Summary
The conserved Translin-TRAX complexes, also known as C3PO, have been implicated in many biological processes, but how they function remains unclear. Recently, C3PO was shown to be an endoribonuclease that promotes RNA interference in animal cells. Here we show that C3PO does not play a significant role in RNAi in the filamentous fungus Neurospora crassa. Instead, the Neurospora C3PO functions as a ribonuclease that removes the 5′ pre-tRNA fragments after the processing of pre-tRNAs by RNase P. In addition, the translin and trax mutants have elevated levels of tRNA and protein translation and are more resistant to a cell-death inducing agent. Finally, we showed that C3PO is also involved in tRNA processing in mouse embryonic fibroblast cells. Together, this study identified the endogenous RNA substrates of C3PO and provides a potential explanation for its roles in seemingly diverse biological processes.
Introduction
Translin and its partner protein TRAX (Translin Associated Protein X) are highly conserved from fission yeast to human 1–3. The two form a heteromeric complex and are proposed to be involved in many biological processes in different organisms, including normal cell growth, RNA metabolism, genome stability, neuronal development and spermatogenesis 4–8. Although the translin and trax genes are not essential for cell survival, studies of translin/trax mutants suggested that they are involved in cell proliferation in the fission yeast, motor response in Drosophila, and spermatogenesis and behavior in mouse 4,9–11. The Translin-TRAX complex is able to bind DNA and RNA 1,2,7,12, but how it acts to regulate diverse biological processes is unknown.
Recently, the Translin-TRAX complex was biochemically purified as C3PO (component 3 promoter of RISC) from Drosophila and human cells 13,14. C3PO is necessary for efficient RNA interference (RNAi) mediated by small interfering RNAs (siRNAs) 13,14. RNAi is a conserved eukaryotic gene silencing mechanism mediated by small noncoding RNAs 15–18. In RNAi pathways, small interfering RNA (siRNAs), which are generated by Dicer cleavage of double-stranded RNA (dsRNA), associate with and guide Argonaute family proteins to their RNA targets to regulate gene expression. Genetic depletion of C3PO impairs RNAi efficiency in both Drosophila and human cells. Importantly, C3PO was demonstrated to be an RNA-specific endonuclease that can promote RNAi by removing the passenger strand of siRNA duplex. Crystal structures of C3PO revealed that six Translin and two TRAX subunits form an asymmetric octamer with the RNase catalytic residues located on the TRAX subunits 14,19. However, it is unclear whether Translin and TRAX also have a similar role in RNAi in other eukaryotic organisms. In addition, the functions of Translin and TRAX in different biological processes suggest that the complex has other cellular functions, but the endogenous RNA substrates of the Translin-TRAX complex as an endonuclease are not known.
The filamentous fungus Neurospora crassa is an important eukaryotic model system for RNAi studies 20,21. We previously showed that QIP, an exonuclease, interacts with the Argonaute protein QDE-2 and removes the nicked passenger strand from the siRNA duplex 22. Therefore, the role of C3PO in Drosophila and human is similar that of QIP in Neurospora. In addition to siRNAs, several other type of small RNAs have been identified in Neurospora 23,24.
There is one Translin (NCU06664, 239aa) and one TRAX homolog (NCU06059, 349aa) in the Neurospora genome, which are highly conserved to the counterparts in Drosophila and human. In this study, we set out to determine the function of C3PO in Neurospora by identifying its endogenous RNA substrates. We showed that the Neurospora Translin and TRAX do not have a significant role in RNAi. Instead, we discovered that the lack of the Neurospora Translin-TRAX complex (nC3PO) resulted in dramatic accumulation of pre-tRNA fragments in Neurospora. Genetic, molecular and biochemical experiments showed that the Translin-TRAX complex acts as an RNase that removes 5′ pre-tRNA fragments after the processing of pre-tRNAs by RNase P. Moreover, translin and trax mutants have elevated tRNA levels, increased protein translation efficiency and increased resistance to a programmed cell death inducing agent. This study revealed the endogenous substrates of the RNase activity of the Translin-TRAX complex and provides a potential mechanism that explains its roles in many biological processes.
Results
Neurospora Translin and TRAX play no significant role in RNAi
To determine whether the Neurospora Translin and TRAX form a complex in vivo, a construct that expresses Myc-His-tagged TRAX (TRX) was transformed into a wild-type (WT) strain and the Myc-His-tagged TRAX from this strain was purified by a nickel column followed by immunoprecipitation using c-Myc monoclonal antibody. The same purification procedure was performed using a wild-type strain that lacks the Myc-His-tagged TRAX. As shown in Figure 1a, two major specific protein bands (indicated by the arrows) were observed in the purification products of the Myc-TRX strain. Although there were several other minor bands that show differences between the two samples, they did not appear consistently in independent purification experiments, suggesting that they are either degradation products of the two major bands or non-specific proteins. Mass spectrometry analysis identified the top band as the Myc-tagged TRAX and the lower band as the Neurospora Translin, indicating that as in other eukaryotic organisms, the Neurospora Translin and TRAX form a complex.
Figure 1. Neurospora Tranlin and TRAX are not required for RNAi and siRNA passenger strand removal.
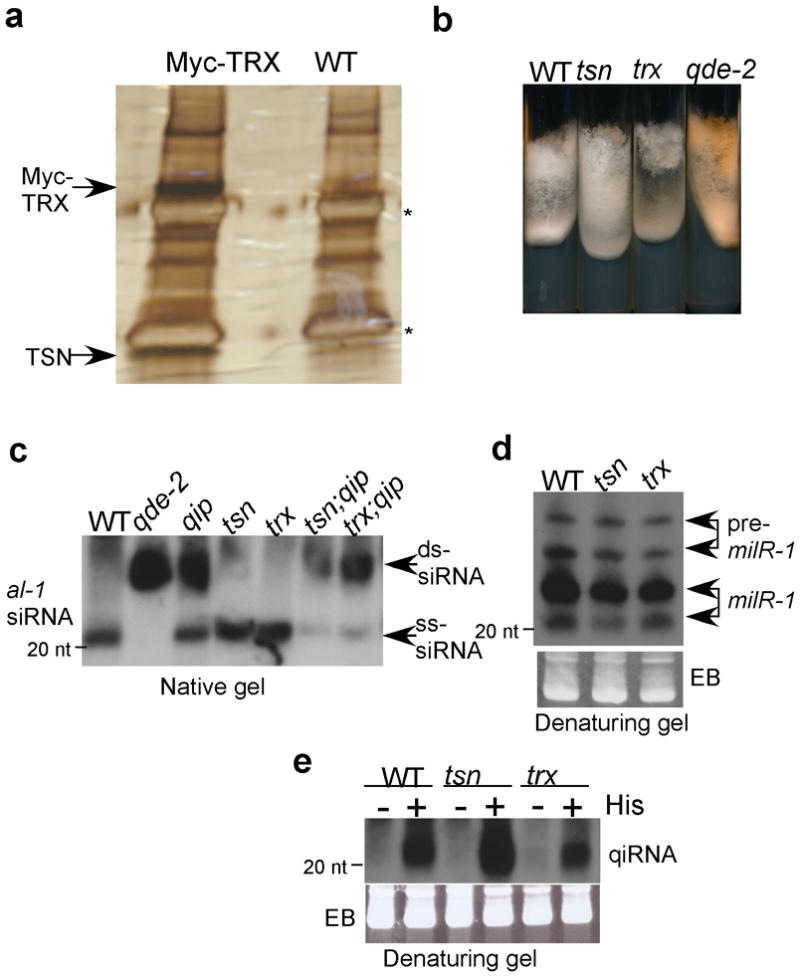
(a) Purification of the Neurospora Translin-TRAX complex. SDS-PAGE gel shows the c-Myc immunoprecipitation products from the protein extracts of the wild-type and Myc-TRX strains. The two arrows indicate the bands identified by mass spectrometry as Myc-TRX and TSN, respectively. The two asterisks indicate the IgG bands. (b) A photograph of Neurospora slants showing the gene silencing of al-1 by the expression of the dsal-1 construct in the indicated strains. Cultures were grown with QA (1 × 10−3 M). In the qde-2 strain, the dsal-1-induced gene silencing was abolished. (c) Northern blot analyses of al-1 siRNA of the indicated strains by a native gel. Cultures were grown with QA (1 × 10−3 M) and RNA samples from the indicated strains were used. The two arrows indicate the siRNA duplex (ds-siRNA) and single-stranded siRNA (ss-siRNA) respectively. (d and e) Northern blot analyses showing the expression profiles of (d) milR-1 milRNAs and (e) qiRNA in the indicated strains. His: histidine.
To determine the role of Translin and TRAX in dsRNA-triggered RNAi, a construct that can inducibly express dsRNA specific for albino-1 (al-1) gene in the presence of quinic acid (QA)22,25 was introduced into wild-type, translin knock-out (tsnKO) and trax knock-out (trxKO) strains26 at the his-3 locus by homologous recombination. The resulted transformants were purified to obtain homokaryon strains. The silencing of al-1 expression by RNAi blocks carotenoid biosynthesis, resulting in a color change from orange to white. As shown in Figure 1b, the expression of the dsRNA specific for al-1 resulted in a color change that was similar in the wild-type, tsn and trx strains, indicating that the nC3PO does not play a major role for the dsRNA-induced RNAi in Neurospora.
Because of the role of C3PO in removing the siRNA passenger strand in animals, we compared the production and nature of al-1 siRNA in the tsnKO and trxKO strains with those in the wild-type, qde-2 and qip mutants. As shown previously 22, almost all al-1 siRNA produced in the wild-type strain was single stranded, and only siRNA duplex but no single-stranded siRNA was present in the qde-2 mutant (Figure 1c). The vast majority of siRNA in the qip strain was in duplex form, confirming the role of QIP in siRNA passenger strand removal. In contrast to the siRNA profile in the qip strain, all siRNA was single-stranded in both tsnKO and trxKO mutants. To examine the possibility that QIP and the nC3PO may play a redundant role in siRNA passenger strand removal, we created tsnKO;qip and trxKO;qip double mutants and introduced the dsal-1 construct into the strains by co-transformation. As shown in Figure 1c and supplementary Figure 1, although the levels of total al-1 siRNA were lower in the transformants of the double mutants (likely to due to that co-transformation triggered a partial silencing of the dsal-1 construct)27, the ratios of the single-stranded siRNA/total siRNA were not statistically different between the qip single mutant and the tsnKO;qip and trxKO;qip double mutants. Although we cannot exclude the possibility that nC3PO plays a minor role in RNAi, these results indicate that Translin and TRAX do not play a significant role in the siRNA-mediated gene silencing in Neurospora.
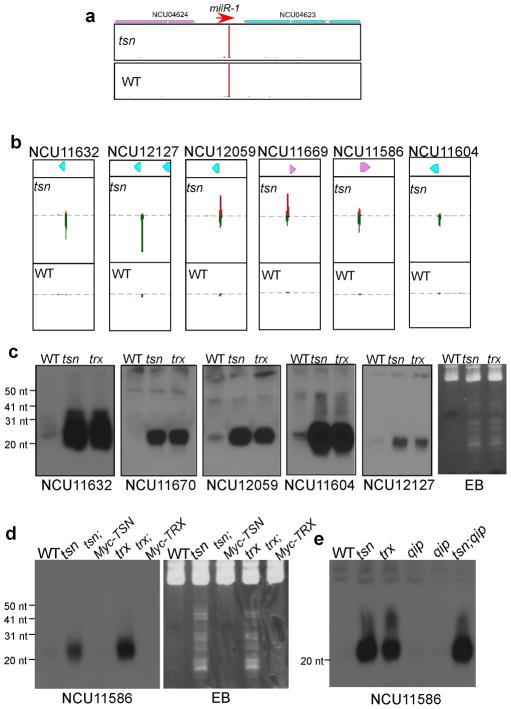
We also examined the role of Translin and TRAX in the production of the Neurospora miRNA-like small RNAs 24. As shown in Figure 1d, although the maturation of milR-1 milRNA was completely blocked in the qip mutant 24, the milRNA production was normal in the tsnKO and trxKO mutants. Similarly, the amounts of DNA damage-induced qiRNA23 in the tsnKO and trxKO mutants were comparable to that in the wild-type strain (Figure 1e). These results indicate that Translin and TRAX are not involved in the production of known types of Neurospora sRNAs.
Accumulation of small RNA species in the tsn and trx mutants
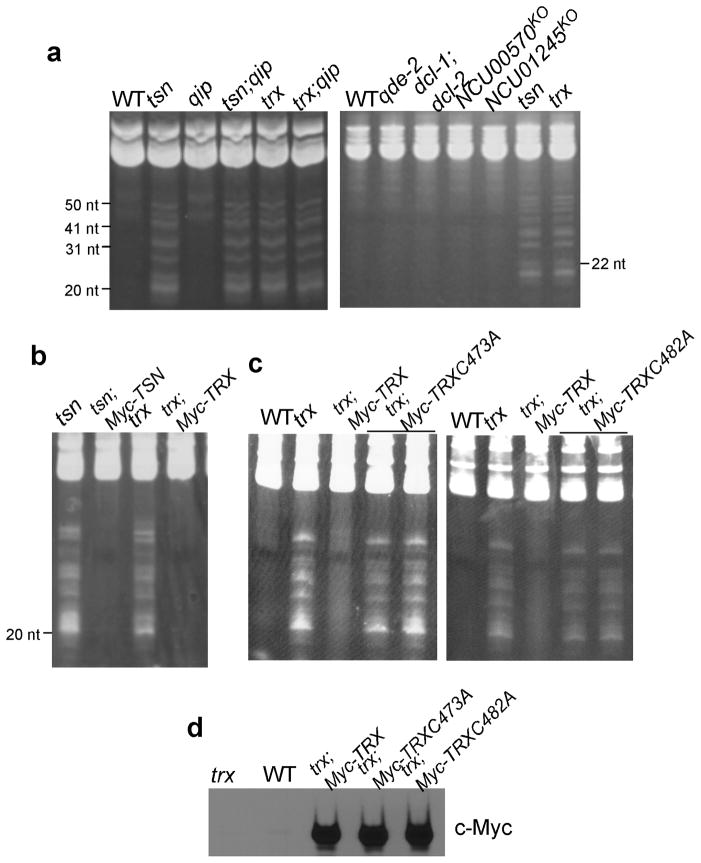
The lack of a significant role for Translin and TRAX in RNAi and small RNA production in Neurospora and conservation of the Translin-TRAX complex in eukaryotes suggest that Translin and TRAX have other roles in Neurospora. To investigate this possibility, we compared the total small RNA profile of a wild-type Neurospora strain to those in the tsnKO and trxKO mutants in gels stained with ethidium bromide (EB) of the enriched small RNA samples. As shown in Figure 2a, a ladder of ~18–50 nt small RNA species accumulated to high levels in both the tsn and trx mutants but not in the wild-type and RNAi-mutant strains. The most abundant species that accumulated in tsn and trx mutants were around 20 nt in length. To examine whether the lack of the Translin-TRAX complex in the tsn and trx mutants is responsible for the accumulation these RNAs, constructs for expression of Myc-tagged wild-type Translin (Myc-TSN) or Myc-tagged TRAX was transformed into the tsnKO and trxKO mutants, respectively. As shown in Figure 2b, the expression of the Translin or TRAX rescued the phenotype in the tsn and trx mutants, indicated by the disappearance of the RNA ladder in these strains.
Figure 2. Accumulation of small RNA species in the tsnKO and trxKO strains is dependent on the catalytic activity of the Translin-TRAX complex.
(a–c) Ethidium bromide-stained denaturing PAGE gels showing the small RNA profiles in the indicated strains. Total RNA was used in b and c. (d) Western blot analysis by the anti-c-Myc antibody showing the expression of Myc-TRX in the indicated strains. The wild-type (WT) and the trxko (trx) strains were used as negative controls.
The accumulation of small RNA species and the proposed function of C3PO as an RNA-specific nuclease raise the possibility that these RNAs are the substrates of the Translin-TRAX complex. To test this hypothesis, we created two Myc-TRX constructs in which RNase catalytic cysteine residues (Cys473 or Cys482) 13,14,19 were mutated to alanine (C473A or C482A) and introduced these constructs separately into the trx mutant. As shown in Figure 2c and 2d, despite similar expression levels of the mutant proteins to that of the wild-type protein, neither mutant construct rescued the phenotype of the trx mutant, suggesting that the accumulated small RNA species in the tsn and trx mutants are the endogenous RNA substrates of the Translin-TRAX complex in Neurospora.
Accumulation of pre-tRNA fragments in tsn and trx mutants
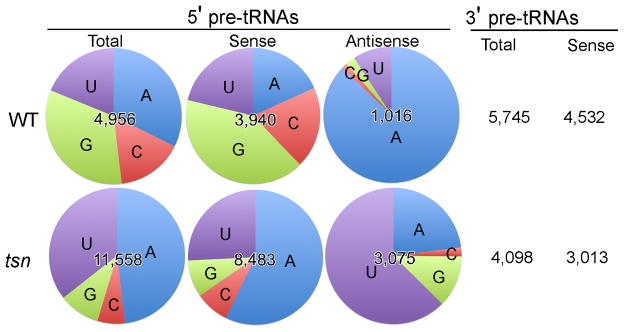
To identify the accumulated small RNA species in the tsn and trx mutants, we size-fractioned the small RNA samples of the wild-type and tsnKO strains, generated cDNA libraries of small RNAs (17–30 nt) and sequenced them using an Illumina/Solexa Genome Analyzer. We obtained 1,572,492 and 1,658,457 reads from the wild-type and tsn libraries, respectively, that perfectly match to the Neurospora genome. After the small RNA sequences were mapped to the assembled Neurospora genome sequences, we compared the profiles of small RNA distributions between the wild-type strain and the tsn mutant. Although the distribution of small RNAs appeared to be comparable in both strains for most of the Neurospora genome (Supplementary Figure 2), there were significantly more small RNA reads in the tsn mutants than the wild-type that were mapped to the regions immediately upstream of the predicted Neurospora tRNA genes (Figure 3). There were two times more sRNA reads within 100 nt upstream of the 5′end of predicted tRNAs in the tsn mutant than in the wild-type strain. It is important to note that there is significantly more small RNA in the tsn mutant than the wild-type strain (as indicated by the EB stained gels), but similar total sRNA reads were obtained from the two strains. Thus, the relative numbers of the sRNA reads from the tsn mutant were underestimated. Therefore, the actual differences of the 5′ tRNA small RNA reads between the tsn mutant and the wild-type strains should be greater. Although the 5′ ends of the wild-type small RNAs mapped to these regions showed no significant nucleotide preference, the small RNAs in the tsn mutant exhibited a preference for 5′ A (48%) and U (36%). Most of these small RNAs matched the sense strand of the predicted pre-tRNAs, but a significant fraction also matched the antisense strand. In the tsn mutant, the antisense but not sense small RNAs had a strong preference for 5′ U (63%). The 5′ U preference of the antisense small RNAs is reminiscent of the Neurospora small RNAs produced by RNAi pathways. Detailed comparison of the small RNA distribution over the entire genome between the wild-type and tsnKO strains revealed that there was a significant accumulation of small RNAs upstream of 56 predicted tRNA genes (Supplementary Table 1) in the tsnKO strain, including those for 15 lysine tRNAs, 14 glutamic acid tRNAs and 11 aspartic acid tRNAs. In addition to the tRNA loci, there are 18 other loci with significant increases of small RNAs in the tsn mutant (Supplementary Table 2). These small RNAs matched to predicted gene open-reading frame regions, intergenic regions and repeat regions. For these non-tRNA loci originated sRNAs, no common functional and sequence motifs are observed.
Figure 3. Deep sequencing of small RNAs reveals the accumulation of 5′ pre-tRNA fragments in the tsnKO strain.
Pie charts showing the numbers of small RNA reads and their 5′ nucleotide preferences from the 5′ and 3′ ends of the predicted Neurospora pre-tRNAs. Small RNAs matching within 100 nt upstream (5′) or downstream (3′) of predicted mature tRNA were included in the analyses.
tRNA genes are transcribed in eukaryotes by RNA polymerase III to produce tRNA precursors with 5′ and 3′ sequences that are removed to yield functional tRNAs 28–30. The 5′ leader sequences of pre-tRNAs are cleaved by the endonuclease RNase P to generate mature 5′ends of tRNAs and the 3′ sequences are processed by tRNase Z and other enzymes. Supplementary Figure 3a shows the accumulation of small RNA in one representative tRNA locus (NCU1217, Lys tRNA). The secondary structure of the NCU12127 tRNA and the predicted pre-tRNA cleavage sites by RNase P and tRNase Z are shown in Supplementary Figure 3b. The accumulated RNAs mapped immediately upstream of the predicted mature Lys tRNA sequence. The exact match of the small RNA species to those of the 5′ leader sequences of the pre-tRNAs indicates that these RNAs are mostly pre-tRNA fragments that have been processed by RNase P.
As shown in Figures 4a and 4b, levels of milR-1 milRNAs were comparable in both the wild-type and tsn strains; however, the numbers of small RNA (sense and antisense) reads that matched to 5′ ends of pre-tRNAs for some tRNA genes were dramatically increased in the tsn mutant. In addition, the levels of antisense small RNA, although lower than those of the sense RNAs, were similarly increased in the tsn mutant. To confirm the deep sequencing results, we performed northern blot analysis on a few randomly selected tRNAs using probes that specifically detect the 5′ leader sequences of pre-tRNA transcripts (Figure 4c and 4d). As expected, small RNAs containing the 5′ leader sequences accumulated to high levels in both tsn and trx mutants but were barely detectable in the wild-type strain. In addition, the expression of the Myc-tagged Translin or Myc-tagged TRAX in the tsn or trx mutants, respectively, resulted in the decrease of pre-tRNA fragments to wild-type levels (Figure 4d). The levels of pre-tRNA fragments in the qip mutants were comparable to levels in the wild-type strain and were not further increased in the tsn;qip double mutant (Figure 4e), further indicating that QIP and the nC3PO have distinct roles in different cellular processes. Although most of the pre-tRNA signals were seen around 20–22 nts, higher molecular weight signals were observed for some probes, suggesting that pre-tRNA fragments are the major sRNA species seen on the EB-stained gels in the tsn and trx strains.
Figure 4. High levels of 5′ pre-tRNA fragments in the tsnKO and trxKO strains.
(a and b) Comparison of small RNA distributions at the (a) milR-1 and (b) representative tRNA loci between the wild-type and tsnKO strains. Numbers of small RNAs (logarithmic scale) in 10-nt sliding windows are shown. Small RNAs matched to either Watson (red) or Crick (green) strand of DNA are shown. (c–e) Northern blot analyses of 5′ pre-tRNA fragments of selected tRNAs in the indicated strains.
The antisense-specific small RNAs are products of RNAi
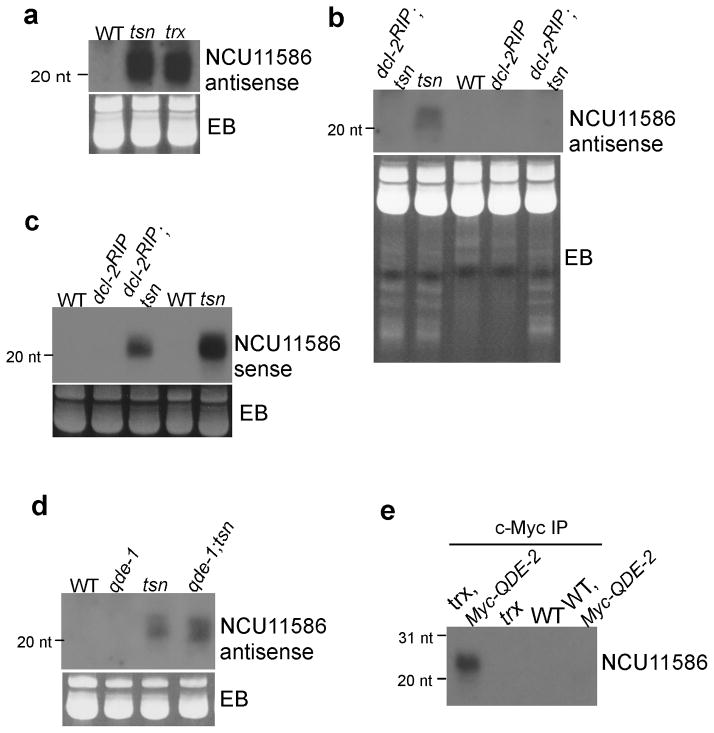
The antisense small RNAs complementary to some of the pre-tRNA fragments were also dramatically increased in the tsn and trx mutants (Figure 5a). To examine whether the antisense sRNAs are the products of RNAi pathway, we generated dcl-2; tsn double mutants. DCL-2 is the major Dicer in Neurospora 31. As shown in Figure 5b, the antisense-specific sRNA was completely abolished in the dcl-2; tsn mutant. In addition, a high molecular weight RNA smear accumulated in the dcl-2; tsn mutant (Supplementary Figure 4), suggesting that it is dsRNA. Furthermore, the sense-specific sRNA was also modestly reduced in the dcl-2; tsn mutant (Figure 5c), indicating that the antisense sRNA are small RNAs produced by Dicer. To examine whether QDE-1, an RdRP that is essential for quelling 32,33, is required for the production of antisense sRNAs, a qde-1; tsn double mutant was generated. As shown in Figure 5d, the levels of the antisense specific sRNA were comparable in both the wild-type and qde-1; tsn strains, indicating that QDE-1 is not required for the production of antisense specific RNA. It is likely that either SAD-1 and RRP-3 25,34, the two other RdRPs in Neurospora, are involved in converting the pre-tRNA fragments into dsRNAs or these RdRPs may play redundant roles. Furthermore, immunoprecipitation of Myc-QDE-2 from a trx; Myc-QDE-2 strain showed that the NCU11586 specific sRNA is associated with Myc-QDE-2 indicating that at least some of the sRNAs are incorporated into the Argonaute protein (Figure 5e). Together, these results demonstrate that the antisense sRNAs are produced by RNAi using the sense sRNA as templates. It is important to note that the small ladders shown by EB-stained gel (Figure 5b, lower panel) in the tsn and dcl-2; tsn double mutants were similar, indicating that the majority of small RNAs accumulated in the tsn mutant are cleavage products of pre-tRNAs by RNase and not Dicer products.
Figure 5. Dicer is required for the production of the antisense-specific small RNAs.
(a–d) Northern analyses showing the levels of the antisense and sense sRNAs that match to the 5′ pre-tRNA fragment of NCU11586 in the indicated strains. Ethidium bromide-stained gels of the small RNAs are shown below. (e) Immunoprecipitation of by the c-Myc antibody followed by Northern blot analysis showing that Myc-QDE-2 associated with the 5′ pre-tRNA fragments.
5′ pre-tRNA fragments are substrates of nC3PO
Both the Drosophila and human Translin-TRAX complexes act as single-stranded RNA endonucleases in vitro 13,14, suggesting that the pre-tRNA fragments accumulated in the Neurospora tsn and trx mutants are the in vivo substrates of the Translin-TRAX complex. To test this hypothesis, we co-expressed the Neurospora TRAX and the His-tagged Translin in E. coli and purified the recombinant Neurospora nC3PO by a TALON-Metal affinity column. In addition, we also expressed and purified the mutant complex that contains the C473A TRAX mutation. As expected, the recombinant Translin and TRAX form a complex in E. coli (Figure 6a). We then examined the ability of recombinant nC3PO to cleave the Neurospora pre-tRNA fragments in vitro. As shown in Figure 6b, the addition of the wild-type nC3PO to RNA sample of the tsn mutant resulted in the disappearance of the small RNAs on the EB-stained gel without significant effects on the high molecular weight RNA bands. In addition, Northern blot analysis showed that the level of 5′ tRNA fragment of NCU11586 was dramatically decreased by the treatment. In contrast, the C473A mutant nC3PO had no effects on the small RNA ladder seen on the EB-stain gel and on the 5′ pre-tRNA fragment level. This result demonstrates that the nC3PO is indeed an ribonuclease that can degrade the pre-tRNA fragments.
Figure 6. The pre-tRNA fragments are substrates of the Neurospora Translin-TRAX complex.
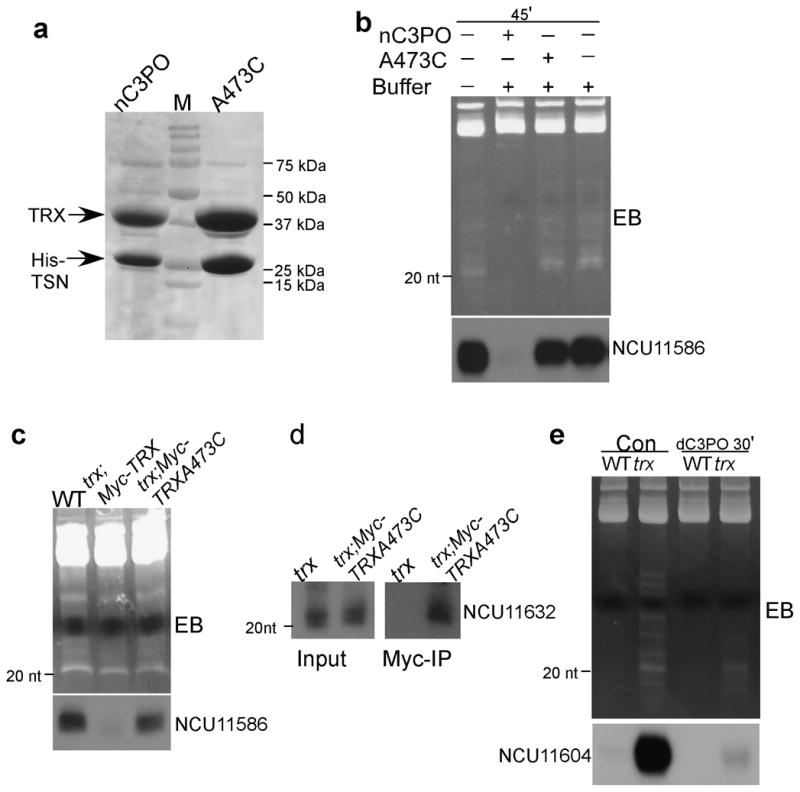
(a) A Coomassie Blue-stained gel showing the purified recombinant wild-type Neurospora Translin-TRX complex (nC3PO) and the nC3PO with the TRX catalytic point mutation (C473A). (b) The recombinant nC3PO can digest the 5′ pre-tRNA fragments in vitro. Ethidium bromide-stained gel of the small RNAs (top panel) and Northern blot analysis (bottom panel) of RNA treated in vitro with nC3PO/nC3PO(C473A) are shown. The small RNA preparation from the trxKO strain was used as the RNA substrate. (c) The nC3PO partailly purified from Neurospora can digest 5′ pre-tRNA fragments in vitro. Ethidium bromide-stained gel of the small RNAs and Northern blot analysis of trax sRNA samples treated with the c-Myc immunoprecipitates from the indicated strains are shown. (d) Immunoprecipitation of Myc-TRX (C473A) from the trx; Myc-TRXC473A strain showing that the catalytically dead nC3PO associated with 5′ pre-tRNA. The trx strain expressing no Myc-tagged protein was used as a negative control. (e) The recombinant Drosophila C3PO can digest 5′ pre-tRNA fragments in vitro. Ethidium bromide-stained gel of the small RNAs (top panel) and Northern blot analyses (bottom panel) of sRNA treated in vitro with Drosophila C3PO (dC3PO) are shown. The small RNA preparation from the trxKO strain was used as the RNA substrate.
To show that the endogenous nC3PO from Neurospora has a similar activity, we immunoprecipitated Myc-TRX and Myc-TRXC473A from the trx; Myc-TRX and trx; Myc-TRXC473A strains, respectively. A wild-type strain that does not express Myc-tagged protein was used as a negative control for the immunoprecipitation. The immunoprecipitates from the three strains were separately incubated with the RNA samples of the tsn mutant. As shown in Figure 6c, the immunoprecipitates from the trx; Myc-TRX strain, but not from the wild-type and trx; Myc-TRXC473A strains, reduced the small RNA ladder on the EB-strained gel and the level of 5′ pre-tRNA fragment.
We then examined whether the nC3PO is associated with 5′ pre-tRNA fragments in vivo. Because the C473A mutation of TRAX is catalytically inactive, we reasoned that the nC3PO-RNA association can be stabilized in the trx; Myc-TRXC473A mutant. After Myc-immunoprecipitation, the Myc-TRXC473A associated small RNA was examined by Northern blot analysis. As shown in Figure 6d, the pre-tRNA fragments was specifically pulled-down by the immunoprecipitation in the trx; Myc-TRXC473A strain, indicating that nC3PO associates with pre-tRNA fragments in vivo. Taken together, our results demonstrate that the 5′ pre-tRNA fragments are the in vivo substrates of nC3PO.
Because of the conservation of Translin-TRAX complexes in eukaryotes, we also examined whether the recombinant Drosophila C3PO (dC3PO) has a similar activity towards the 5′ pre-tRNA fragments. As shown in Figure 6e, the addition of the recombinant dC3PO to RNA samples of the tsn or trx mutants also resulted in a rapid disappearance of the small RNA ladder and the 5′ pre-tRNA fragments, suggesting that both the Drosophila and Neurospora Translin-TRAX complexes have similar activities towards 5′ pre-tRNA fragments.
tsn and trx mutant phenotypes
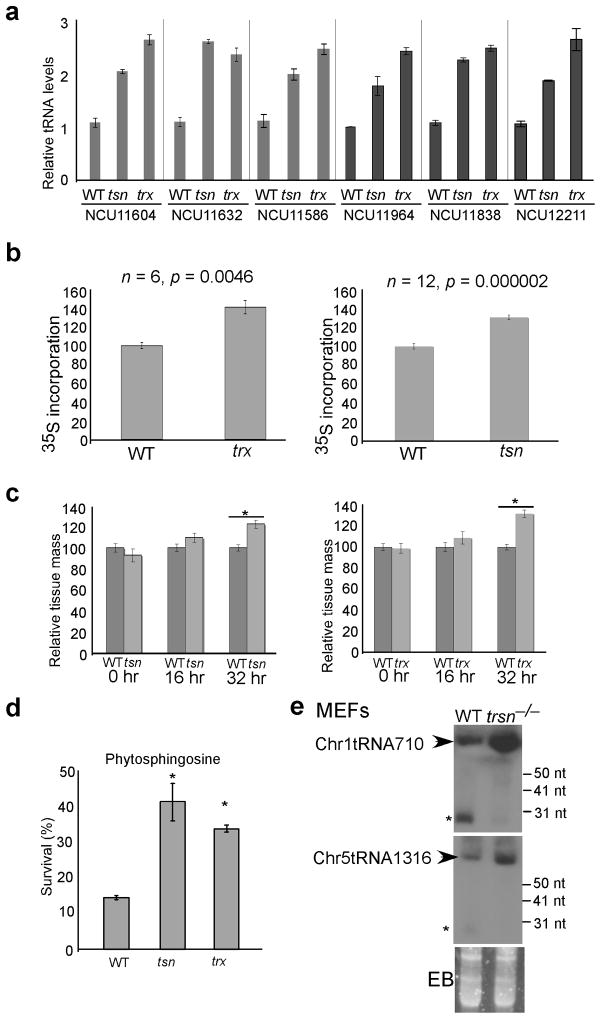
The developmental phenotypes of the tsn and trx knock-out strains were similar to the wild-type strain under normal culture conditions. Because the nC3PO regulates the processing of tRNAs in Neurospora, we examined the tRNA expression levels of genes that are regulated by the complex. As shown in Figure 7a, the tRNA levels of six selected genes were all significantly elevated in both the tsn and trx mutants, suggesting that the nC3PO suppresses tRNA production in Neurospora. It is possible that in addition to 5′ end pre-tRNA fragments, tRNAs/pre-tRNAs are also substrates for the Translin-TRAX complex.
Figure 7. Tranlin and TRAX regulate levels of tRNA and protein translation.
(a) qRT-PCR results showing the increase of tRNA levels in the tsnKO and trxKO strains of selected tRNAs compared to those in the wild-type strain. Error bars, ± s.e.m.. n=2. (b) The amount of protein synthesis also increased in the tsnKO and trxKO strains relative to levels in the wild-type strain. After a 30-min incubation with 35S methionine, amount of 35S labeled protein in various strains were determined. Error bars, ± s.e.m.. (c) Comparison of cell growth between the wild-type strain and the mutants. The dried cell mass of the wild-type strain at the indicated time points was set at 100. Error bars, ± s.e.m.. n=6–12. The asterisks indicate p value < 0.05. (d) Comparison of survival rates between the wild-type strain and the mutants after treatment with phytosphingosine. Error bars, ± s.e.m.. n=3. The asterisks indicate p value < 0.05 when the mutant is compared to the wild-type strain. (e) Northern blot analysis showing the levels of the mouse tRNAs (indicated by the arrows) in the wild-type and trsn−/− MEF cells. The asterisks indicate a small RNA species in the wild-type but not in the trsn−/− MEF cells.
Due to the essential role of tRNAs in general protein translation, we then compared the amounts of protein synthesized during a 30-min pulse of 35S-methionine in the wild-type strains and the mutants. Compared to the wild-type strain, a modest (~30%) but significant increase of 35S incorporation was observed in both the tsn and trx mutant strains (Figure 7b). Consistent with the elevated protein translation efficiency, both mutants also displayed a modest increase in cell growth comparing to the wild-type strain (Figure 7c). Similarly, cell proliferation rates was previously found to be elevated in the tsn and trx mutants of fission yeast 4. We also examined the sensitivities of the tsn and trx mutant strains to a series of stress-inducing agents and found that both mutants showed increased resistance to the programmed cell death-inducing agent phytosphingosine 35 (Figure 7d). Together, these results suggest that the nC3PO regulates general protein translation efficiency and cell proliferation through its role in tRNA processing.
Elevation of tRNA levels in translin knockout MEFs
The 5′ pre-tRNA fragments in mammalian tRNAs are generally much smaller than those in Neurospora 36,37 and could not be detected by conventional analyses (data not shown). To examine whether the role of C3PO in tRNA processing is conserved in mammals, we performed Northern blot analyses and qRT-PCR analyses to compare the levels of some mouse tRNAs between the wild-type and translin knockout (trsn−/−) mouse embryonic fiboblast (MEF) cells 14 (Figure 7e and Supplementary Figure 5). We found that the levels of six mouse tRNA levels were significantly elevated in the trsn−/− cells. In addition, Northern blot analysis showed that a ~25 nt tRNA-derived small RNA species in the wild-type cells was abolished in the trsn−/− cells, suggesting that it was a C3PO cleavage product from the corresponding tRNA. These results suggest that as in Neurospora, C3PO is also involved in tRNA processing in mammals.
Discussion
tRNAs play essential roles in protein synthesis. The maturation of tRNAs requires the accurate processing of pre-tRNAs at both 5′ and 3′ ends. RNase P has long been known to be responsible for the processing of pre-tRNAs at 5′end 28–30, but how the cleaved 5′ RNA fragments are further processed was not known. In this study, we identified the highly conserved Translin-TRAX complex as a major ribonuclease that digests the 5′ pre-tRNA fragments in Neurospora. This conclusion is supported by genetic and biochemical evidence. First, the RNase P-cleaved 5′ pre-tRNA fragments accumulated to very high levels in both tsn and trx mutants. Second, the catalytic residues of the Translin-TRAX complex were essential for processing of pre-tRNA fragments. Third, the recombinant nC3PO degraded the pre-tRNA fragments in vitro and associate with pre-tRNA fragments in cells. Because of the conservation of Translin-TRAX complexes and tRNA processing pathways in eukaryotes, our results suggest that Translin-TRAX complexes may have a similar function in other organisms. Supporting this notion, we showed that C3PO is also involved in tRNA processing in MEFs.
Although we found the accumulation of 5′ pre-tRNA fragments in the tsn and trx mutants for 56 predicted Neurospora tRNAs (out of 413)38, this is likely an underestimation of tRNAs regulated by Translin-TRAX because the coverage of our small RNA deep sequencing was limited to RNAs from 17–30 nts. Thus, 5′ pre-tRNAs that are larger or smaller would be missed in our analyses. Nonetheless, it is also likely that other ribonucleases are also involved in degradation of 5′ pre-tRNA fragments. It is also worth noting that some tRNAs can be transcribed without 5′ leader sequences and do not require RNase P for maturation 39. In addition to the pre-tRNA fragments, small RNA accumulation was found for 18 other non-tRNA loci (ORFs, intergenic and repeat regions), suggesting that nC3PO has additional non-tRNA substrates.
tRNA-derived RNA fragments or small RNAs are a major class of small RNA species in small RNA sequencing studies from fungi to animals 24,40,41. While most of these small RNAs are generated by RNAi-independent processing events, some had been demonstrated to be the products of Dicer processing 42. Interestingly, one of the human tRNA-derived RNA fragment was found to be highly expressed in cancer cell lines and its down-regulation impaired cell proliferation 40. On the other hand, the Angiogenin-induced tRNA fragments were shown to inhibit protein translation 43. These results indicate that the tRNA-derived small RNAs are not random degradation products and can have important biological functions.
Tranlin and TRAX are highly conserved in eukaryotic organisms from fungi to human. In addition to the role of the Translin-TRAX complex in human and Drosophila dsRNA-triggered RNAi pathways 13,14,19, both proteins have been implicated in many biological processes, including normal cell growth, genome stability, neuronal development and spermatogenesis, in different organisms 4–8,44. These functions of Translin-TRAX complex suggest that it have functions other than dsRNA-triggered RNAi and have endogenous RNA targets. Our study identified 5′ pre-tRNA fragments as the major substrates for the Translin-TRAX complex in Neurospora. The lack of a signficant RNAi function in Neurospora for Translin and TRAX suggests that their role in pre-tRNA processing is more ancient than their role in RNAi. In addition, we found that in the tsn and trx mutants, tRNA levels, protein translation efficiency and cell growth were elevated. The increase in levels of protein translation we observed in the tsn and trx mutants is consistent with the increase of cell proliferation rates of tsn and trx mutant cells observed in fission yeast 4. In addition, both Translin and TRAX are also known to be required for normal cell proliferation of mouse embryonic stem cells 5. Because the changes in tRNA levels can differentially affect protein expression for different proteins, the role of Translin and TRAX in tRNA and other RNA processing may provide a potential explanation for its many biological roles in different organisms.
Online Methods
Strains and growth conditions
The wild-type strain used in this study was FGSC 4200(a). Mutant strains, including qde-1, qde-2, qde-2 (D664A), qde-3, qip, dcl-2 and a dcl-1 dcl-2 double mutant (dclDKO), were generated in previous studies 22,25. tsnKO (FGSC12632, A; FGSC 12633, a) and trxKO (NCU06059, FGSC13557, a) were obtained from the Fungal Genetic Stock Center. Myc-tagged protein expressing strains, including Myc-His-TRANSLIN, Myc-HIS-TRAX, and Myc-His-TRAX (C473A, C482A) strains, were created in this study. The protein expression constructs were transformed into the indicated Neurospora strains by homologous recombination at the his-3 locus and homokaryon strains were obtained by microconidial isolation 45. The expression of transgenes at the his-3 locus is under the control of the qa-2 inducible promoter 46. The tsnKO and trxKO his-3 recipient strains were created by crossing a tsnKO/trxKO strain to a his-3 wild-type strain (FGSC6103). The double and triple mutants were obtained by crossing the corresponding single or double mutants. The dsal-1 construct was introduced into the tsnKO;qip and trxKO;qip double mutant strains by co-transformation with pBT6 plasmid (bearing the benomyl-resistant beta- tubulin gene).
Liquid cultures were grown in minimal medium (1x Vogel’s, 2% glucose) at 30 °C overnight and then at room temperature with shaking at 130 rpm for 24 hrs 47. For liquid cultures containing QA, 0.01 M QA (pH 5.8) was added to the liquid culture medium containing 1xVogel’s, 0.1% glucose and 0.17% arginine.
Purification and cloning of sRNAs
Small RNAs (~17–30 nt) were purified on 16% PAGE/7 M urea gels from wild-type and tsnko total RNA samples 22. RNAs from regions of interest were eluted from the gels in elution buffer containing 10 mM Tris-Cl (pH7.5), 1 mM EDTA and 0.3 M NaCl. RNAs were precipitated with 20 μg glycogen and 1 volume of isopropanol. RNA was incubated with 1 U/μl Calf Intestinal Phosphatase (NEB) and 1 U/μl SUPERase•In, in Buffer 3 (NEB) at 37°C for 1 hr, extracted twice with phenol and precipitated with ethanol. RNA and two 18-nt and 26-nt synthetic RNA markers were incubated with 10 μM of pre-adenylated DNA oligo (5′-AppCTGTAGGCACCATCAAT/ddC-3′), 1 unit/μl of SUPERase•In, 10% DMSO and 2 unit/μl T4 RNA ligase (Takara Bio Inc., 2050A) in 10 μl ligation buffer (50 mM Tris-Cl pH 7.5, 10 mM MgCl2, 0.06 μg/μl BSA, 10 mM DTT). The 3′-ligated-RNAs were phosphorylated with 1 unit/μl Polynucleotide Kinase in buffer 3 (NEB) containing 1 unit/μl SUPERase•In and 2 mM ATP at 37°C for 1 hr, extracted with phenol, precipitated with ethanol and 5′-ligated with 2 unit/μl T4 RNA ligase and 30 μM 5′ linker 5′-GTT CTA CAG TCC GAC GAT C-3′ in 10 μl 1X buffer containing 1 unit/μl SUPERase•In, 0.1 μg/μl BSA and 10% DMSO. cDNA was synthesized using SuperScript III with RT oligo 5′-ATT GAT GGT GCC TAC AG-3′. The cDNA was PCR-amplified with oligos containing Illumina/Solexa linkers. Deep sequencing was performed on HiScanSQ (Illumina).
All small RNA analyses were performed with custom Perl (5.8.6) scripts on a chromosome genome assembly based on N. crassa assembly 7 48 (available upon request from Michael Fretaig)24, rDNA sequence 23 and Version 3 gene predictions for the N. crassa assembly 7 release including mitochondria sequence (Broad Institute). Gene annotations, structural RNAs and repeats in version 3 gene predictions were mapped and converted to sequence coordinates in the chromosome genome assembly. Small RNA reads with perfect matches were mapped to the genome, mitochondrial DNA sequence and rDNA sequence. A single match returned a unique genomic locus, which was defined by a chromosome number, the start and the end of the matched sequence on the chromosome and its orientation. To calculate small RNA reads derived from a single genomic locus, the small RNA reads were normalized to the total number of matched genomic loci (repeat-normalized reads). The density of small RNA is presented using non-overlapping 10-nt sliding windows along the Watson or Crick strands of each chromosome (Figures 4a and 4b).
Immunopurification of the Myc-TRX, Myc-TRX(C473A) and Myc-QDE-2 ribonucleoprotein complexes was performed as previously described 22.
Northern blot analyses and quantitative RT-PCR assay
Equal amounts of enriched Neurospora small RNA22 (12.5–25 μg) or total RNA of MEFs14 (40μg) from different RNA samples were separated on a 16% denaturing polyacrylamide/7 M urea gels and transferred onto a Hybond-NX membrane (GE Healthcare). An RNA ladder (Ambion) or a mixture of primers of different sizes were used as size markers. Crosslinking of RNA to Hybond-NX was performed using a carbodiimide-mediated cross-linking method at 60°C for 2 hours as described 49, followed by baking at 80°C for 2 hours. For native gel blots, UV cross-linking was used. Hybridization was performed according to the manufacturer’s instructions in ULTRAhyb™ oligo hybridization buffer (Ambion) for StareFire® (IDT) labeled DNA probes or in ULTRAhyb™ hybridization buffer (Ambion) for T7-transcribed riboprobes. The Starfire probe sequences and primer sequences for PCR fragments used in T7 transcribed riboprobes are listed in Supplementary Table 3.
qRT-PCR were performed as previously described 25, and primers used are listed in Supplementary Table 3.
Preparation of recombinant Neurospora C3PO
The cDNAs of Translin and TRAX/TRAX (C473A) were cloned in pETDuet-1 (Duet expression vector, Novagen) with modifications. The expression constructs were transformed into E. coli BL21 (DE3), which were grown in LB medium containing 100ug/ml Ampicillin to OD600 of 0.6 at 37°C, followed by induction of 0.2–0.4 mM IPTG induction. The cells were resuspended in Buffer A (10mM Hepes pH 7.4, 10mM KOAc, 2mM Mg(OAc)2, 5mM β-mercaptoethanol) with freshly added protease inhibitors and 5 mM imidazole, and sonicated on ice. The supernatant was then run through TALON Metal Affinity Resins (Clontech) at 4°C, sequentially washed with Buffer B (Buffer A with 1M NaCl) with 5 mM imidazole, Buffer A with 5 mM imidazole, 10 to 20mM imidazole, and then eluted by 150mM imidazole containing Buffer A. The nC3PO fraction was further purified by a Superose 6 gel filtration column.
Nuclease cleavage assay
The RNA cleavage assay was carried out as previously described 13. Reactions containing recombinant Drosophila C3PO (0.4 to 1.8 μg) and the enriched Neurospora small RNA (25 μg) were incubated at 30 °C for the indicated time.
Measurement of protein translation
Conidia from 5 to 7 day-old cultures were isolated and inoculated into 150-mm Petri dishes containing 1x Vogel’s minimum medium with 2% glucose and were incubated at room temperature for 2 days under constant light. Mycelial discs of 11–13 mm were cut from the mycelial mats, cultured in 1x Vogel’s minimum medium with 2% glucose for 16 hrs at room temperature and then metabolically labeled with 1 μCi/ml EXPRE35S35S protein labeling mix (PerkinElmer) for 30 min. The protein extracts were then prepared as previously described 50,51, and 50 μg of total protein was precipitated by 10% TCA on filter paper 413 (VWR) for 30 min. The filter papers were then washed with 10% TCA twice for 5 to 15 min each and then with 1:1 ethanol/diethyl ether followed drying and a second wash with diethyl ether or 100% ethanol and drying. The dried filter papers were immersed in 5 ml scintillation fluid and 35S signals were counted. For control cultures, the protein synthesis inhibitor cycloheximide (10 μg/ml) was added before the labeling.
Measurement of cell growth
Conidia from 5 to 7-day-old Neurospora slants were inoculated into 150mm petri dishes containing 1xVogel’s minimum medium with 2% glucose and were incubated at room temperature for 2 days under constant light. Mycelial discs of 7 mm were cut from the mycelial mats and cultured in 1xVogel’s minimum medium with 2% glucose at room temperature. Cultures were collected at the indicated time points (Figure 7c) and dried in oven at 80oC for 1 day before the weight of the dried cell mass was determined.
Measurement of sensitivity to phytosphingosine
Measurement of phytosphingosine resistance was performed as previously described with modifications 35. 1×106 conidia/ml were germinated at room temperature shaker for about 5 hours, followed by phytosphingosine (Santa Cruz Biotechnology) treatment (10μg/ml in ethanol) for two hours, with ethanol treatment as the control. Aliquots of the cultures were then diluted and spread onto fresh sorbose-containing plates and incubated at 30°C until colonies appeared, and the colonies were counted and compared with the control to determine the survival rates.
Supplementary Material
Acknowledgments
We thank Haiyan Yuan in Yi Liu’s lab at UT Southwestern Medical Center for technical assistance and Drs. Ying Liu and Xuecheng Ye in Qinghua Liu’s lab for providing the recombinant Drosophila C3PO and for advice on C3PO purification. This work was supported by grants from the National Institutes of Health [R01 GM062591(YL), R01 GM084283(YL), R01GM084010 (QL) and R01GM058800 (CM)] and from the Welch Foundation [I-1560 (YL) and I-1608 (QL)]. Craig Mello is a Howard Hughes Medical Institute Investigator.
Footnotes
Author Contributions
L.L. designed, conducted and interpreted experiments. W.G. performed sRNA sequencing and analyses. C. L. prepared RNA from MEFs. C.M. and Q. L. interpreted experimental results. Y.L. designed and interpreted experiments and wrote the paper.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Jaendling A, McFarlane RJ. Biological roles of translin and translin-associated factor-X: RNA metabolism comes to the fore. Biochem J. 2010;429:225–34. doi: 10.1042/BJ20100273. [DOI] [PubMed] [Google Scholar]
- 2.Jaendling A, Ramayah S, Pryce DW, McFarlane RJ. Functional characterisation of the Schizosaccharomyces pombe homologue of the leukaemia-associated translocation breakpoint binding protein translin and its binding partner, TRAX. Biochim Biophys Acta. 2008;1783:203–13. doi: 10.1016/j.bbamcr.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 3.Li Z, Wu Y, Baraban JM. The Translin/Trax RNA binding complex: clues to function in the nervous system. Biochim Biophys Acta. 2008;1779:479–85. doi: 10.1016/j.bbagrm.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laufman O, Ben Yosef R, Adir N, Manor H. Cloning and characterization of the Schizosaccharomyces pombe homologs of the human protein Translin and the Translin-associated protein TRAX. Nucleic Acids Res. 2005;33:4128–39. doi: 10.1093/nar/gki727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang S, et al. Translin-associated factor X is post-transcriptionally regulated by its partner protein TB-RBP, and both are essential for normal cell proliferation. J Biol Chem. 2004;279:12605–14. doi: 10.1074/jbc.M313133200. [DOI] [PubMed] [Google Scholar]
- 6.Aoki K, et al. A novel gene, Translin, encodes a recombination hotspot binding protein associated with chromosomal translocations. Nat Genet. 1995;10:167–74. doi: 10.1038/ng0695-167. [DOI] [PubMed] [Google Scholar]
- 7.Kasai M, et al. The translin ring specifically recognizes DNA ends at recombination hot spots in the human genome. J Biol Chem. 1997;272:11402–7. doi: 10.1074/jbc.272.17.11402. [DOI] [PubMed] [Google Scholar]
- 8.Wang J, Boja ES, Oubrahim H, Chock PB. Testis brain ribonucleic acid-binding protein/translin possesses both single-stranded and double-stranded ribonuclease activities. Biochemistry. 2004;43:13424–31. doi: 10.1021/bi048847l. [DOI] [PubMed] [Google Scholar]
- 9.Suseendranathan K, et al. Expression pattern of Drosophila translin and behavioral analyses of the mutant. Eur J Cell Biol. 2007;86:173–86. doi: 10.1016/j.ejcb.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 10.Chennathukuzhi V, et al. Mice deficient for testis-brain RNA-binding protein exhibit a coordinate loss of TRAX, reduced fertility, altered gene expression in the brain, and behavioral changes. Mol Cell Biol. 2003;23:6419–34. doi: 10.1128/MCB.23.18.6419-6434.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stein JM, et al. Behavioral and neurochemical alterations in mice lacking the RNA-binding protein translin. J Neurosci. 2006;26:2184–96. doi: 10.1523/JNEUROSCI.4437-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu XQ, Xu L, Hecht NB. Dimerization of the testis brain RNA-binding protein (translin) is mediated through its C-terminus and is required for DNA- and RNA-binding. Nucleic Acids Res. 1998;26:1675–80. doi: 10.1093/nar/26.7.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y, et al. C3PO, an endoribonuclease that promotes RNAi by facilitating RISC activation. Science. 2009;325:750–3. doi: 10.1126/science.1176325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye X, et al. Structure of C3PO and mechanism of human RISC activation. Nat Struct Mol Biol. 2011;18:650–7. doi: 10.1038/nsmb.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buhler M, Moazed D. Transcription and RNAi in heterochromatic gene silencing. Nat Struct Mol Biol. 2007;14:1041–1048. doi: 10.1038/nsmb1315. [DOI] [PubMed] [Google Scholar]
- 16.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Q, Paroo Z. Biochemical principles of small RNA pathways. Annu Rev Biochem. 2010;79:295–319. doi: 10.1146/annurev.biochem.052208.151733. [DOI] [PubMed] [Google Scholar]
- 18.Siomi H, Siomi MC. On the road to reading the RNA-interference code. Nature. 2009;457:396–404. doi: 10.1038/nature07754. [DOI] [PubMed] [Google Scholar]
- 19.Tian Y, et al. Multimeric assembly and biochemical characterization of the Trax-translin endonuclease complex. Nat Struct Mol Biol. 2011;18:658–64. doi: 10.1038/nsmb.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Catalanotto C, Nolan T, Cogoni C. Homology effects in Neurospora crassa. FEMS Microbiol Lett. 2006;254:182–9. doi: 10.1111/j.1574-6968.2005.00037.x. [DOI] [PubMed] [Google Scholar]
- 21.Li L, Chang SS, Liu Y. RNA interference pathways in filamentous fungi. Cell Mol Life Sci. 2010;67:3849–63. doi: 10.1007/s00018-010-0471-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maiti M, Lee HC, Liu Y. QIP, a putative exonuclease, interacts with the Neurospora Argonaute protein and facilitates conversion of duplex siRNA into single strands. Genes Dev. 2007;21:590–600. doi: 10.1101/gad.1497607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee HC, et al. qiRNA is a new type of small interfering RNA induced by DNA damage. Nature. 2009;459:274–7. doi: 10.1038/nature08041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee HC, et al. Diverse pathways generate microRNA-like RNAs and Dicer-independent small interfering RNAs in fungi. Mol Cell. 2010;38:803–14. doi: 10.1016/j.molcel.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choudhary S, et al. A double-stranded-RNA response program important for RNA interference efficiency. Mol Cell Biol. 2007;27:3995–4005. doi: 10.1128/MCB.00186-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colot HV, et al. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci U S A. 2006;103:10352–7. doi: 10.1073/pnas.0601456103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romano N, Macino G. Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol Microbiol. 1992;6:3343–53. doi: 10.1111/j.1365-2958.1992.tb02202.x. [DOI] [PubMed] [Google Scholar]
- 28.Hartmann RK, Gossringer M, Spath B, Fischer S, Marchfelder A. The making of tRNAs and more - RNase P and tRNase Z. Prog Mol Biol Transl Sci. 2009;85:319–68. doi: 10.1016/S0079-6603(08)00808-8. [DOI] [PubMed] [Google Scholar]
- 29.Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev. 2010;24:1832–60. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walker SC, Engelke DR. Ribonuclease P: the evolution of an ancient RNA enzyme. Crit Rev Biochem Mol Biol. 2006;41:77–102. doi: 10.1080/10409230600602634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Catalanotto C, et al. Redundancy of the two dicer genes in transgene-induced posttranscriptional gene silencing in Neurospora crassa. Mol Cell Biol. 2004;24:2536–45. doi: 10.1128/MCB.24.6.2536-2545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cogoni C, Macino G. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature. 1999;399:166–9. doi: 10.1038/20215. [DOI] [PubMed] [Google Scholar]
- 33.Lee HC, et al. The DNA/RNA-dependent RNA polymerase QDE-1 generates aberrant RNA and dsRNA for RNAi in a process requiring replication protein A and a DNA helicase. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shiu PK, Raju NB, Zickler D, Metzenberg RL. Meiotic silencing by unpaired DNA. Cell. 2001;107:905–16. doi: 10.1016/s0092-8674(01)00609-2. [DOI] [PubMed] [Google Scholar]
- 35.Castro A, Lemos C, Falcao A, Glass NL, Videira A. Increased resistance of complex I mutants to phytosphingosine-induced programmed cell death. J Biol Chem. 2008;283:19314–21. doi: 10.1074/jbc.M802112200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zasloff M, Santos T, Romeo P, Rosenberg M. Transcription and precursor processing of normal and mutant human tRNAiMet genes in a homologous cell-free system. J Biol Chem. 1982;257:7857–63. [PubMed] [Google Scholar]
- 37.Nashimoto M, Wesemann DR, Geary S, Tamura M, Kaspar RL. Long 5′ leaders inhibit removal of a 3′ trailer from a precursor tRNA by mammalian tRNA 3′ processing endoribonuclease. Nucleic Acids Res. 1999;27:2770–6. doi: 10.1093/nar/27.13.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Galagan JE, et al. The genome sequence of the filamentous fungus Neurospora crassa. Nature. 2003;422:859–68. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
- 39.Randau L, Schroder I, Soll D. Life without RNase P. Nature. 2008;453:120–3. doi: 10.1038/nature06833. [DOI] [PubMed] [Google Scholar]
- 40.Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–49. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nunes CC, et al. Diverse and tissue-enriched small RNAs in the plant pathogenic fungus, Magnaporthe oryzae. BMC Genomics. 2011;12:288. doi: 10.1186/1471-2164-12-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cole C, et al. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA. 2009;15:2147–60. doi: 10.1261/rna.1738409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell. 2011;43:613–23. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ishida R, et al. A role for the octameric ring protein, Translin, in mitotic cell division. FEBS Lett. 2002;525:105–10. doi: 10.1016/s0014-5793(02)03095-8. [DOI] [PubMed] [Google Scholar]
- 45.Ebbole D, Sachs MS. A rapid and simple method for isolation of Neurospora crassa homokaryons using microconidia. Fung Gen Newsl. 1990;37:17–18. [Google Scholar]
- 46.He Q, Cheng P, He Q, Liu Y. The COP9 signalosome regulates the Neurospora circadian clock by controlling the stability of the SCFFWD-1 complex. Genes & Dev. 2005;19:1518–1531. doi: 10.1101/gad.1322205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davis RL, deSerres D. Genetic and microbial research techniques for Neurospora crassa. Meth Enzymol. 1970;27A:79–143. [Google Scholar]
- 48.Lewis ZA, et al. Relics of repeat-induced point mutation direct heterochromatin formation in Neurospora crassa. Genome Res. 2009;19:427–37. doi: 10.1101/gr.086231.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pall GS, Codony-Servat C, Byrne J, Ritchie L, Hamilton A. Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by northern blot. Nucleic Acids Res. 2007;35:e60. doi: 10.1093/nar/gkm112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheng P, Yang Y, Heintzen C, Liu Y. Coiled-coil domain mediated FRQ-FRQ interaction is essential for its circadian clock function in Neurospora. EMBO J. 2001;20:101–108. doi: 10.1093/emboj/20.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garceau N, Liu Y, Loros JJ, Dunlap JC. Alternative initiation of translation and time-specific phosphorylation yield multiple forms of the essential clock protein FREQUENCY. Cell. 1997;89:469– 476. doi: 10.1016/s0092-8674(00)80227-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.