservoir species relationships in South America might be more complex than the typically predominant association of one hantavirus strain and one rodent reservoir species.
The most abundant rodent species captured during this study were Akodon spp., followed by O. nigripes and Oxymycterus sp., both of the latter two have been previously implicated as potential pathogenic hantavirus reservoirs.7,15 The mean hantavirus antibody seroprevalence found in rodents in this study was 2.5% (with a regional range of 0% to 5.6%). Similar seroprevalence values have been reported for other American countries, such as 3.5% in Mexico,16 1.4% in Chile,17 5.6% in Argentina18 and 2.1% in Colombia.19 Recently, Armien and others20 reported a rodent prevalence of antibodies against Choclo virus ranging from 3% to 33% in neighborhoods where HPS cases have occurred in western Panamá.
The mean value for hantavirus antibody seroprevalence in humans in Paraná was 2.4%, but ranging from 0% to 8%, depending on the region. The highest seroprevalence was from individuals living in areas where HPS cases have been reported. Seroprevalence results from other South American countries and other regions in Brazil revealed different frequencies of hantavirus exposure, such as 32.9% in western region of Panama, 13.5% in Colombia,21 1.7% in Venezuela,22 4.7% in the Anajatuba municipality, state of Maranhão, Brazil,23 10.9% in rural residents of the Anajatuba municipality,24 and 14.3% in Jardinópolis, southeastern Brazil.25 Such differences in anti-hantavirus seroprevalence show high levels of virus exposure in these populations and draw attention to the possibility of asymptomatic or less virulent infections. In agreement, a high incidence rate of hantavirus infections without HPS was observed in four communities of western Panama, where 70 individuals seroconverted during the surveyed period (2001–2007) without HPS classical signs.20
The results from this study suggest a positive correlation between the percentage of seropositive rodents and HPS cases. On the other hand, anti-hantavirus IgG-positive rodent reservoirs were identified in areas without reported cases, suggesting that the geographic distribution of reservoir rodent species alone is not enough to link a particular area with disease risk. Other factors, such as seasonality, population density, and human behavior must be included in risk analyses. Furthermore, HPS outbreaks have been linked to precipitation; however, climatic factors alone have not been sufficient to predict the spatial-temporal dynamics of the environment-reservoir-virus system.26
In all studied areas, two hantavirus genotypes were identified: Araucária and Jaborá. Phylogenetic analysis showed that six rodent-borne hantaviruses cluster together with high statistical support with Araucaria (Juquitiba-like) virus identified in HPS cases and rodents from Brazil and Paraguay. It is a homogenous group, sharing a 97%/100% of similarity at nucleotide/amino acidic level. It can be considered as a sister clade to a group formed by several South American hantaviruses, including pathogenic viruses like Andes, Lechiguanas, and Araraquara, as well as non-pathogenic hantaviruses like Maciel or Pergamino. Another five rodent-borne hantaviruses from Paraná state form a monophyletic group with Jaborá hantaviruses from Paraguay and Brazil (Santa Catarina state). This group shares a 92%/99.9% identity at nucleotide/amino acidic level, respectively, and contains viruses collected mostly from A. montensis, and one from A. serrensis. The partial S sequences analyzed here are not informative enough to determine the relationships of this genotype with other South American hantaviruses. It is noteworthy that the hantaviruses identified in different Akodon species (A. montensis and A. serrensis) from General Carneiro are more closely related than the A. montensis-borne hantavirus from Jaguariaíva (10299), raising the possibility of a spillover infection from A. montensis to A. serrensis. The phylogenetic tree displayed in Figure 3 was constructed using a subset of sequences encompassing nucleotides 978 to 1,250 from Araucaria virus (AY740633). In this analysis a hantavirus genomic sequence retrieved from an A. paranaensis specimen collected in General Carneiro was placed into the Araucaria group with high statistical support (posterior probability of 1). The intra-genotypic identity was 99.5%/100% at nucleotide/amino acidic level, respectively.
In some regions, viral co-circulation was observed in trapped rodents and both genotypes have been previously described in South Brazil.7,9 Nonetheless, only the Araucaria hantavirus has been detected in patients with HPS and the Jaborá virus has not yet been associated with human diseases.9 Generally, sigmondontine rodents have been captured in disturbed habitats, such as cultivated fields, reforested areas, and peridomestic settings. However, host population and pathogen prevalence may vary locally and many rodent species show distinct habitat preferences. Thus, additional local studies are essential to define the risk of human disease.27
The presence of different rodent species within the same natural setting and are infected by the same hantavirus genotype, has been previously reported.7,9,17,28 In this study, A. paranaensis was found for the first time to be infected with Araucaria hantavirus, making a total of four different rodent species that are associated with the same hantavirus genotype. The impact of this number of host species on the natural transmission cycle remains unclear. Jaborá virus was found in the previously reported host (A. montensis) and also in A. serrensis, which has not been previously identified as a hantavirus reservoir. Interestingly, both species were captured in the same locality (General Carneiro) but in different seasons (March 2010 and December 2009, respectively).
Hantavirus infections have been described with a wide variety of clinical forms, from asymptomatic to the classical clinical manifestations of HFRS and HPS29,30 Although serologic evidence indicated that hantavirus circulated in human and rodents populations in all regions of the Paraná state, classical HPS cases have not been reported in all regions (i.e., Northwestern and Northeastern Paraná), suggesting that either oligo/asymptomatic infections occurs. It cannot be ruled out that occasionally hantavirus infection are not recognized or reported. It seems likely that pathogen exposure varies among these different areas, which could have practical implications on disease transmission.
Recently, several novel hantaviruses with unknown pathogenic potential were identified in widely separated geographical regions in a variety of soricomorpha mammals. Nothing is known about the pathogenicity of these hantaviruses at the moment, but finding these viruses among such evolutionary divergent species, predicts that other groups of mammals could also carry these viruses.31 Notably, in the Itambaracá region, 27% of the animals captured was Didelphis albiventris. Although serologically positive rodents were detected in this region, RT-PCR analyses of lung samples from these animals were negative for hantavirus.
It is important to mention that the main objective of this study was to determine the serological status of the human population and of the potential rodent reservoirs regarding hantavirus exposure in different biomes of Paraná State. For this reason, some expeditions were performed only once per area. Hence, we are aware that these particular data relate only to one period and reflect the landscape only for that time. However, human seroprevalence data (from individuals without previous HPS classical clinical presentations) and the distributions of seropositive reservoir species will assist health authorities in adjusting their prevention policies and defining a permanent surveillance program for the entire region.
Although the impacts of environmental change and degradation on the dispersion dynamics of the seropositive rodent reservoir have not been evaluated, it has been suggested that habitat fragmentation and the loss of species diversity may be altering hantavirus infection dynamics.32 In the last several decades, Paraná has witnessed high rates of environmental degradation and human population growth. The impacts of the consequential losses in biodiversity on human health should be monitored and the complex factors involved in the dynamics of hantaviruses in nature.
ACKNOWLEDGMENTS
We thank the Public Health Secretary of the State of Paraná for their assistance in the rodent capture expeditions and human blood collection.
Disclaimer: The authors have no competing financial interests.
Footnotes
Financial support: Fiocruz, CNPq, Fundação Araucária, CAPES. SMR and CNDS are sponsored by a CNPq fellowship.
Authors' addresses: Sonia M. Raboni, Infectious Disease Department, Universidade Federal do Paraná, Instituto Carlos Chagas, Fiocruz, Paraná, Brazil, E-mail: sraboni@ufpr.br. Adriana Delfraro, Facultad de Ciencias, Universidad de la Republica, Montevideo, Uruguay, E-mail: adelfraro@gmail.com. Luana de Borba, Vanessa Stella, Marina R. de Araujo, Suzana Carstensen, and Claudia N. Duarte dos Santos, Instituto Carlos Chagas, Fiocruz, Paraná, Brazil, E-mails: luturtle@onda.com.br, vastella1@hotmail.com, marinaaraujo@hotmail.com, suzana_carstensen@yahoo.com.br, and clsantos@tecpar.br. Bernardo R. Teixeira, Programa de Pós graduação em Biologia Parasitária, Fiocruz, Rio de Janeiro; and Instituto Oswaldo Cruz, Fiocruz, Rio de Janeiro, Brazil, E-mail: brt@ioc.fiocruz.br. Giselia Rubio and Angela Maron, Secretaria de Saúde do Estado do Paraná, Brazil, E-mails: giseliarubio@sesa.pr.gov.br and angela.maron@gmail.com. Elba R. S. Lemos and Paulo S. D'Andrea, Instituto Oswaldo Cruz, Fiocruz, Rio de Janeiro, Brazil, E-mails: elemos@ioc.fiocruz.br and dandrea@ioc.fiocruz.br.
References
- 1.Meyer BJ, Schmaljohn CS. Persistent hantavirus infections: characteristics and mechanisms. Trends Microbiol. 2000;8:61–67. doi: 10.1016/s0966-842x(99)01658-3. [DOI] [PubMed] [Google Scholar]
- 2.Peters CJ, Simpson GL, Levy H. Spectrum of hantavirus infection: hemorrhagic fever with renal syndrome and hantavirus pulmonary syndrome. Annu Rev Med. 1999;50:531–545. doi: 10.1146/annurev.med.50.1.531. [DOI] [PubMed] [Google Scholar]
- 3.Nelson R, Canate R, Pascale JM, Dragoo JW, Armien B, Armien AG, Koster F. Confirmation of Choclo virus as the cause of hantavirus cardiopulmonary syndrome and high serum antibody prevalence in Panama. J Med Virol. 2010;82:1586–1593. doi: 10.1002/jmv.21864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Puerta H, Cantillo C, Mills J, Hjelle B, Salaza-Bravo J, Mattar S. Hantavirus del Nuevo mundo – ecologia y epidemiologia de un virus emergente en latinoamerica. Medicina (B Aires) 2006;66:343–356. [PubMed] [Google Scholar]
- 5.Mir MA. Hantaviruses. Clin Lab Med. 2010;30:67–91. doi: 10.1016/j.cll.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raboni SM, Rubio G, DE Borba L, Zeferino A, Skraba I, Goldenberg S, Dos Santos CN. Clinical survey of hantavirus in southern Brazil and the development of specific molecular diagnosis tools. Am J Trop Med Hyg. 2005;72:800–804. [PubMed] [Google Scholar]
- 7.Raboni SM, Hoffmann FG, Oliveira RC, Teixeira BR, Bonvicino CR, Stella V, Carstensen S, Bordignon J, D'Andrea PS, Lemos ER, Duarte Dos Santos CN. Phylogenetic characterization of hantaviruses from wild rodents and hantavirus pulmonary syndrome cases in the state of Parana (southern Brazil) J Gen Virol. 2009;90:2166–2171. doi: 10.1099/vir.0.011585-0. [DOI] [PubMed] [Google Scholar]
- 8.Raboni SM, Levis S, Rosa ES, Bisordi I, Delfraro A, Lemos E, Correia DC, Duarte Dos Santos CN. Hantavirus infection in Brazil: development and evaluation of an enzyme immunoassay and immunoblotting based on N recombinant protein. Diagn Microbiol Infect Dis. 2007;58:89–97. doi: 10.1016/j.diagmicrobio.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 9.Oliveira RC, Padula PJ, Gomes R, Martinez VP, Bellomo C, Bonvicino CR, Lima DI, Bragagnolo C, Caldas AC, D'Andrea PS, Lemos ER. Genetic characterization of hantaviruses associated with sigmodontine rodents in an endemic area for hantavirus pulmonary syndrome in southern Brazil. Vector Borne Zoonotic Dis. 2011;11:301–314. doi: 10.1089/vbz.2010.0001. [DOI] [PubMed] [Google Scholar]
- 10.Mills JN, Childs JE, Ksiazek T, Peters C. Methods for Trapping and Sampling Small Mammals for Virologic Testing. Atlanta, GA: Centers for Disease Control and Prevention; 1995. [Google Scholar]
- 11.Smith MF, Patton JL. The diversification of South American murid rodents: evidence from mitochondrial DNA sequence data for the akodontine tribe. Biol J Linn Soc Lond. 1993;50:149–177. [Google Scholar]
- 12.Johnson AM, Bowen MD, Ksiazek TG, Williams RJ, Bryan RT, Mills JN, Peters CJ, Nichol ST. Laguna Negra virus associated with HPS in western Paraguay and Bolivia. Virology. 1997;238:115–127. doi: 10.1006/viro.1997.8840. [DOI] [PubMed] [Google Scholar]
- 13.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 14.Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- 15.Delfraro A, Tome L, D'Elia G, Clara M, Achaval F, Russi JC, Rodonz JR. Juquitiba-like hantavirus from 2 nonrelated rodent species, Uruguay. Emerg Infect Dis. 2008;14:1447–1451. doi: 10.3201/eid1409.080455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castro-Arellano I, Suzan G, Leon RF, Jimenez RM, Lacher TE., Jr Survey for antibody to hantaviruses in Tamaulipas, Mexico. J Wildl Dis. 2009;45:207–212. doi: 10.7589/0090-3558-45.1.207. [DOI] [PubMed] [Google Scholar]
- 17.Medina RA, Torres-Perez F, Galeno H, Navarrete M, Vial PA, Palma RE, Ferres M, Cook JA, Hjelle B. Ecology, genetic diversity, and phylogeographic structure of andes virus in humans and rodents in Chile. J Virol. 2009;83:2446–2459. doi: 10.1128/JVI.01057-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mills JN, Schmidt K, Ellis BA, Calderon G, Enria DA, Ksiazek TG. A longitudinal study of hantavirus infection in three sympatric reservoir species in agroecosystems on the Argentine Pampa. Vector Borne Zoonotic Dis. 2007;7:229–240. doi: 10.1089/vbz.2006.0614. [DOI] [PubMed] [Google Scholar]
- 19.Aleman A, Iguaran H, Puerta H, Cantillo C, Mills J, Ariz W, Mattar S. First serological evidence of hantavirus infection in rodents in Colombia. Rev Salud Publica (Bogota) 2006;8((Suppl 1)):1–12. [PubMed] [Google Scholar]
- 20.Armien B, Pascale J, Munoz C, Lee SJ, Kook L, Choi MA, Broce C, Armien A, Gracia F, Hjelle B, Koster F. Incidence rate for hantavirus infections without pulmonary syndrome, Panama. Emerg Infect Dis. 2011;17:1936–1939. doi: 10.3201/eid1710.101717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mattar S, Parra M. Serologic evidence of hantavirus infection in humans, Colombia. Emerg Infect Dis. 2004;10:2263–2264. doi: 10.3201/eid1012.040821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rivas YJ, Moros Z, Moron D, Uzcategui MG, Duran Z, Pujol FH, Liprandi F, Ludert JE. The seroprevalences of anti-hantavirus IgG antibodies among selected Venezuelan populations. Ann Trop Med Parasitol. 2003;97:61–67. doi: 10.1179/000349803125002788. [DOI] [PubMed] [Google Scholar]
- 23.Mendes WS, da Silva AA, Neiva RF, Costa NM, de Assis MS, Vidigal PM, da GL Leite M, da Rosa ES, de A Medeiros DB, de B Simith D, da C Vasconcelos PF. Serologic survey of hantavirus infection, Brazilian Amazon. Emerg Infect Dis. 2010;16:889–891. doi: 10.3201/eid1605.090766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Travassos da Rosa ES, Sampaio de Lemos ER, de Almeida Medeiros DB, Simith DB, de Souza Pereira A, Elkhoury MR, Mendes WS, Vidigal JR, de Oliveira RC, D'Andrea PS, Bonvicino CR, Cruz AC, Nunes MR, da Costa Vasconcelos PF. Hantaviruses and hantavirus pulmonary syndrome, Maranhao, Brazil. Emerg Infect Dis. 2010;16:1952–1955. doi: 10.3201/eid1612.100418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campos GM, Moro de Sousa RL, Badra SJ, Pane C, Gomes UA, Figueiredo LT. Serological survey of hantavirus in Jardinopolis County, Brazil. J Med Virol. 2003;71:417–422. doi: 10.1002/jmv.10489. [DOI] [PubMed] [Google Scholar]
- 26.Goodin DG, Paige R, Owen RD, Ghimire K, Koch DE, Chu YK, Jonsson CB. Microhabitat characteristics of Akodon montensis, a reservoir for hantavirus, and hantavirus seroprevalence in an Atlantic forest site in eastern Paraguay. J Vector Ecol. 2009;34:104–113. doi: 10.1111/j.1948-7134.2009.00013.x. [DOI] [PubMed] [Google Scholar]
- 27.Mills JN, Yates TL, Ksiazek TG, Peters CJ, Childs JE. Long-term studies of hantavirus reservoir populations in the southwestern United States: rationale, potential, and methods. Emerg Infect Dis. 1999;5:95–101. doi: 10.3201/eid0501.990111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heyman P, Vaheri A, Lundkvist A, Avsic-Zupanc T. Hantavirus infections in Europe: from virus carriers to a major public-health problem. Expert Rev Anti Infect Ther. 2009;7:205–217. doi: 10.1586/14787210.7.2.205. [DOI] [PubMed] [Google Scholar]
- 29.Butler JC, Peters CJ. Hantaviruses and hantavirus pulmonary syndrome. Clin Infect Dis. 1994;19:387–394. doi: 10.1093/clinids/19.3.387. quiz 395. [DOI] [PubMed] [Google Scholar]
- 30.McCaughey C, Hart C. Hantaviruses. J Med Microbiol. 2000;47:587–599. doi: 10.1099/0022-1317-49-7-587. [DOI] [PubMed] [Google Scholar]
- 31.Klempa B. Hantaviruses and climate change. Clin Microbiol Infect. 2009;15:518–523. doi: 10.1111/j.1469-0691.2009.02848.x. [DOI] [PubMed] [Google Scholar]
- 32.Suzan G, Marce E, Giermakowski JT, Armien B, Pascale J, Mills J, Ceballos G, Gomez A, Aguirre AA, Salazar-Bravo J, Armien A, Parmenter R, Yates T. The effect of habitat fragmentation and species diversity loss on hantavirus prevalence in Panama. Ann N Y Acad Sci. 2008;1149:80–83. doi: 10.1196/annals.1428.063. [DOI] [PubMed] [Google Scholar]