Abstract
Par-4 is a pro-apoptotic, tumor suppressor protein that induces apoptosis selectively in cancer cells. Endoplasmic reticulum-stress and higher levels of protein kinase A in tumor cells confer the coveted feature of cancer selective response to extracellular and intracellular Par-4, respectively. Recent studies have shown that systemic Par-4 confers resistance to tumor growth in mice, and that tumor-resistance is transferable by bone marrow transplantation. Moreover, recombinant Par-4 inhibits the growth of tumors in mice. As systemic Par-4 induces apoptosis via cell surface GRP78, strategies that promote GRP78 trafficking to the cell surface are expected sensitize cancer cells to circulating levels of Par-4. This review illustrates the domains and mechanisms by which Par-4 orchestrates the apoptotic process in both cell culture models and in physiological settings.
Keywords: Par-4, GRP78, Apoptosis
Introduction
The tumor suppressor protein, prostate apoptosis response-4 (Par-4) was originally discovered in rat prostate cancer cells, and is now known to be ubiquitously expressed in myriad tissues across different species (El-Guendy and Rangnekar, 2003). Par-4 can selectively cause apoptosis in a wide variety of cancers, leaving normal cells unaffected. This selective nature of Par-4 makes it an attractive therapeutic option. Since its identification, Par-4 function has been investigated in diverse tumors to tease out its immediate targets and the precise mechanism underlying its ability to induce apoptosis. This review illustrates the domains and mechanisms by which Par-4 orchestrates the apoptotic process and confers host resistance to the growth of tumors.
Domains of Par-4
The Par-4 gene was first identified as an immediate early apoptotic gene in prostate cancer cells. This discovery was part of an effort involving differential screening to find pro-apoptotic genes that were induced in response to Ca2+ elevation in cells. It was also found that androgen ablation was equally effective in inducing Par-4 expression in androgen-dependent cells of the prostate (Sells et al., 1994). The Par-4 gene is located on an unstable region 12q21 of the human chromosome that is often deleted in gastric and pancreatic cancer (Johnstone et al., 1998). Par-4 is known to be down-regulated in over 70% of renal cancers (Cook et al., 1999), neuroblastoma (Kogel et al., 2001), acute and chronic forms of leukemia (Boehrer et al., 2001). Oncogenes like Ras, Raf and Src have been found to down-regulate Par-4 along with other proteins to promote cellular transformation; nevertheless, restoration of Par-4 in the cells inhibits the neoplastic progression (Qiu et al., 1999).
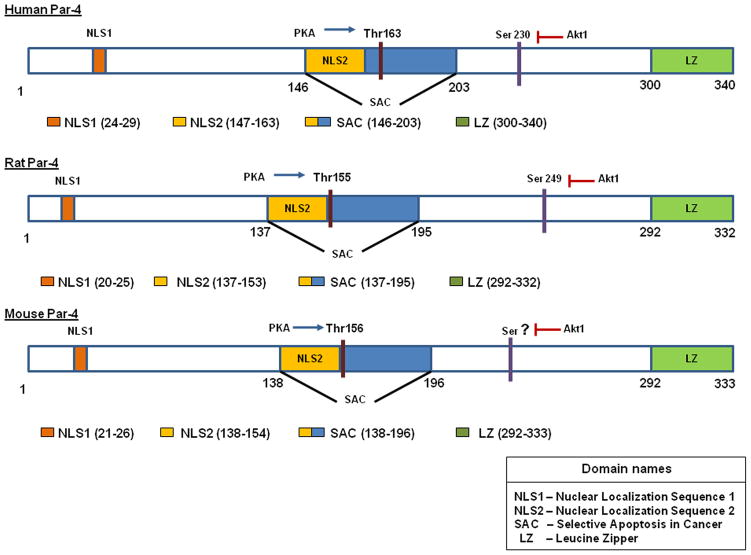
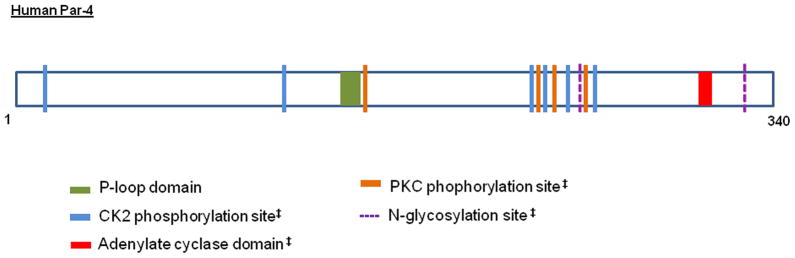
The protein encoded by the Par-4 gene is a multi-domain protein comprising of 343 amino acids. The key domains that are 100% conserved across the human, mouse and rat genomes are the leucine zipper domain (LZ) at the carboxyl terminal region, two nuclear localization sequences (NLS1,NLS2) and a nuclear export sequence (El-Guendy et al., 2003). Par-4 contains the SAC (Selective for Apoptosis of Cancer Cells) domain, which is unique to Par-4 and is also 100% conserved in rat, mouse and humans (Figure 1). Although the functions of some of the domains have been partially elucidated, there are other domains in Par-4 that have no known function in the context of Par-4. These include the casein kinase 2 (CK2) phosphorylation site, an ATP-GTP binding motif, protein kinase C (PKC) phosphorylation site, also sites for N-glycosylation and N-myristoylation (Figure 2) (http://expasy.org/tools/). These domains may collectively contribute to the activity of Par-4 and may also have a role in protein conformation.
Figure 1. Functional domains of Par-4.
Functionally active domains of Par-4 that are conserved across the human, rat and mouse species are depicted. The numbers depict their relative amino acid positions.
Figure 2. Putative domains of human Par-4.
Putative domains of human Par-4 with functions yet to be confirmed experimentally are shown. Domains marked by ‡ are conserved across the human, mouse and rat species.
Par-4 exhibits two nuclear localization sequences NLS1 and NLS2 at the N-terminal region. These domains are fully conserved across rat, mouse and human Par-4. The presence of a nuclear localization sequence led to the belief that Par-4 must have a functional role in the nucleus. It is now known that NLS2, a bipartite sequence, is essential for the translocation of Par-4 from the cytoplasm to the nucleus, and is necessary to cause apoptosis (El-Guendy and Rangnekar, 2003). This was confirmed using a Par-4 deletion mutant lacking the NLS2 sequence that was unable to translocate to the nucleus, thereby losing its ability to block NF-κB activity and induce apoptosis. The NLS1 sequence is relatively shorter in length (six amino acids long), and has no known function assigned thus far. Experiments with ΔNLS1 mutant of Par-4 had no effect on its ability to translocate to the nucleus or induce apoptosis (El-Guendy et al., 2003). These findings suggest that although NLS1 is evolutionarily conserved, its function may be eclipsed by the NLS2 domain or it may have lost its functional relevance.
A critical domain found at the carboxyl terminal region of Par-4 is the leucine zipper domain (LZ). Par-4 contains five leucine repeats at its carboxyl terminus. The primary function of a leucine zipper domain is to allow protein-protein interactions. The leucine zipper domain contains about 40 amino acids found in the form of heptad repeats, where the fourth position of each repeat is occupied by a leucine (El-Guendy and Rangnekar, 2003). It forms a scissor-like structure made of parallel coiled coil α-helices that are held in place by hydrophobic interactions. These helices can form hetero- or homo-dimers, which diverge and can bind the major groove of the DNA using a region of adjacent basic residues (Miller, 2009). Par-4 is found to homo- and hetero-dimerize with almost all of its binding partners through its leucine zipper domain. Such partner proteins include the Wilm’s tumor protein WT1 (Johnstone et al., 1996), Dlk and ζPKC (Diaz-Meco et al., 1996; Kogel et al., 1998). Phosphorylation and inactivation of Par-4 by AKT1 is also facilitated by the binding of AKT1 to the LZ domain (Goswami et al., 2005).
One of the domains designated SAC, is both unique and indispensible for the pro-apoptotic activity of Par-4. SAC is a core domain that is 59 amino acids in length. It was identified in our laboratory by serially deleting the Par-4 sequence from both the amino-and carboxyl-ends and constitutes the fundamental pro-apoptotic unit of Par-4. The SAC domain includes the NLS2 domain and the Thr residue, which is the target for PKA phosphorylation. The SAC domain gives Par-4 the ability to selectively kill cancer cells, while leaving normal cells unaffected (Zhao and Rangnekar, 2008). When PKA activity was up-regulated in normal cells using stimulants like cAMP or through ectopic over-expression of the catalytic subunit of PKA, it resulted in sensitization of the cells to SAC mediated apoptosis (Gurumurthy et al., 2005). The cancer selectivity of SAC and Par-4 was due to the higher endogenous PKA levels exhibited by cancer cells. The ability of SAC to cause apoptosis extends to in vivo conditions as well. Intra-tumoral injection of an adenoviral construct expressing SAC resulted in significant reduction in tumor volume. This effect was seen in xenografts derived from Par-4-resistant as well as sensitive cells (Zhao and Rangnekar, 2008)
Intracellular Par-4 mediated apoptosis
Depending on the nature of stimulus, apoptosis can occur via two different pathways, extrinsic and intrinsic. The extrinsic pathway involves binding of extracellular ligands such as Fas/FasL and TNF to the corresponding death receptors. The intrinsic pathway or the mitochondrial pathway on the other hand, is activated by intracellular stimuli such as oxidative stress, endoplasmic reticulum (ER) stress, hypoxia, DNA damage. Although the two pathways are activated by different mechanisms, they are interlinked by common signaling factors like NF-κB and proteolytic enzymes like caspases (Burz et al., 2009; Sayers, 2011). Apoptosis by Par-4 involves an intracellular, as well as an extrinsic mechanism. Intracellular Par-4 is phosphorylated by protein kinase A (PKA) at a specific T155 residue found in the SAC (selective for apoptosis induction in cancer cells) domain of Par-4 (Figure 1). The phosphorylation of the Thr155 residue allows trafficking of the Fas/FasL to the plasma membrane. The Fas/FasL interacts with FADD causing activation of the Fas/FasL-FADD-Caspase 8 apoptotic death pathway.
One of the features that make Par-4 unique among most tumor suppressors is its ability to induce apoptosis selectively in cancer cells. This ability to selectively cause apoptosis in cancer cells has been attributed to the difference in PKA levels between normal and cancer cells (El-Guendy et al., 2003; Gurumurthy et al., 2005). PKA is a heterotrimeric holoenzyme which activates a number of pro-survival pathways like the Raf/MEK/ERK or androgen receptor (AR) pathway. Studies have shown the PKA levels are often elevated in a variety of cancers (Merkle and Hoffmann, 2011; Tortora and Ciardiello, 2002). The basal level of PKA in normal cells is insufficient to cause T155 phosphorylation, thereby making normal cells resistant to Par-4 mediated apoptosis. It is also seen that endogenous level of Par-4 does not by itself cause apoptosis, unless accompanied by another stimulus. However ectopic Par-4 expression kills cancer cells quite effectively.
Par-4 also causes inhibition of NF-κB transcription function. NF-κB is an important cytokine-induced transcriptional regulator that generally promotes cell survival. NF-κB is known to be up-regulated in a wide variety of cancers. Par-4 modulates NF-κB function in a number ways, both in the cytoplasm as well as in the nucleus. Moreover, in the cytoplasm Par-4 blocks the activity ofζPKC, an atypical PKC which activates NF-κB and AP-1, thereby promoting cell survival. Par-4 is able to prevent the ζPKC–mediated phosphorylation of IκB that is crucial for the nuclear translocation and activity of NF-κB (Diaz-Meco et al., 1999). NF-κB inducers often act through reactive oxygen intermediates/reactive oxygen species (ROI/ROS). The resulting activation and nuclear translocation of NF-κB leads to transcription of genes encoding inflammatory proteins, as well as cell survival proteins (LaCasse et al., 1998; Lee and Burckart, 1998; Kim et al., 2006).
The role of Par-4 in blocking the NF-κB inflammatory pathway is not known, however there is evidence showing that inflammatory factors can promote cell survival and tumorigenesis. It is also known that cancer cells exhibit high levels of oxidative stress resulting from elevated levels (Valko et al., 2006). Hence, it would be interesting to study if ectopic Par-4 can effectively induce apoptosis in cells with high levels of ROI/ROS. If such an effect is observed, it could provide therapeutic benefit by preventing tumor formation in patients with diminished activity of antioxidant enzymes.
In addition to sending the Fas/FasL to the cell membrane, the T155 phosphorylation promotes the translocation of Par-4 from the cytoplasmic compartment to the nucleus. Entry into the nuclear compartment allows Par-4 to inhibit the transcriptional activity of NF-κB. Par-4 contains a nuclear localization sequence NLS2, which is critical for its translocation into the nuclear compartment (El-Guendy et al., 2003). PKA phosphorylation facilitates this process but is not enough to send Par-4 into the nucleus; the precise regulation of the nuclear transport remains to uncovered. Although we know that nuclear Par-4 can inhibit the transcription activity of NF-κB, the nature of its interaction with the NF-κB transcription machinery has not been uncovered. There could be a number of possibilities involving interactions with co-repressors or by direct DNA binding by which Par-4 could block NF-κB activity in the nucleus.
Par-4 is known to down-regulate transcription by enhancing the repression activity of the tumor suppressor protein Wilms’ Tumor 1 (WT1). WT1 is known to repress cell proliferation in several cancers (Johnstone et al., 1996). As WT1 is expressed in a tissue-specific manner, repression in those tissues may occur via an NF-κB dependent mechanism. Further studies are necessary to explore the possibility that Par-4 blocks NF-κB activity through WT1 in the nucleus. Par-4 is also known to sequester topoisomerase 1 (TOP1), thereby attenuating its ability to relax supercoiled DNA. Sequestration of TOP1 prevents DNA unwinding, thus making the DNA template inaccessible to transcription factors (Goswami et al., 2008). TOP1 sequestration by Par-4 may also result in inhibition of the transcriptional activity of NF-κB. The combined effect of Fas/FasL translocation and the down-regulation of NF-κB activity is essential for the pro apoptotic action of intracellular Par-4. Blocking either of these processes leads to increased resistance to apoptosis by Par-4.
Par-4 secretion and the role of GRP78
The extrinsic effect of Par-4 is a relatively new yet significant finding. It was discovered that Par-4 is secreted by normal and cancer cells. Cell culture experiments using Brefeldin A (BFA), an antiviral antibiotic that blocks protein trafficking from the ER to the Golgi complex (Fujiwara et al., 1988), showed that secretion of Par-4 occurs through the classical ER-Golgi pathway (Burikhanov et al., 2009). Treatment of cells with ER stress inducers greatly increased Par-4 secretion. Both, normal and cancer cells are able to secrete Par-4 however only cancer cells are susceptible to it pro-apoptotic effects (Burikhanov et al., 2009). This effect of Par-4 was attributed to a protein called glucose-regulated protein 78 (GRP78), which under normal conditions is an endoplasmic reticulum (ER) resident protein which functions as a chaperone involved in protein folding and a regulator of ER stress signaling (Lee, 2007).
Under conditions of ER stress, intracellular Par-4 binds to GRP78 and facilitates its translocation from the ER to the plasma membrane, where GRP78 acts as a receptor for Par-4 at the cell surface. Once it is extracellular, Par-4 binds to GRP78 and uses FADD as the adaptor to recruit caspase-8 to the membrane. Activated caspase-8 then triggers the basic apoptotic machinery involving caspase 3 and other downstream effector proteins (Shrestha-Bhattarai and Rangnekar, 2010).
Tumor Resistance conferred by Par-4
In addition to its effects in cell culture, Par-4 is also active in mouse models. Par-4 null mice are prone to tumor development and there is a much higher incidence of prostate hyperplasia in male mice. Par-4 knockout mice that are homozygous or heterozygous for the gene develop spontaneous tumors and are also highly sensitive to carcinogen induced tumors in a number of different tissues (Garcia-Cao et al., 2005). Consistent with its role as a tumor suppressor, down-regulation and silencing of Par-4 has been observed in a number of cancers due to promoter hypermethylation. There are studies showing a point mutation in the SAC domain that leads to premature termination of Par-4 in endometrial cancers (Moreno-Bueno et al., 2007). This evidence underscores the importance of Par-4 as a tumor suppressor protein and its physiological impact on cancer development and progression.
The SAC domain of Par-4 is responsible for its ability to selectively cancer kill cancer cells (El-Guendy et al., 2003). Over-expression of SAC domain induces apoptosis in a variety of cancer cell lines and xenografts, including those which were previously resistant to apoptosis by full length Par-4 (Zhao et al., 2007). The selectivity towards cancer versus normal cells arose from the fact that cancer cells exhibit high PKA levels leading to increased Thr155 phosphorylation necessary for SAC activation (Zhao and Rangnekar, 2008). Lack of the leucine zipper domain meant that SAC could escape binding to Akt1 and consequently inhibition by Akt1 phosphorylation, which explained the increased ability of SAC to kill cells that were otherwise resistant to full length Par-4 (Goswami et al., 2005).
Researchers in the field of cancer therapy are constantly in search of treatments that can target cancer cells and leave the normal cells unaffected. This much desired trait made the discovery of SAC particularly significant, but also posed the question as to whether this effect could be observed in vivo, which lead to the development of the SAC transgenic mouse (Zhao et al., 2007). The mouse helped establish that SAC is well tolerated by the animal and that it did not affect developmental processes. This was important because apoptosis is a crucial part of normal development. The mice were fertile and had a normal lifespan, indicating that the ubiquitously expressed SAC domain did not target normal tissues.
Upon testing the mouse embryonic fibroblasts (MEFs) for oncogenic transformation, the control MEFs transduced with oncogenic Ras or c-Myc adenovirus developed significantly higher number of foci compared to the SAC MEFs. Also the SAC MEFs showed a 10 fold increase in apoptosis compared to the control MEFs post infection with Ras or c-Myc adenovirus (Zhao et al., 2007). The littermate controls showed enlarged and disorganized architecture in several tissues and a high incidence of hepatocarcinomas (liver) and lymphomas (spleen). On the other hand, the SAC transgenic mice showed increased resistance to the development of spontaneous tumors and had a normal preserved tissue architecture (Zhao et al., 2007).
SAC domain transgenic mice were also resistant to oncogene-induced tumors (Zhao et al., 2007). Male TRAMP (transgenic adenocarcinoma of the mouse prostate) mice develop uniform spontaneous autochthonous prostate tumors that mimic the pathogenesis of human prostate cancer (Hurwitz et al., 2001). Crosses between the tumor resistant SAC domain mice and the tumor prone TRAMP mice provided strong evidence supporting the ability of the SAC domain to impart tumor resistance. All of the SAC−/−/TRAMP+/− control mice developed adenocarcinoma of the prostate in 6 months, while only ~25% of the SAC+/−/TRAMP+/− mice progressed to develop adenocarcinoma (Zhao et al., 2007). Moreover, in these SAC+/−/TRAMP+/− mice that developed adenocarcinoma, there were prostatic intraepithelial neoplasia (PIN) areas that contained pockets of loss of the SAC-transgene. Accordingly, all of the tumors from these mice showed loss of the SAC-transgenic. These findings indicate that the SAC domain functioned as a tumor suppressor, which had to be eliminated for adenocarcinoma development in mice.
Extracellular Par-4/SAC induces apoptosis by binding to GRP78 at the cell surface and activating the caspase-8/caspase-3 pathway (Burikhanov et al., 2009). The functional importance of this finding was supported by the observation that SAC transgenic protein is secreted in the serum of the transgenic mouse. This secreted SAC is found systemically and retains its activity. The conditioned medium from the SAC MEFs was able to kill a wide range of cancer cells leaving normal cells unaffected. Notably, this activity was seen in the mouse model as well. Injection of aggressive Lewis lung carcinoma1 (LLC1) cells into the flanks of the control mice resulted in tumor growth but the SAC-transgenic mice were highly resistant to the growth of tumors (Zhao et al., 2011). This emphasized the relevance of the secreted SAC domain and it systemic activity outside the cells.
A decisive experiment, which marked the functional and possibly clinical relevance of secreted SAC, was the ability to transfer its activity to another animal by bone marrow transplantation. The bone marrow of SAC-GFP transgenic or control GFP transgenic mice was transplanted into the littermate control mice. It was quite exciting to see that the serum from the recipient animals transplanted with SAC-GFP bone marrow was able to successfully induce apoptosis in LLC1 cells in culture, while the serum from control mice had no significant effect (Zhao et al., 2011). Our laboratory has also reported that recombinant TRX-Par-4 and TRX-SAC can selective cause cancer cell apoptosis in cell culture models and can also significantly inhibit lung metastasis in vivo (Burikhanov et al., 2009; Zhao et al., 2011). This finding could have a profound clinical impact because it shows that the pro-apoptotic activity of Par-4/SAC is not only systemic in nature but can also be transferred to patients lacking the required levels of the protein. Importantly, the bone marrow transplantation experiment indicated that Par-4/SAC is stable and functionally active for over 4 months in mice. This implies that Par-4/SAC could be produced as recombinant proteins using bacteria in a manner similar to insulin (Keefer et al., 1981), and could be used in cancer patients as an injectable therapy. Although the results in mouse models do not always translate to humans, the 100% homology of the SAC domain in mice and humans does significantly improve the chances of success.
Role of dietary compounds in Par-4 mediated apoptosis
Dietary agents containing bioactive compounds such as genistein, curcumin, and resveratrol, are known to have pro-apoptotic, antioxidant and anti-inflammatory properties. Combinations of some dietary compounds such as curcumin with standard chemotherapy drugs have shown synergistic effects in clinical trials (Khan et al., 2010). These compounds are found in natural food products, and exhibit very low toxicity in patients (Patel et al., 2010). A Phase IIa clinical trial showed that systemically administered curcumin caused significant reduction in aberrant crypt foci (ACF) of the colon that serves as marker for the assessment of the potency of anti-cancer agents (Carroll et al., 2011). Curcumin is known to cause radiosensitization of prostate cancer cells and squamous cell carcinoma cells coupled with radioprotective effects in normal cells (Nambiar et al., 2011). Preliminary studies in our laboratory showed that treatment with curcumin resulted in both, enhanced secretion of Par-4 and increased sensitivity to Par-4 mediated apoptosis (Bhattarai and Burikhanov, data not shown). Flow cytometry analysis revealed that cells treated with curcumin exhibit elevated levels of cell surface GRP78 compared to vehicle treated cells (Bhattarai and Burikhanov, data not shown), which might explain the sensitization to Par-4. These observations suggest the synergistic nature of the combination of dietary compounds and Par-4 based therapy for cancer. Based on these initial observations, we are currently investigating the combinatorial effects of Par-4 and other dietary compounds that have shown promise in clinical trials.
Future perspectives
Par-4 is often down-regulated in cancer, yet the exact nature of it regulation at the transcriptional or translational level is understudied. The sequence of Par-4 contains a number of phosphorylation sites for cellular kinases that are known to play a role in cancer cell survival and disease progression. Casein kinase 2 (CK2) is one such enzyme that can potentially phosphorylate human Par-4 at six different sites. CK2, a serine/threonine protein kinase, is known to be elevated in a number of cancers and its over-expression is associated with reduced patient survival. CK2 is known to have a pro-survival and antiapoptotic effect and can make cells more metastatic (Ruzzene and Pinna, 2010). There is no evidence suggesting the negative regulation of Par-4 by CK2, however, there is a likelihood that elevated CK2 levels might result in Par-4 phosphorylation thereby inactivating it. If this hypothesis holds true, it can have valuable therapeutic implications because some CK2 inhibitors have shown promising anti-proliferative effects and are already undergoing clinical trials (Meek and Cox, 2011). It would also help to better understand the post translational regulation of Par-4 in cancer.
Par-4 is known to be a tumor suppressor protein that selectively induces apoptosis in cancer cells. However, Par-4 contains a conserved ‘GGDEF’ motif (Figure 2), which is intriguingly homologous to the adenylyl cyclase domain (Pei and Grishin, 2001). Adenylyl cyclases are enzyme that convert adenosine triphosphate (ATP) to cyclic adenosine -3′,5′-monophosphate (cAMP), which acts as a second messenger (Pavan et al., 2009). cAMP can regulate a number downstream processes through the activation of PKA that is known to phosphorylate Par-4 at the T155 site thereby activating it. This poses the question about the function of Par-4 as an adenylyl cyclase, and the ability to activate itself in tumor cells. What makes this question even more compelling is the observation that Par-4 possesses a phosphate binding loop domain (P-loop domain with the sequence GXXXXGKS), which is commonly found in proteins that bind nucleotides. It is known to interact with the phosphate groups of nucleotides and the Mg2+ (Priya et al., 2011). This domain is found in human Par-4 but is not conserved in the rat and mouse species, suggesting that it might have been acquired during the course of evolution. Could the acquisition of this P-loop domain indicate that Par-4 plays additional roles other than tumor suppression in humans? The P-loop sequence is followed by a DxxGQ motif that is known to stabilize the bound nucleotide. Thus, Par-4 possesses all the elements that would be essential for the making of an adenylyl cyclase-like protein; this observation requires functional validation.
The activity of a number of Ras-like GTPases and kinases such as Abl is known to depend on the P-loop domain. Point mutations in this domain are known to make chronic myeloid leukemia (CML) cells more oncogenic and also resistant to the BCR-ABL inhibitor Imatinib (Cang and Liu, 2008). Non small cell lung carcinoma (NSCLC) tumors bearing point mutations in the P-loop domain within the epidermal growth factor receptor (EGFR) are known to become more aggressive. However, patients with these mutations seem to respond better to tyrosine kinase inhibitor (TKI) therapy as compared to those without the mutations (Yoshikawa et al., 2012). Although these observations clearly underscore the importance of the P-loop domain in cancer progression and therapy, it must be noted that most of the evidence available is directed toward proteins that promote cell survival and cancer progression. Although these observations clearly underscore the importance of the P-loop domain in cancer progression and therapy, it must be noted that most of the evidence available is directed toward proteins that promote cell survival and cancer progression. Investigating the role of the P-loop domain in a pro-apoptotic scenario could provide new insights into the regulation of Par-4.
Besides recombinant Par-4/SAC made in bacteria, Par-4/SAC prepared from mammalian cells in order to preserve the posttranslational modifications in Par-4/SAC, could provide an effective way to battle cancer. The normal development, lifespan and fertility exhibited by the SAC transgenic mice indicate that secreted Par4/SAC is well tolerated by the animals and there is no apparent toxicity associated with its systemic expression and release. One the major problems encountered while considering recombinant proteins as a therapeutic option is the delivery, stability and bioavailabilty of the protein if interest. It would be interesting to explore new delivery options like nanobins, which have shown to increase the potency of the anticancer drugs (Lee et al., 2009). Such nanotechnology based systems could protect Par-4/SAC from degradation and effectively deliver it to the site of the tumor. Moreover, monotherapy is rarely encountered during cancer treatment. Usually treatment involves a combination of drugs as well as treatment modalities. Combining Par-4/SAC based treatment with standard chemoptherapy drugs like cisplatin has shown promise. A recent report indicated that cisplatin causes apoptosis in a Par-4-dependent manner and that Par-4 over-expression sensitizes cells to cisplatin-induced apoptosis (Chaudhry et al., 2012).
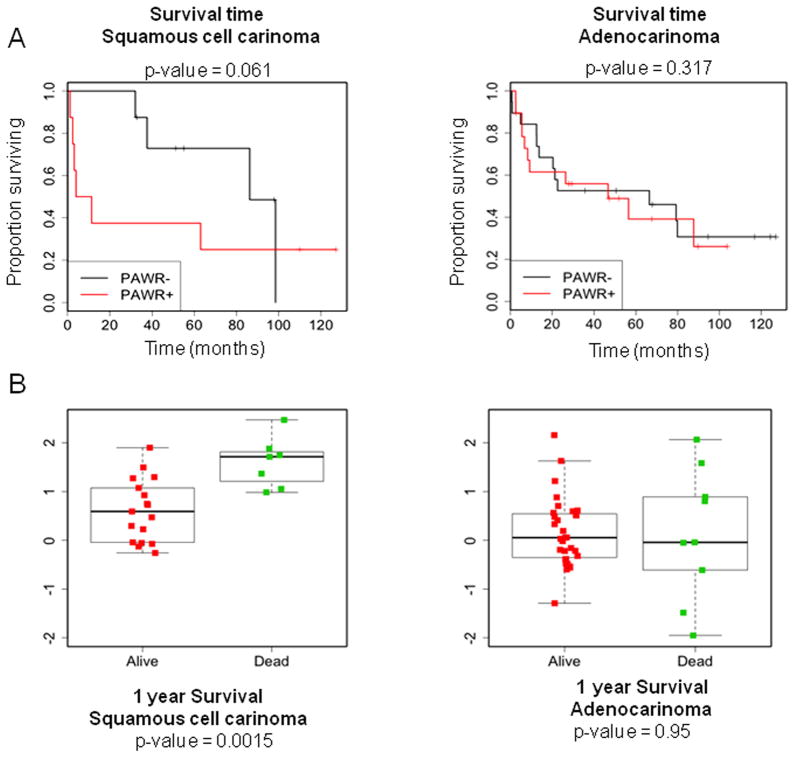
Data mining, a relatively new but very useful technology, involves compilation of data from a wide range of sources and using it to observe the patterns which develop and analyze them to yield useful information. Oncomine is one such cancer microarray database and data mining platform developed specifically for cancer research (Rhodes et al., 2004). Recently, we performed a statistical study using multiple data sets (obtained from Oncomine) on Par-4 mRNA levels in tumor and normal lung tissues. This study revealed some intriguing findings that might seem counter-intuitive to the expected outcome. The data showed that patients with squamous cell lung carcinoma had increased Par-4 mRNA levels which correlated with decreased overall survival in patients (Figure 3). However, high levels of Par-4 mRNA in patients with lung adenocarcinoma showed no such effect on overall survival, indicating that this observation might be pertinent only to specific types of cancer and may reflect the effect of downstream apoptosis-resistance mechanisms that are elevated in cancer.
Figure 3. Relationship between mRNA levels of Par-4 and patient survival.
(A) Oncomine anaylsis indicated increased Par-4 mRNA levels correlate with decreased survival in squamous cell lung carcinoma patients (logrank P = 0.061). Survival time in patients with lung adenocarcinoma shows no significant correlation with mRNA levels of Par-4 (logrank P = 0.94).
(B) A composite of multiple data sets from Oncomine depicting 1 year survival status in patients with squamous cell carcinoma and adenocarcinoma of the lung. The colored squares represent data from different studies.
A multitude of reports have underlined the therapeutic value of Par-4. However much work remains to be done on using Par-4 or target its receptor GRP78. The interaction of Par-4 with GRP78 at the cell surface provides an attractive model for the development of small molecules or monoclonal antibodies that can target GRP78 and elicit a Par-4-like apoptotic response. A bio-available formulation of 3,3 diindolylmethane (B-DIM), a chemopreventive agent currently used to treat recurring respiratory papillomatosis, was shown to induce Par-4 expression and apoptosis in cancer cells. Treatment with DIM also increased sensitivity to standard chemotherapeutic drugs like cisplatin and gemcitabine (Azmi et al., 2008). Using FDA approved chemotherapeutic drugs to stimulate GRP78 translocation to the cell surface in combination with agents like B-DIM would be another way to effectively sensitize the tumors to circulating levels of Par-4. These alternatives need to be developed and tested in mouse models to better characterize the interaction of Par-4 with GRP78 and provide new targeted anti-cancer treatments.
Acknowledgments
This study was supported by KLCR grant, and NIH/NCI grant CA060872 (to VMR). We thank Li, University of Kentucky, for the Oncomine analysis, and Tripti Shrestha-Bhattarai, University of Kentucky, for a critical reading on the article.
References
- Azmi AS, Ahmad A, Banerjee S, Rangnekar VM, Mohammad RM, Sarkar FH. Chemoprevention of pancreatic cancer: characterization of Par-4 and its modulation by 3,3′ diindolylmethane (DIM) Pharm Res. 2008;25(9):2117–2124. doi: 10.1007/s11095-008-9581-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehrer S, Chow KU, Puccetti E, Ruthardt M, Godzisard S, Krapohl A, Schneider B, Hoelzer D, Mitrou PS, Rangnekar VM, Weidmann E. Deregulated expression of prostate apoptosis response gene-4 in less differentiated lymphocytes and inverse expressional patterns of par-4 and bcl-2 in acute lymphocytic leukemia. Hematol J. 2001;2(2):103–107. doi: 10.1038/sj/thj/6200089. [DOI] [PubMed] [Google Scholar]
- Burikhanov R, Zhao Y, Goswami A, Qiu S, Schwarze SR, Rangnekar VM. The tumor suppressor Par-4 activates an extrinsic pathway for apoptosis. Cell. 2009;138(2):377–388. doi: 10.1016/j.cell.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burz C, Berindan-Neagoe I, Balacescu O, Irimie A. Apoptosis in cancer: key molecular signaling pathways and therapy targets. Acta Oncol. 2009;48(6):811–821. doi: 10.1080/02841860902974175. [DOI] [PubMed] [Google Scholar]
- Cang S, Liu D. P-loop mutations and novel therapeutic approaches for imatinib failures in chronic myeloid leukemia. J Hematol Oncol. 2008;1:15. doi: 10.1186/1756-8722-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll RE, Benya RV, Turgeon DK, Vareed S, Neuman M, Rodriguez L, Kakarala M, Carpenter PM, McLaren C, Meyskens FL, Jr, Brenner DE. Phase IIa clinical trial of curcumin for the prevention of colorectal neoplasia. Cancer Prev Res (Phila) 2011;4(3):354–364. doi: 10.1158/1940-6207.CAPR-10-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry P, Singh M, Parent S, Asselin E. Prostate apoptosis response 4 (Par-4), a novel substrate of caspase-3 during apoptosis activation. Mol Cell Biol. 2012;32(4):826–839. doi: 10.1128/MCB.06321-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook J, Krishnan S, Ananth S, Sells SF, Shi Y, Walther MM, Linehan WM, Sukhatme VP, Weinstein MH, Rangnekar VM. Decreased expression of the pro-apoptotic protein Par-4 in renal cell carcinoma. Oncogene. 1999;18(5):1205–1208. doi: 10.1038/sj.onc.1202416. [DOI] [PubMed] [Google Scholar]
- Diaz-Meco MT, Lallena MJ, Monjas A, Frutos S, Moscat J. Inactivation of the inhibitory kappaB protein kinase/nuclear factor kappaB pathway by Par-4 expression potentiates tumor necrosis factor alpha-induced apoptosis. J Biol Chem. 1999;274(28):19606–19612. doi: 10.1074/jbc.274.28.19606. [DOI] [PubMed] [Google Scholar]
- Diaz-Meco MT, Municio MM, Frutos S, Sanchez P, Lozano J, Sanz L, Moscat J. The product of par-4, a gene induced during apoptosis, interacts selectively with the atypical isoforms of protein kinase C. Cell. 1996;86(5):777–786. doi: 10.1016/s0092-8674(00)80152-x. [DOI] [PubMed] [Google Scholar]
- El-Guendy N, Rangnekar VM. Apoptosis by Par-4 in cancer and neurodegenerative diseases. Experimental cell research. 2003;283(1):51–66. doi: 10.1016/s0014-4827(02)00016-2. [DOI] [PubMed] [Google Scholar]
- El-Guendy N, Zhao Y, Gurumurthy S, Burikhanov R, Rangnekar VM. Identification of a unique core domain of par-4 sufficient for selective apoptosis induction in cancer cells. Mol Cell Biol. 2003;23(16):5516–5525. doi: 10.1128/MCB.23.16.5516-5525.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Oda K, Yokota S, Takatsuki A, Ikehara Y. Brefeldin A causes disassembly of the Golgi complex and accumulation of secretory proteins in the endoplasmic reticulum. J Biol Chem. 1988;263(34):18545–18552. [PubMed] [Google Scholar]
- Garcia-Cao I, Duran A, Collado M, Carrascosa MJ, Martin-Caballero J, Flores JM, Diaz-Meco MT, Moscat J, Serrano M. Tumour-suppression activity of the proapoptotic regulator Par4. EMBO Rep. 2005;6(6):577–583. doi: 10.1038/sj.embor.7400421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami A, Burikhanov R, de Thonel A, Fujita N, Goswami M, Zhao Y, Eriksson JE, Tsuruo T, Rangnekar VM. Binding and phosphorylation of par-4 by akt is essential for cancer cell survival. Mol Cell. 2005;20(1):33–44. doi: 10.1016/j.molcel.2005.08.016. [DOI] [PubMed] [Google Scholar]
- Goswami A, Qiu S, Dexheimer TS, Ranganathan P, Burikhanov R, Pommier Y, Rangnekar VM. Par-4 binds to topoisomerase 1 and attenuates its DNA relaxation activity. Cancer Res. 2008;68(15):6190–6198. doi: 10.1158/0008-5472.CAN-08-0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurumurthy S, Goswami A, Vasudevan KM, Rangnekar VM. Phosphorylation of Par-4 by protein kinase A is critical for apoptosis. Mol Cell Biol. 2005;25(3):1146–1161. doi: 10.1128/MCB.25.3.1146-1161.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwitz AA, Foster BA, Allison JP, Greenberg NM, Kwon ED. The TRAMP mouse as a model for prostate cancer. Curr Protoc Immunol. 2001;Chapter 20(Unit 20):25. doi: 10.1002/0471142735.im2005s45. [DOI] [PubMed] [Google Scholar]
- Johnstone RW, See RH, Sells SF, Wang J, Muthukkumar S, Englert C, Haber DA, Licht JD, Sugrue SP, Roberts T, Rangnekar VM, Shi Y. A novel repressor, par-4, modulates transcription and growth suppression functions of the Wilms’ tumor suppressor WT1. Mol Cell Biol. 1996;16(12):6945–6956. doi: 10.1128/mcb.16.12.6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone RW, Tommerup N, Hansen C, Vissing H, Shi Y. Mapping of the human PAWR (par-4) gene to chromosome 12q21. Genomics. 1998;53(2):241–243. doi: 10.1006/geno.1998.5494. [DOI] [PubMed] [Google Scholar]
- Keefer LM, Piron MA, De Meyts P. Human insulin prepared by recombinant DNA techniques and native human insulin interact identically with insulin receptors. Proc Natl Acad Sci U S A. 1981;78(3):1391–1395. doi: 10.1073/pnas.78.3.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan N, Adhami VM, Mukhtar H. Apoptosis by dietary agents for prevention and treatment of prostate cancer. Endocr Relat Cancer. 2010;17(1):R39–52. doi: 10.1677/ERC-09-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Hawke N, Baldwin AS. NF-kappaB and IKK as therapeutic targets in cancer. Cell Death Differ. 2006;13:738–747. doi: 10.1038/sj.cdd.4401877. [DOI] [PubMed] [Google Scholar]
- Kogel D, Plottner O, Landsberg G, Christian S, Scheidtmann KH. Cloning and characterization of Dlk, a novel serine/threonine kinase that is tightly associated with chromatin and phosphorylates core histones. Oncogene. 1998;17(20):2645–2654. doi: 10.1038/sj.onc.1202204. [DOI] [PubMed] [Google Scholar]
- Kogel D, Reimertz C, Mech P, Poppe M, Fruhwald MC, Engemann H, Scheidtmann KH, Prehn JH. Dlk/ZIP kinase-induced apoptosis in human medulloblastoma cells: requirement of the mitochondrial apoptosis pathway. Br J Cancer. 2001;85(11):1801–1808. doi: 10.1054/bjoc.2001.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCasse EC, Baird S, Korneluk RG, MacKenzie AE. The inhibitors of apoptosis (IAPs) and their emerging role in cancer. Oncogene. 1998;17(25):3247–3259. doi: 10.1038/sj.onc.1202569. [DOI] [PubMed] [Google Scholar]
- Lee AS. GRP78 induction in cancer: therapeutic and prognostic implications. Cancer Res. 2007;67(8):3496–3499. doi: 10.1158/0008-5472.CAN-07-0325. [DOI] [PubMed] [Google Scholar]
- Lee JI, Burckart GJ. Nuclear factor kappa B: important transcription factor and therapeutic target. J Clin Pharmacol. 1998;38(11):981–993. doi: 10.1177/009127009803801101. [DOI] [PubMed] [Google Scholar]
- Lee SM, Chen H, O’Halloran TV, Nguyen ST. “Clickable” polymer-caged nanobins as a modular drug delivery platform. J Am Chem Soc. 2009;131(26):9311–9320. doi: 10.1021/ja9017336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek DW, Cox M. Induction and activation of the p53pathway: a role for the protein kinase CK2? Mol Cell Biochem. 2011;356(1–2):133–138. doi: 10.1007/s11010-011-0966-3. [DOI] [PubMed] [Google Scholar]
- Merkle D, Hoffmann R. Roles of cAMP and cAMP-dependent protein kinase in the progression of prostate cancer: cross-talk with the androgen receptor. Cell Signal. 2011;23(3):507–515. doi: 10.1016/j.cellsig.2010.08.017. [DOI] [PubMed] [Google Scholar]
- Miller M. The importance of being flexible: the case of basic region leucine zipper transcriptional regulators. Curr Protein Pept Sci. 2009;10(3):244–269. doi: 10.2174/138920309788452164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Bueno G, Fernandez-Marcos PJ, Collado M, Tendero MJ, Rodriguez-Pinilla SM, Garcia-Cao I, Hardisson D, Diaz-Meco MT, Moscat J, Serrano M, Palacios J. Inactivation of the candidate tumor suppressor par-4 in endometrial cancer. Cancer Res. 2007;67(5):1927–1934. doi: 10.1158/0008-5472.CAN-06-2687. [DOI] [PubMed] [Google Scholar]
- Nambiar D, Rajamani P, Singh RP. Effects of phytochemicals on ionization radiation-mediated carcinogenesis and cancer therapy. Mutat Res. 2011;728(3):139–157. doi: 10.1016/j.mrrev.2011.07.005. [DOI] [PubMed] [Google Scholar]
- Patel VB, Misra S, Patel BB, Majumdar AP. Colorectal cancer: chemopreventive role of curcumin and resveratrol. Nutr Cancer. 2010;62(7):958–967. doi: 10.1080/01635581.2010.510259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavan B, Biondi C, Dalpiaz A. Adenylyl cyclases as innovative therapeutic goals. Drug Discov Today. 2009;14(19–20):982–991. doi: 10.1016/j.drudis.2009.07.007. [DOI] [PubMed] [Google Scholar]
- Pei J, Grishin NV. GGDEF domain is homologous to adenylyl cyclase. Proteins. 2001;42(2):210–216. doi: 10.1002/1097-0134(20010201)42:2<210::aid-prot80>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Priya R, Kumar A, Manimekalai MS, Gruber G. Conserved glycine residues in the P-loop of ATP synthases form a doorframe for nucleotide entrance. J Mol Biol. 2011;413(3):657–666. doi: 10.1016/j.jmb.2011.08.045. [DOI] [PubMed] [Google Scholar]
- Qiu SG, Krishnan S, el-Guendy N, Rangnekar VM. Negative regulation of Par-4 by oncogenic Ras is essential for cellular transformation. Oncogene. 1999;18(50):7115–7123. doi: 10.1038/sj.onc.1203199. [DOI] [PubMed] [Google Scholar]
- Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzzene M, Pinna LA. Addiction to protein kinase CK2: a common denominator of diverse cancer cells? Biochim Biophys Acta. 2010;1804(3):499–504. doi: 10.1016/j.bbapap.2009.07.018. [DOI] [PubMed] [Google Scholar]
- Sayers TJ. Targeting the extrinsic apoptosis signaling pathway for cancer therapy. Cancer Immunol Immunother. 2011;60(8):1173–1180. doi: 10.1007/s00262-011-1008-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sells SF, Wood DP, Jr, Joshi-Barve SS, Muthukumar S, Jacob RJ, Crist SA, Humphreys S, Rangnekar VM. Commonality of the gene programs induced by effectors of apoptosis in androgen-dependent and -independent prostate cells. Cell Growth Differ. 1994;5(4):457–466. [PubMed] [Google Scholar]
- Shrestha-Bhattarai T, Rangnekar VM. Cancer-selective apoptotic effects of extracellular and intracellular Par-4. Oncogene. 2010;29(27):3873–3880. doi: 10.1038/onc.2010.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortora G, Ciardiello F. Protein kinase A as target for novel integrated strategies of cancer therapy. Ann N Y Acad Sci. 2002;968:139–147. doi: 10.1111/j.1749-6632.2002.tb04332.x. [DOI] [PubMed] [Google Scholar]
- Valko M, Rhodes CJ, Moncol J, Izakovic M, Mazur M. Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem Biol Interact. 2006;160(1):1–40. doi: 10.1016/j.cbi.2005.12.009. [DOI] [PubMed] [Google Scholar]
- Yoshikawa S, Kukimoto-Niino M, Parker L, Handa N, Terada T, Fujimoto T, Terazawa Y, Wakiyama M, Sato M, Sano S, Kobayashi T, Tanaka T, Chen L, Liu ZJ, Wang BC, Shirouzu M, Kawa S, Semba K, Yamamoto T, Yokoyama S. Structural basis for the altered drug sensitivities of non-small cell lung cancer-associated mutants of human epidermal growth factor receptor. Oncogene. 2012 doi: 10.1038/onc.2012.21. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Burikhanov R, Brandon J, Qiu S, Shelton BJ, Spear B, Bondada S, Bryson S, Rangnekar VM. Systemic Par-4 inhibits non-autochthonous tumor growth. Cancer Biol Ther. 2011;12(2):152–157. doi: 10.4161/cbt.12.2.15734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Burikhanov R, Qiu S, Lele SM, Jennings CD, Bondada S, Spear B, Rangnekar VM. Cancer resistance in transgenic mice expressing the SAC module of Par-4. Cancer Res. 2007;67(19):9276–9285. doi: 10.1158/0008-5472.CAN-07-2124. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Rangnekar VM. Apoptosis and tumor resistance conferred by Par-4. Cancer Biol Ther. 2008;7(12):1867–1874. doi: 10.4161/cbt.7.12.6945. [DOI] [PMC free article] [PubMed] [Google Scholar]