Abstract
Thrombin-catalyzed activation of factor VIII (FVIII) occurs through proteolysis at three P1 Arg residues: Arg372 and Arg740 in the FVIII heavy chain and Arg1689 in the FVIII light chain. Cleavage at the latter two sites is relatively fast compared with cleavage at Arg372, which appears rate limiting. Examination of the P3-P3′ residues flanking each P1 site revealed that those sequences at Arg740 and Arg1689 are more optimal for thrombin cleavage than at Arg372, suggesting these sequences may impact reaction rates. Recombinant FVIII variants were prepared with mutations swapping scissile bond flanking sequences in the heavy chain individually and in combination with a second swap or with a P1 point mutation. Rates of generation of A1 and A3-C1-C2 subunits were determined by Western blotting and correlated with rates of cleavage at Arg372 and Arg1689, respectively. Rates of thrombin cleavage at Arg372 were increased ~10-fold and ~3-fold compared with WT FVIII when replaced with P3-P3′ residues flanking Arg740 or Arg1689, respectively, and these values paralleled increased rates of A2 subunit generation and procofactor activation. Positioning of more optimal residues flanking Arg372 abrogated the need for initial cleavage at Arg740 to facilitate this step. These results show marked changes in cleavage rates correlate with the extent of cleavage-optimal residues flanking the scissile bond and modulate the mechanism for procofactor activation.
Factor VIII (FVIII1) is an inactive procofactor that is essential in blood coagulation as mutations or splicing defects in the FVIII gene lead to hemophilia A. FVIII is synthesized as an ~300 kDa single chain molecule corresponding to 2332 amino acids in six distinct domains ordered as: NH2-A1-A2-B-A3-C1-C2-COOH (1, 2). Segments rich in acidic residues border the A domains of FVIII and are designated a1 (residues 337–372) and a2 (residues 711–740) which directly follow the A1 and A2 domains, respectively, and a3 (residues 1649–1689) preceding the A3 domain. FVIII circulates primarily as a heterodimer composed of a heavy chain (A1-a1-A2-a2-B domains) and a light chain (a3-A3-C1-C2 domains) as a result of proteolytic processing at the B-a3 junction (3).
The activated cofactor, factor VIIIa (FVIIIa) binds to factor IXa (FIXa) on a membrane surface and this complex activates factor X (FX). FVIIIa increases the catalytic efficiency of FIXa by several orders of magnitude in a primarily kcat-dependent manner (4). Conversion of the procofactor to activated FVIIIa occurs through limited proteolysis by either thrombin or FXa (5, 6) via cleavage of three P1 residues at Arg740 (a2-B junction), Arg372 (a1-A2 junction), and Arg1689 (a3-A3 junction) (5). Thrombin is considered the physiological activator of FVIII inasmuch as von Willebrand factor inhibits the membrane-dependent procofactor activation catalyzed by FXa (5, 7). An essential step in activation of the procofactor is cleavage at Arg372 as this cleavage allows exposure of FIXa-interactive sites in the A2 domain that are otherwise masked (8). Cleavage at Arg1689 is also critical as it not only increases cofactor specific activity, but releases FVIIIa from von Willebrand factor (9–11). Inactivation of FVIIIa resulting in down-regulation of the intrinsic factor Xase complex (3) occurs through two mechanisms: spontaneous inactivation via dissociation of the weak electrostatic interaction between the A1/A3-C1-C2 dimer and the A2 subunit (12, 13), and proteolytic inactivation via cleavage at Arg336 and Arg562 by activated protein C (5, 12, 14) and FXa (5,15).
Substrate recognition by thrombin occurs through interaction with one or both anion binding exosites (ABE1 and ABE2) followed by active site docking (16). This docking primarily affects the Vmax for substrate activation by thrombin, while it is believed that exosite binding exclusively determines substrate affinity (17). Examination of the P3-P3′2 region of thrombin substrates reveals a variety of residues that can be accommodated by thrombin and it is speculated that P1-P3′ flanking sites primarily affect substrate docking independent of binding affinity (16, 18, 19).
Previous studies on the proteolytic activation of FVIII by thrombin have indicated that both ABE1 and ABE2 contribute to the activation mechanism (20). Furthermore, there appears to be a competition between heavy chain and light chain for this exosite-dependent binding, with binding to and cleavage of FVIII heavy chain preferred (21). Additionally, this mechanism is further complicated by the presence of three P1 cleavage sites which differ in P3-P3′ sequences flanking the P1 Arg372, Arg740, and Arg1689, which are represented by 370QIR↓SVA375, 738EPR↓SFS743, and 1687SPR↓SFQ1692, respectively. Residues flanking Arg372 are considered non-optimal for thrombin cleavage with only two residues optimal (bold) for cleavage in the P3-P3′ sequence, while residues flanking at the two other P1 sites are considered near-optimal with four out of six residues optimal (bold) (22). Inasmuch as the rate limiting cleavage occurs at Arg372 (3, 23), there is a possible role for residues flanking the P1 site in the overall reaction mechanism. In the current study, the effects of P3-P3′ residues on thrombin-catalyzed cleavage and activation of FVIII were investigated using recombinant FVIII variants possessing P3-P3′ sequence swaps of residues flanking P1 residues Arg372, Arg740, and Arg1689. Results from this study show marked changes in site-specific cleavage rates that correlate with the degree of cleavage-optimal residues present flanking the scissile bond.
EXPERIMENTAL PROCEDURES
Reagents
The coagulation proteins FIXa, FXa, FX, and human α-thrombin, were obtained from Enzyme Research Laboratories. Chromogenic FXa substrate Pefachrome® FXa (CH3OCOD-CHA-Gly-Arg-pNA) (Pentapharm), horseradish peroxidase-labeled streptavidin (Calbiochem), and recombinant non-sulfated hirudin-(55–65) (Calbiochem) were purchased from the indicated suppliers. Pete Lollar and John Healey (Emory University) generously provided the Bluescript FVIII vector (pBS FVIII) and B-domainless FVIII expression construct RENeo FVIII. Monoclonal antibodies C5 (recognizing the a1 region) and 2D2 (recognizing the A3 domain) were gifts from Drs. Zaverio Ruggeri and Lisa Regan (Bayer), respectively. The monoclonal FVIII antibodies R8B12 (GMA-012) (recognizing the A2 domain, (12) and GMA-8003 (recognizing the C2 domain) were obtained from Green Mountain Antibodies.
Phospholipid Vesicles
N-octyl glucoside was used to prepare phospholipid vesicles containing 20% phosphatidylserine, 40% phosphatidylcholine and 40% phosphatidylethanolamine (Avanti Polar Lipids) (25).
FVIII-deficient Plasma
FVIII-deficient plasma was prepared by incubating pooled human plasma with 25 mM EDTA for ~18 h at 4 °C followed by addition of 25 mM CaCl2 (24). Aliquots were snap-frozen in liquid N2 and stored at −80 °C.
Construction, Expression, and Purification of Recombinant Proteins
The FVIII expression construct FVIIIHSQ-MSAB-NotI-RENeo was restricted using the endonucleases XhoI and NotI to create B-domainless FVIII cDNA and cloned into the pBluescript II K/S-vector. The B-domainless FVIII cDNA was then further restricted using endonucleases SacII/ApaI to create the Arg1689 and Arg740 P3-P3′ mutants and endonucleases SpeI/SacII to create the Arg372 P3-P3′ mutants. All P3-P3′ mutants were subcloned into the pBluescript II K/S-vector and introduced into the construct using the Stratagene QuikChange site-directed mutagenesis kit as previously described (26, 27). Dideoxy sequencing was used to confirm the presence of only the desired mutation. The mutated FVIII cDNA was then ligated back into the FVIII expression shuttle construct and subjected to a second round of dideoxy sequencing, again to confirm that only the desired mutation was present. The FVIII expression construct was transfected to BHK cells using FuGENE6 (Roche). Geneticin® antibiotic selection, subcloning, and cloning of stable transfectants were performed by standard methods, and the cloned cells were cultured in roller bottles for protein expression (27). Approximately 300 mL of conditioned media was collected daily for 4 days and the expressed proteins were purified by SP-Sepharose (Amersham Biosciences) column chromatography as previously described (27). Fractions were assayed using FVIII-deficient plasma in a one-stage clotting assay in order to determine fractions containing FVIII. Yields of purified wild-type (WT) and variant FVIII ranged from ~0.2 to 1.0 mg per liter of conditioned media. Resultant FVIII was >90% pure as judged by SDS-PAGE with the main contaminant being albumin. FVIII samples were snap-frozen in liquid N2 and stored at −80 °C.
One-stage Clotting Assay
FVIII-deficient pooled human plasma was incubated with activated partial thromboplastin reagent (Trinity Biotech) for 7 min at 37 °C. A dilution of FVIII was added to the cuvette and after incubation at 37 °C for 1 min, CaCl2 (6 mM final concentration) was added and the time to form a clot was measured using an ST4 coagulation analyzer (Diagnostica Stago) and compared with a pooled normal plasma standard (George King Biomedical, Inc.).
Enzyme-linked Immunosorbent Assay
A sandwich ELISA was used to determine the final concentration of purified FVIII proteins as previously described (26). Briefly, GMA-8003 was used as the capture antibody and biotinylated R8B12 was used as the detection antibody. A chromogenic assay was used to determine the amount of FVIII bound to the plate utilizing streptavidin-linked horseradish peroxidase (Calbiochem) with the chromogen o phenylenediamine dihydrochloride (Sigma). Purified commercial recombinant FVIII (Kogenate; Bayer) was used as the standard. FVIII specific activity values were determined using one-stage clotting and ELISA (26).
Thrombin Cleavage of FVIII
WT or mutant FVIII (100 nM) was incubated at 22°C with indicated concentrations of thrombin (see Legends) in a buffer containing 20 mM HEPES, 0.14 M NaCl, 5 mM CaCl2, and 0.01% Tween 20, pH 7.2. Samples were removed at indicated time points during the time course and terminated by addition of sodium dodecyl sulfate-polyacrylamide-gel electrophoresis (SDS-PAGE) sample buffer (0.04 M Tris-HCl, 2% (w/v) SDS, 4% (v/v) glycerol, 0.05% bromophenol blue, pH 6.8) containing 3% (v/v) 2-mercaptoethanol and boiled for 3 min. Cleavage rates were assessed by rates of A1, A2, and/or A3-C1-C2 subunit generation.
Electrophoresis and Western Blotting
Thrombin cleavage of WT and mutant FVIII was evaluated by SDS-PAGE (28) on 8% polyacrylamide gels using a BioRad mini-gel apparatus at 175 V for 1 h. Protein transfer occurred on polyvinylidene fluoride membrane for 1 h at 100 V in ice cold transfer buffer (10 mM 3-(cyclohexylamino)propane-1-sulfonic acid, 10% (v/v) methanol, pH 11).Western blotting was performed by probing the membranes with anti-FVIII monoclonal antibodies as indicated in the figure legends, followed by addition of an alkaline phosphatase-linked goat anti-mouse (Sigma) as the secondary antibody. An enhanced chemifluorescence (ECF®) system (Amersham Biosciences) was used to detect the signal and the blots were scanned over a range of 650–740 nm with a Storm 860 instrument (GE Healthcare). Densitometry was used to quantitate the linear density regions of the blots using Image Quant software (GE Healthcare).
Determination of Rates of Subunit Generation
Rates of subunit generation following thrombin cleavage of WT and mutants were calculated from the blots based upon summing all FVIII band density values. The intensity of each band was determined by volume integration and corrected for variability in background staining by using densities in the areas immediately adjacent to each band. The concentrations of A1, A2, and A3-C1-C2 subunits were normalized to the total density in each lane to correct for any differences in loading. Initial time points were fit using nonlinear least-squares regression to the single exponential equation:
| (1) |
Where Ao is the total nanomolar concentration of A1, A2, or A3-C1-C2 and t is the time in minutes. The absolute value of kobs represents the rate of the A1, A2, or A3-C1-C2 subunit generation that was normalized by the thrombin concentration and expressed in nM A1, A2, or A3-C1-C2 subunit/min/nM thrombin, respectively. All experiments were performed at least three separate times and the average values with standard deviations are shown. The quality of the fits was acceptable with R squared values ranging from 0.946–0.995. Data were also subjected to analysis using a quadratic equation (21) and yielded rates that were typically within 30% of those obtained with the single exponential equation (data not shown). Rate values are presented using the single exponential since the overall quality of the fitted curves was superior with this method.
FXa Generation Assay
A purified protein system was used to measure the rate of FX conversion to FXa (29). In order to assess thrombin activation of FVIII over a time course, FVIII (100 nM) was reacted with 5 nM or 0.5 nM thrombin in a buffer containing 20 mM HEPES, 0.14 M NaCl, 5 mM CaCl2, and 0.01% Tween 20, pH 7.2. Samples were removed at indicated times and thrombin activity was inhibited by the addition of hirudin (2 U/mL). The dilute FVIIIa solution (2 nM, final) was added to FIXa (20 nM) in the presence of phospholipid vesicles (10 µM). FXa generation was initiated by addition of FX (300 nM). The reactions were terminated with EDTA (50 mM) and rates of FXa generation were determined by the addition of the chromogenic substrate Pefachrome® FXa (0.46 mM final concentration). Reactions were read at 405 nm for 5 minutes using a Vmax microtiter plate reader (Molecular Devices).
RESULTS
Specific Activity of Recombinant FVIII Protein
Thrombin cleaves the FVIII procofactor at Arg740, Arg372, and Arg1689. Recent studies have shown that cleavage at Arg740 appears several-fold faster than Arg1689 and this effect likely results in part from exosite-driven competition between the heavy chain and light chain of FVIII for thrombin binding (21). Furthermore, cleavage at Arg740 facilitates cleavage at Arg372 (30), which appears to represent the rate-limiting step in procofactor activation. The current study investigates the effect of the P3-P3′ residues of FVIII on thrombin proteolysis at the three cleavage sites and, to this end, several recombinant B domain-deleted FVIII mutants were prepared and stably expressed. The variants 372(P3-P3′)740, 372(P3-P3′)1689, and 740(P3-P3′)372, designate variants where the entire P3-P3′ sequences surrounding one scissile bond in FVIII were swapped with another (see Table 1). For example, the 372(P3-P3′)740 variant replaced the P3-P3′ sequence at Arg372 with that from Arg740. In addition, a variant possessing the double swap 372(P3-P3′)740/740(P3-P3′)372 and one possessing the 740 for 372 swap plus a point mutation at Arg740 to Gln to create a non-cleavable site (372(P3-P3′)740/R740Q) were prepared.
TABLE 1.
Specific activity values and sequences of FVIII P3-P3' mutants
| Factor VIII | 372(P3-P3') Sequencea (residues 370–375) |
740(P3-P3') Sequencea (residues 738–743) |
1689(P3-P3') Sequencea (residues 1687–1692) |
Specific Activityb |
|---|---|---|---|---|
| WT | QIR↓SVA | EPR↓SFS | SPR↓SFQ | % 100 ± 6 |
| 372(P3-P3')740 | EPR↓SFS | 98 ± 5 | ||
| 372(P3-P3')1689 | SPR↓SFQ | 14 ± 2 | ||
| 372(P3-P3')740/R740Q | EPR↓SFS | EPQ↓SFS | 38 ± 4 | |
| 372(P3-P3')740/740(P3-P3')372 | EPR↓SFS | QIR↓SVA | 122 ± 18 | |
| 740(P3-P3')372 | QIR↓SVA | 73 ± 2 |
Sequences for the P3-P3' residues flanking Arg372, Arg740, and Arg1689 are shown using single-letter amino acid code and specific mutations are indicated in bold type. Arrows indicate the position of the scissile bond.
Specific activity values are presented as a percentage ± standard deviation of the WT value.
Specific activity values of WT and variant FVIII were measured using one-stage clotting and ELISA assays (Table 1). The specific activity values suggest an essentially normal phenotype for the 372(P3-P3′)740, 372(P3-P3′)740/740(P3-P3′)372, and 740(P3-P3′)372 variants. The 372(P3-P3′)740/R740Q variant showed a modestly reduced specific activity (~40% of WT), whereas the value for the 372(P3-P3′)1689 variant (~15% of WT) was consistent with a mild hemophilic phenotype. The observed reduction in the specific activity for FVIII 372(P3-P3′)740/R740Q can be attributed solely to the Gln mutations at Arg740 since this value was similar to the previous specific activity determined previously for the single P1 site mutation alone (30).
Thrombin-Catalyzed Cleavage of 372(P3-P3′) and 740(P3-P3′) Recombinant FVIII Mutants
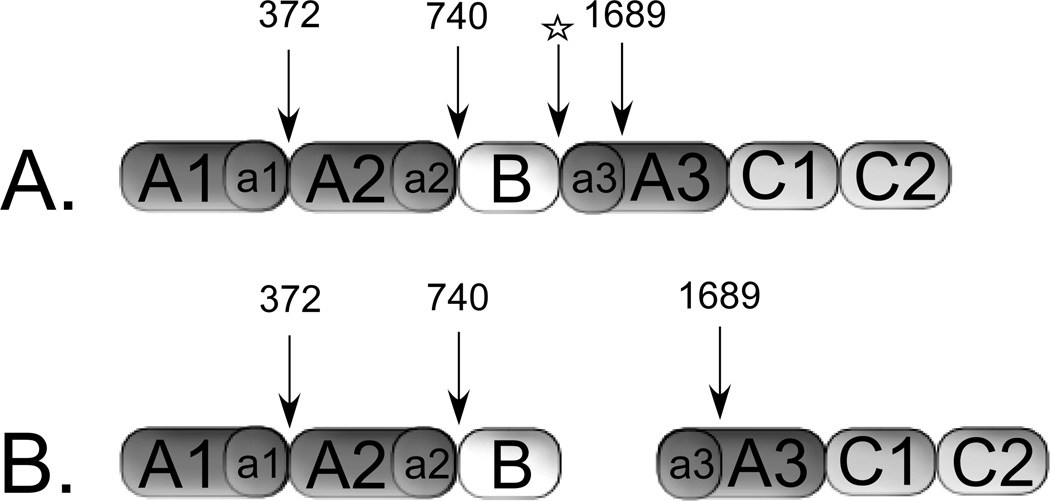
The variants 372(P3-P3′)740 and 372(P3-P3′)1689 were used to assess whether the more thrombin-optimal P3-P3′ residues that surround Arg740 and Arg1689 accelerate thrombin-catalyzed cleavage at Arg372. These cleavage sites are schematically represented in Figure 1, which also illustrates the FVIII single chain (A) and heterodimer (B). The B-domainless recombinant FVIII used in this study is expressed in near equal amounts of single chain and heterodimer. Furthermore, a B domain remnant of 14 amino acid residues remains and separates the a2 and a3 segments in the single chain form or represents the C-terminal end of the heavy chain of the heterodimer.
Figure 1. Forms of B-domainless FVIII Secreted from BHK cells.
Secreted B-domainless FVIII single chain (A) and heterodimer forms (B). Thrombin cleavage sites indicated by arrows and (☆) represents a furin cleavage site. The B-domain in B-domainless FVIII represents a 14 amino acid remnant connecting the a2 and a3 acidic regions.
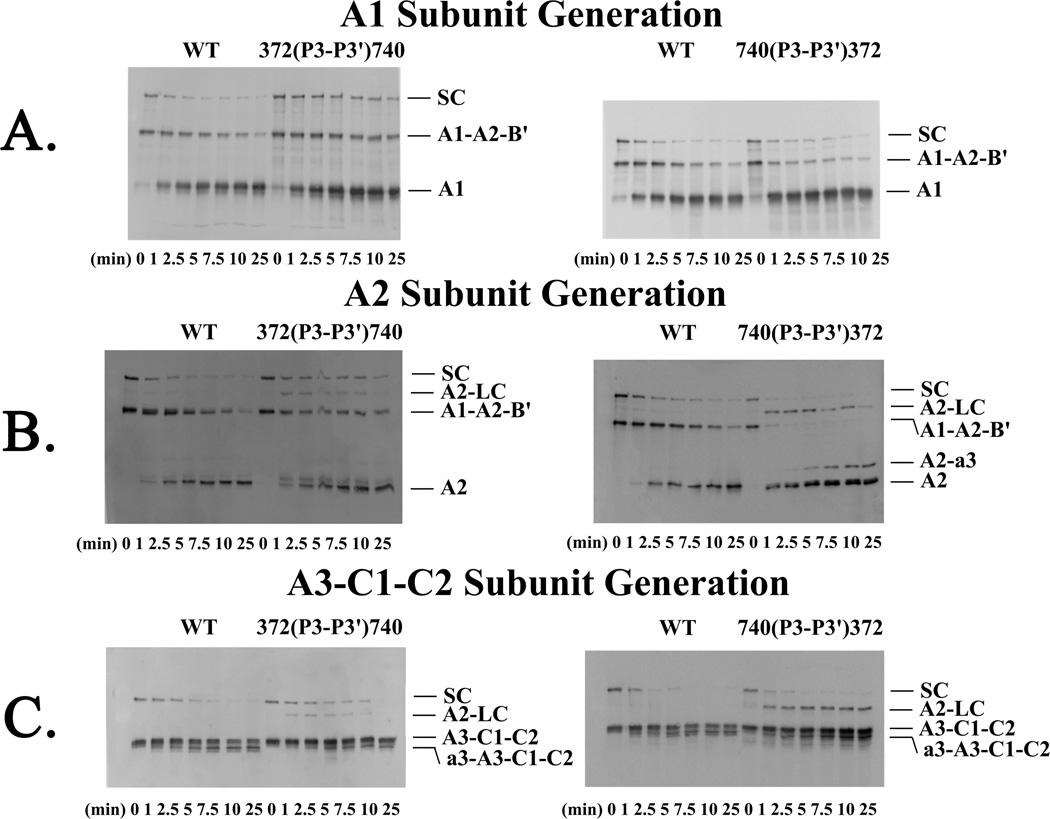
Reactions were run over a 25-minute time course, and results were visualized using SDS-PAGE and Western blotting to evaluate rates of FVIIIa subunit generation by thrombin as described in “Experimental Procedures.” Reaction of WT FVIII with thrombin showed efficient cleavage of single chain, and heavy and light chains resulting in the generation of the A1, A2 and A3-C1-C2 FVIIIa subunits (Figure 2). These blots were quantitated by scanning densitometry and subjected to nonlinear least-squares regression analysis (Figure 3) to calculate rates of generation of each FVIIIa subunit (Table 2). Generation of the A1 subunit results from a single cleavage at Arg372, therefore the rate of cleavage at that residue is equivalent to the rate of A1 subunit generation. The A2 subunit is derived from two cleavages; one between the A1 and A2 domains at Arg372 and the other between the a2 segment and B domain remnant at Arg740 (Figure 1). While the A2 subunit released from the heavy chain of the heterodimer requires only the single cleavage at Arg372, both cleavages at Arg372 and Arg740 are necessary to excise this subunit from single chain FVIII. Since the presence or absence of the remnant is not discernable using SDS-PAGE, it is not possible to directly determine the rate of cleavage at Arg740 in recombinant B-domainless FVIII. However, rates of A2 subunit generation in the variants are useful to estimate this parameter relative to the WT FVIII value. A single cleavage at Arg1689 in both light chain and single chain yields the A3-C1-C2 subunit, therefore the rate of generation of this subunit represents the rate of cleavage at Arg1689.
Figure 2. Thrombin Cleavage of the Single Swap P3-P3′ FVIII mutants.
WT and the 372(P3-P3′)740 and 740(P3-P3′)372 FVIII variants (100 nM) were reacted with thrombin over a 25 min time course and samples were subjected to Western blotting using C5, R8B12, and 2D2 monoclonal antibodies for detection of A1, A2 and A3-C1-C2 subunits, respectively, as described in “Experimental Procedures.” (A) Representative blots for rates of A1 subunit generation (top) for reactions using 0.75 nM thrombin for WT and 740(P3-P3′)372 and 0.075 nM thrombin for 372(P3-P3′)740; (B) A2 subunit generation (center) using 0.75 nM thrombin for WT and 740(P3-P3′)372 and 0.075 nM thrombin for 372(P3-P3′)740; and (C) A3-C1-C2 subunit generation (bottom) using 0.13 nM thrombin for WT and variants. SC refers to single chain and LC to light chain.
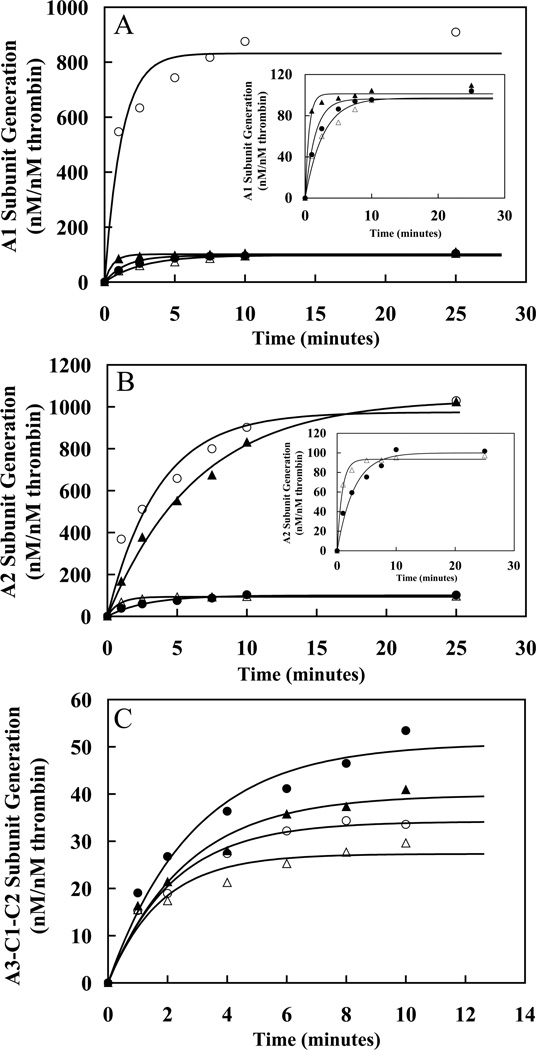
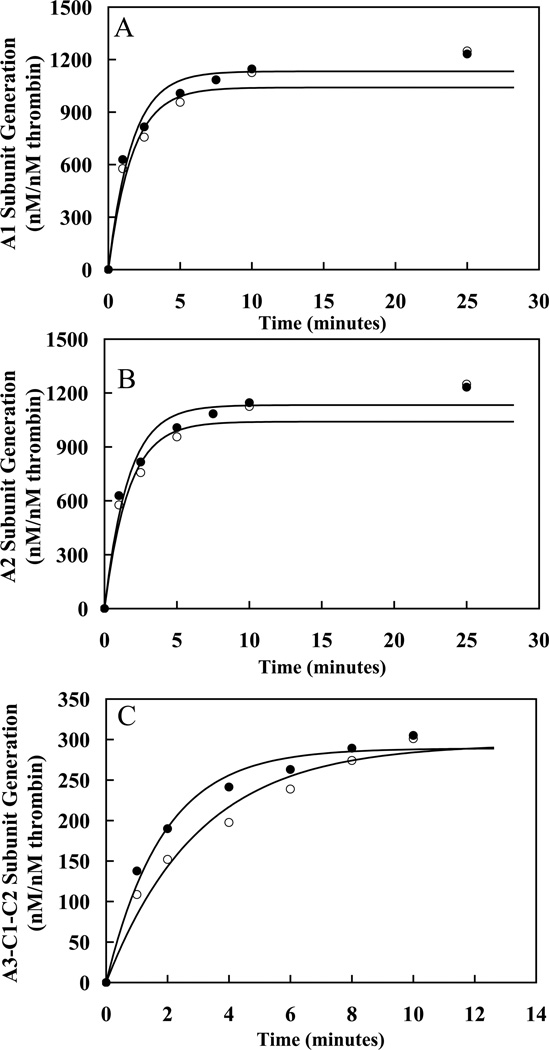
Figure 3. Analysis of the Western Blots of the Single Swap P3-P3′ FVIII mutants.
Panels A–C plot rates derived from densitometry scans of the blots in Figure 2 and for Western blot data of 372(P3-P3′)1689 (data not shown) for rates of generation of A1 (A), A2 (B) and A3-C1-C2 (C) subunits for WT (●), 372(P3-P3′)740 (○), 372(P3-P3′)1689 (▲), and 740(P3-P3′)372 (△) FVIII. Lines are drawn representing the initial time points of the data fit to a single exponential equation using non-linear least squares regression as described in “Experimental Procedures.” Experiments were performed at least three separate times and average values are shown.
TABLE 2.
Rates A1, A2, and A3-C1-C2 subunit generation during WT and P3-P3′ mutant FVIII activation by thrombin
| Factor VIII | A1 subunit generationa | A2 subunit generationa | A3-C1-C2 subunit generationa |
|---|---|---|---|
| nM A1 min−1 (nM IIa)−1 | nM A2 min−1 (nM IIa)−1 | nM A3-C1-C2 min−1 (nM IIa)−1 | |
| WT | 63 ± 0.9 | 21 ± 1.1 | 199 ± 29 |
| 372(P3-P3′)740 | 652 ± 42 | 236 ± 25 | 121 ± 7.1 |
| 372(P3-P3′)1689 | 175 ± 12 | 164 ± 22 | 123 ± 10 |
| 740(P3-P3′)372 | 147 ± 22 | 168 ± 16 | 124 ± 16 |
| 372(P3-P3′)740/R740Q | 665 ± 19 | 157 ± 15 | 157 ± 13 |
| 372(P3-P3′)740/740(P3-P3′)372 | 576 ± 33 | 216 ± 22 | 99 ± 7.4 |
A1, A2, and A3-C1-C2 subunit generation rates by thrombin cleavage of WT and mutant FVIII were estimated by nonlinear least-squares regression analysis of the data shown in Figures 2–5. The data represent the average ± S.D. values of at least three separate experiments. Western blot assays were performed and data analyzed as described under “Experimental Procedures.”
Evaluation of reactions using the 372(P3-P3′)740 (Figure 2A) and 372(P3-P3′)1689 FVIII (blots not shown) revealed 10-fold and 3-fold increases, respectively, in the rates of cleavage at Arg372 as determined by rates of A1 subunit generation for the variants as compared with WT FVIII (Figure 3A, Table 2). Reactions for the variants used 10% the thrombin concentration employed for WT. Control experiments showed that cleavage rate values were linear over this thrombin concentration range. Although the rate of thrombin cleavage at Arg372 increased for the variants, this increased rate did not appear to be matched by the rate of Arg740 cleavage as suggested by appearance of an A2-LC intermediate which was detectable in the blots using the anti-A2 domain and anti-A3 domain antibodies (Figure 2B and C). Thus the 10-fold and 8-fold increases in A2 subunit generation for the 372(P3-P3′)740 and 372(P3-P3′)1689 mutants, respectively, most likely reflect the increase in the rate of thrombin cleavage at Arg372 (Table 2). The rate of cleavage at Arg1689 for both 372(P3-P3′) mutants was similar to WT FVIII, suggesting little effect of accelerating cleavage at Arg372 on cleavage at Arg1689. These results suggest that placing more optimal residues at Arg372 increases thrombin cleavage at that site. Furthermore, the P3-P3′ residues flanking Arg740 appeared more optimal for Arg372 cleavage than those flanking Arg1689 (Table 2). The P3-P3′ sequences flanking Arg740 and Arg1689 differ only in the P3 and P3′ residues, suggesting the P3 Glu and/or P3′ Ser may be preferred to Ser and/or Gln, respectively.
Examination of the effects of replacing P3-P3′ residues flanking Arg740 on thrombin-catalyzed cleavages of FVIII with those of the less optimal Arg372 flanking sequence employed a similar Western blot approach as described above. Results obtained for the 740(P3-P3′)372 variant revealed an ~2-fold increase in the rate of A1 subunit generation as well as increased rate of A2 subunit generation (Figure 3A and B, Table 2). These results were surprising and, for reasons that are unclear, indicated an accelerated rate of cleavage at the P1 Arg372. However, the persistence of the A2-light chain and A2-a3 intermediates (Figure 2B and C) throughout an extended time course suggested impairment of the rate of cleavage at Arg740 consistent with the less optimal cleavage sequence at this site.
Thrombin-Catalyzed Cleavage of Combined (P3-P3′) Recombinant FVIII Mutants
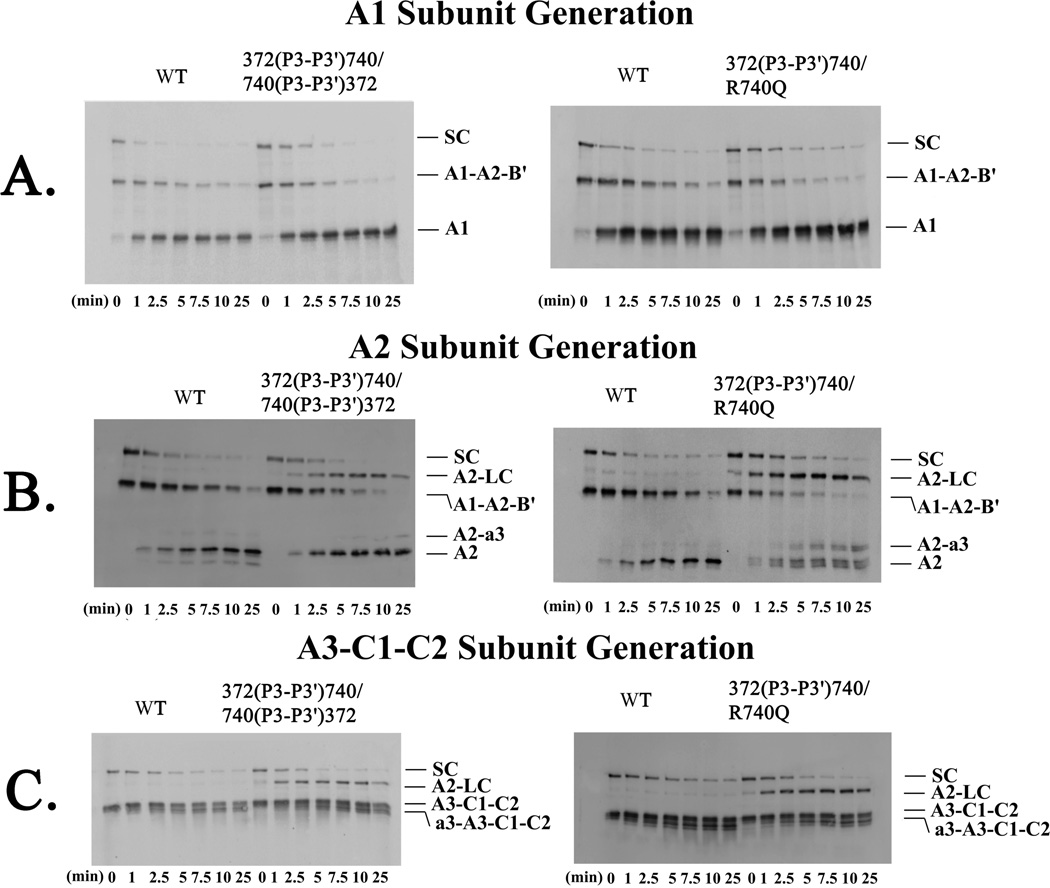
In order to gain insights into the coordination of heavy chain cleavages during the activation process, we prepared a FVIII variant possessing two reciprocal swaps in the sequences flanking Arg372 and Arg740. Thus results obtained with this variant, 372(P3-P3′)740/740(P3-P3′)372, would examine compensatory effects on cleavage within the same chain following replacement of the slow site at Arg372 with the fast site at Arg740 and vice versa. In an earlier study (30), variants showing slower cleavage rates at Arg740 also demonstrated reduced rates of cleavage at the Arg372 site. Results from the Western blotting time course of thrombin activation using the 372(P3-P3′)740/740(P3-P3′)372 variant (Figure 4A and B) showed a 9-fold enhancement in rate of Arg372 cleavage (A1 subunit generation) and a similar increase in the rate of A2 subunit generation (Figure 5A and B, Table 2). Both of these effects were nearly identical to results obtained with the single swap 740(P3-P3′)372 variant. The blotting also revealed two intermediates of A2-LC and A2-a3 (Figure 4B and C) suggesting inefficient cleavage at Arg740 was a result of the swap for the Arg372 flanking sequence. Interestingly, there was a 2-fold decrease in cleavage rate at Arg1689 as seen by a decreased rate of A3-C1-C2 subunit generation (Table 2).
Figure 4. Thrombin Cleavage of the Combination P3-P3′ FVIII mutants.
WT and the 372(P3-P3′)740/R740Q and 372(P3-P3′)740/740(P3-P3′)372 FVIII variants (100 nM) were reacted with thrombin and samples were subjected to Western blotting as described in the legend to Figure 2. (A) Representative blots for rates of A1 subunit generation (top) for reactions using 0.75 nM thrombin for WT and 0.075 nM for the variants; (B) A2 subunit generation (center) using 0.75 nM thrombin for WT and 0.075 nM thrombin for the variants; and (C) A3-C1-C2 subunit generation (bottom) using 0.13 nM thrombin for WT and variants. SC refers to single chain and LC to light chain.
Figure 5. Analysis of the Western Blots of the Combination P3-P3′ FVIII mutants.
Panels A–C plot rates derived from densitometry scans of the blots in Figure 4 for rates of generation of A1 (A), A2 (B) and A3-C1-C2 (C) subunits for 372(P3-P3′)740/R740Q (●) and 372(P3-P3′)740/740(P3-P3′)372 (○) FVIII. Lines are drawn representing the initial time points of the data fit to a single exponential equation using non-linear least squares regression as described in “Experimental Procedures.” Experiments were performed at least three separate times and average values are shown.
The combination mutation variant used above placed the non-optimal P3-P3′ sequence from Arg372 at Arg740 in attempt to slow cleavage at the latter site. In the following experiment, we used an additional combined variant where Arg740 was now replaced with Gln to eliminate this cleavage. This variant, designated 372(P3-P3′)740/Arg740Gln, still retains the optimal flanking residues for cleavage at Arg372 as in the above variant but is now refractory to cleavage at the newly created Gln740 residue. Earlier studies employing the single point mutations where Gln replaced Arg740 (30) showed a markedly reduced rate of cleavage at Arg372 (~700-fold) reflecting a linkage of these catalytic events. Thus the rationale for the current experiment was to assess the influence of the optimal flanking sequence surrounding Arg372 when the alternate site was non-cleavable.
Results using the 372(P3-P3′)740/Arg740Gln FVIII mutant indicated that thrombin cleavage at Arg740 was inhibited as shown by persistence of the A2-LC and A2-a3 intermediates (Figure 4B and C). However, as judged by A1 subunit generation, cleavage at Arg372 was increased by 11-fold compared with WT to a similar value observed for the 372(P3-P3′)740 variant (Figure 5A and Table 2). These results suggest placing the more optimal P3-P3′ residues at Arg372 can abrogate the influence of initial cleavages at Arg740 to yield (near) maximal rates of cleavage at Arg372.
Thrombin Activation of FVIII Proteins
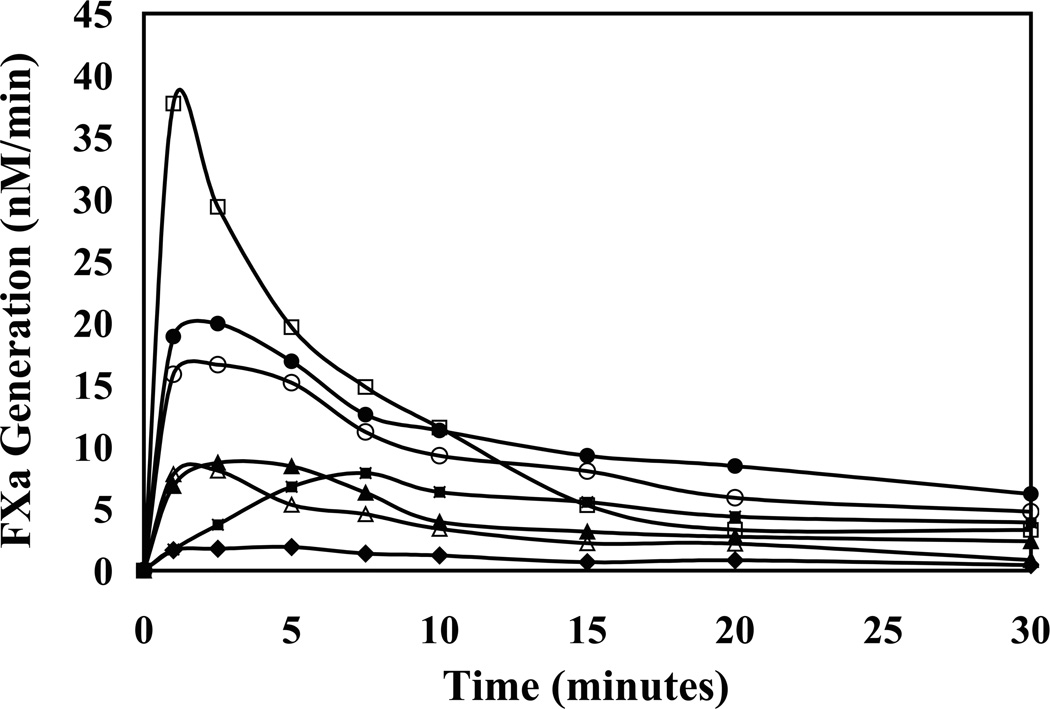
Activation of the FVIII variants was performed to assess the effects of the P3-P3′ mutations on the generation of FVIIIa activity. WT and mutant FVIII (100 nM) were reacted with thrombin over a 30 min time course and activity was monitored using a FXa generation assay as described in “Experimental Procedures.” Two levels of thrombin (5 or 0.5 nM) were used to activate WT FVIII. The rate of activation was directly proportional to the level of thrombin used. At the higher concentration of thrombin (5 nM), WT FVIII demonstrated a peak activity (~40 nM/min) at ~1 min after which cofactor activity steadily declined to ~10% peak activity at 30 minutes (Figure 6). The loss in WT FVIIIa activity during the time course is independent of thrombin and results from the dissociation of A2 subunit from the A1/A3-C1-C2 dimer leading to inactivation of the cofactor and dampening of the intrinsic factor Xase (31).When 0.5 nM thrombin was used to activate WT FVIII, the activation profile yielded a slower activation rate, an ~5-fold reduction in peak activity, and a broader plateau of activation (~5–10 min). The plateau of activity likely reflects a balance in the slower rate of activation coupled with the decay of FVIIIa (31).
Figure 6. Activation of WT and variant FVIII by thrombin.
FXa generation assays were used to monitor rates of activation of WT and variant FVIII (100 nM) by 5 nM and 0.5 nM thrombin as described in “Experimental Procedures.” Solid lines represent WT (□) with 5 nM thrombin and WT (■) and the variants 372(P3-P3′)740/740(P3-P3′)372 (●), 740(P3-P3′)372 (○), 372(P3-P3′)740 (▲), 372(P3-P3′)740/R740Q (△), and 372(P3-P3′)1689 (♦) with 0.5 nM thrombin. All experiments were performed at least three separate times and average values are shown.
FVIII variants were activated using the lower thrombin concentration. In all cases, the generation of FVIIIa occurred at a faster time course than was observed using this thrombin concentration with WT FVIII (Figure 6). Similar activity profiles were observed for the 740(P3-P3′)372 and 372(P3-P3′)740/740(P3-P3′)372 mutants. These variants demonstrated a faster rate of procofactor activation compared to WT (1–2 min versus 5–10 min) with an ~2-fold increase in peak height. The 372(P3-P3′)740 and 372(P3-P3′)740/R740Q variants also showed faster rates of activation (peaking at 1–2 min and 2–5 min, respectively) and had similar peak values of activation compared to WT FVIII. Lastly, the 372(P3-P3′)1689 variant, although showing a faster rate of activation, had an ~8-fold reduction in peak activation. This variant also demonstrated low specific activity that suggested substitution of residues affected cofactor function. However, in all cases these faster activating variants all exhibited increased rates of cleavage at Arg372 consistent with this site representing the rate limiting step in the activation mechanism.
DISCUSSION
In this study we examined thrombin-catalyzed cleavage and activation of FVIII substrates possessing swaps for the P3-P3′ sequences flanking the scissile bonds in the heavy chain. Results obtained from variants possessing single sequence swaps, a double sequence swap, or a single swap combined with point mutations to render the P1 Arg740 non-cleavable yielded several observations regarding the role of the P3-P3′ sequences in the activation mechanism. First, the sequence flanking a particular P1 makes a variable and, in some cases, substantial contribution to the rate of attack at that site, with more optimal residues for thrombin yielding enhanced reaction rates. Second, P3-P3′ sequences at the scissile bonds in the heavy chain appear to modulate the rate of catalysis at distal P1 sites. Third, comparing effects of P3-P3′ sequences flanking residues Arg740 and Arg1689, which possess differences in only the P3 and P3′ positions, suggest changes in individual residues removed from the P1 site can yield marked effects on catalytic rate. Finally, swaps that place more optimal residues flanking the rate-limiting Arg372 site result in more rapid rates of thrombin-catalyzed cleavage and procofactor activation, independent of the sequences flanking the scissile bond at Arg740 or whether that bond can be cleaved. This latter observation is of particular interest in that it reflects a disengagement of the linked pathway for heavy chain cleavage.
Examination of the P3-P3′ sequences flanking the three scissile bonds in FVIII cleaved by thrombin revealed that two of the P1 residues, Arg740 and Arg1689, are flanked by sequences containing four out of six residues deemed optimal for thrombin cleavage, whereas the sequence flanking Arg372 is optimal at only the P1 and P1′ sites (22). Results in this study showing that replacement of the sequence flanking Arg372 with those of Arg740 and Arg1689 yielded 10- and 3-fold increases in rates of cleavage at the P1 Arg. Thus enhancement in rates of catalysis at this site is clearly influenced by sequence. A primary cause for this rate enhancement is likely derived from the P2 Pro residue, which is a preferred residue at this position (16). Since the P2-P2′ sequences flanking Arg740 and Arg1689 are identical, the rate differences observed in comparing the two replacements reflect differences at the P3 and P3′ positions. Studies using libraries of natural thrombin substrates have revealed little influence of the P3 and P3′ positions on catalysis (32). However, in the case of FVIII, the P3 Glu and/or P3′ Ser present in the Arg740 sequence appear somewhat more preferred for cleavage compared with the Ser and/or Gln in the Arg1689 sequence as judged by the ~3-fold difference in cleavage rates at the substituted Arg372 site.
Both of the Arg740 and Arg1689 sequences affected faster cleavage at Arg372 and yielded faster activation kinetics, consistent with this site representing the apparent rate limiting step in the activation mechanism (23). While replacement with the Arg740 site was benign with respect to specific activity, replacement with the Arg1689 flanking sequence yielded a marked (>80%) reduction in this parameter. The reason for this is not clear but may reflect replacement of specific residues, specifically, the P3 Ser and/or P3′ Gln that inhibit FVIII function. Examination of a Hemophilia A database (33) does not reveal any missense mutations at either residue position 370 (P3) or 375 (P3′) affecting phenotype.
Earlier studies from our laboratory examining thrombin-catalyzed cleavage and activation of FVIII when individual P1 sites were rendered non-cleavable by replacement of the P1 Arg with Gln showed mutation at one site had significant effects at unmodified P1 sites. For example, rates of A1 and A2 subunit generation were reduced ~8–10-fold using a FVIII variant possessing an Arg1689Gln mutation (21). Furthermore, an Arg740Gln variant showed an ~140-fold reduced rate of cleavage at Arg1689 (30). Results from this study show an ~2-fold reduction in the rate of cleavage at Arg1689 when the Arg740Gln mutation is coupled with the 372(P3-P3′)740 mutation. The reason for this large disparity in relative reaction rates may reflect the markedly faster cleavage rate at the Arg372 site in the latter variant that in turn modulates the reaction at the light chain.
We have speculated that the apparent cleavage order of Arg740 > Arg1689 > Arg372 leading to procofactor activation represents a competition for active site docking to sites in the two chains of the FVIII heterodimer with initial attack of the heavy chain at Arg740 (followed by subsequent cleavage at Arg372) representing the favored pathway over initial cleavage at Arg1689 (21). Furthermore, the slow cleavage of Arg372 was thought to be a result of the requirement to first cleave Arg740, possibly requiring a ratcheting mechanism (17) to permit subsequent active site docking. Interestingly, mutations that place optimal residues flanking the Arg372 appear to obviate the need for initial cleavage at Arg740 to facilitate Arg372 cleavage. We observed that either placing the non-optimal residues flanking Arg372 at Arg740 to slow cleavage at this site or replacing the P1 Arg740 with Gln resulted in sustained high rates of cleavage at Arg372 (~10-fold compared with WT) when this site was flanked with the P3-P3′ sequence from Arg740. This faster cleavage of Arg372 for these two variants was confirmed following FVIII activation at low thrombin levels which yielded peak activity values during a shorter time course than was observed for the WT protein. Taken together, these results suggested that thrombin bound to FVIII may have access to all three scissile bonds in the FVIII substrate and catalysis at a given bond reflects in large part docking preferences based upon S and P site interactions.
Sequences flanking scissile bonds and their complementary interaction with binding pocket residues of the proteinase make marked contributions to catalytic rate values in a number of coagulation proteinase systems. For example, exchanging the P2 and P3 binding pocket residues of FXa and activated protein C switched the enzyme specificities for chromogenic substrates and inhibitors (34). In another study, replacing the P2 and P3 residues of prethrombin 2 (Asp-Gly) with those from FIXa (Leu-Thr) resulted in ~20- and ~40-fold reductions in kcat for FXa and prothrombinase, respectively (18). Additionally this study also showed that Km values for prothrombin were essentially unaffected by this and other residue replacements, consistent with dependence for substrate recognition and affinity on exosite interactions.
This influence of P1 flanking sequences on the catalytic mechanism of thrombin activation of FVIII appears to represent a common theme for cleavage of FVIII substrates by exosite-driven enzymes. In an earlier study (19) examining the role of P4–P3′ sequences flanking the two scissile bonds in FVIII cleaved by activated protein C, we observed replacing the faster-reacting site at Arg336 in the A1 domain with the slower reacting site at Arg562 in the A2 domain resulted in as much as an ~100-fold rate reduction in cleavage at the P1 Arg336 in FVIIIa, whereas the reciprocal swap accelerated cleavage at Arg562 in the procofactor by ~25-fold. Inasmuch as little information is available regarding S4–S3′ sites in this proteinase, discrimination of optimal and non-optimal P residues could not be distinguished.
Overall, results from the current study provide insights into the mechanism of the specificity of thrombin for the P1 sites of FVIII. Faster cleavage rates at Arg740 and Arg1689 can be attributed in part to more optimal residues in the P3-P3′ region, while the relatively slower cleavage rate at Arg372 can be accelerated by replacement with more optimal residues for thrombin cleavage. Furthermore, this resultant acceleration appears to reflect the uncoupling of the linkage between cleavage of the two scissile bonds in FVIII heavy chain. Thus, the P3-P3′ residues surrounding Arg740, Arg1689, and Arg372 in FVIII impact rates of thrombin proteolysis at each site and contribute to the mechanism for thrombin activation of the procofactor.
ACKNOWLEDGEMENTS
We would like to thank Pete Lollar and John Healey for the gift of the FVIII cloning and expression vectors and Zaverio Ruggeri, Bill Church, and Lisa Regan for the C5, GMA-8003, and 2D2 monoclonal antibodies, respectively.
Footnotes
Funding
This work was supported by grants HL38199 and HL76213 from the National Institutes of Health (to P.J.F.).
Abbreviations used: BHK, baby hamster kidney; HEPES, N-[2-hydroxyethyl]piperazine-N′-[2-ethanesulfonic acid]; SDS, sodium dodecyl sulfate; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; ELISA, enzyme linked immunosorbent assay; WT, wild-type; FVIII, factor VIII; FVIIIa, factor VIIIa; FIXa, factor IXa; FX, factor X; FXa, factor Xa; ABE, anion binding exosite.
Residues flanking the scissile bonds are denoted using the nomenclature of Schecter and Berger (35). Residues preceding the scissile bond are designated P1..Pn extending toward the NH2-terminus and residues following the scissile bond are designated P1'..Pn' extending toward the COOH-terminus.
REFERENCES
- 1.Vehar GA, Keyt B, Eaton D, Rodriguez H, O'Brien DP, Rotblat F, Oppermann H, Keck R, Wood WI, Harkins RN, et al. Structure of human factor VIII. Nature. 1984;312:337–342. doi: 10.1038/312337a0. [DOI] [PubMed] [Google Scholar]
- 2.Wood WI, Capon DJ, Simonsen CC, Eaton DL, Gitschier J, Keyt B, Seeburg PH, Smith DH, Hollingshead P, Wion KL, et al. Expression of active human factor VIII from recombinant DNA clones. Nature. 1984;312:330–337. doi: 10.1038/312330a0. [DOI] [PubMed] [Google Scholar]
- 3.Fay PJ, Anderson MT, Chavin SI, Marder VJ. The size of human factor VIII heterodimers and the effects produced by thrombin. Biochim Biophys Acta. 1986;871:268–278. doi: 10.1016/0167-4838(86)90208-6. [DOI] [PubMed] [Google Scholar]
- 4.Mann KG, Nesheim ME, Church WR, Haley P, Krishnaswamy S. Surface-dependent reactions of the vitamin K-dependent enzyme complexes. Blood. 1990;76:1–16. [PubMed] [Google Scholar]
- 5.Eaton D, Rodriguez H, Vehar GA. Proteolytic processing of human factor VIII. Correlation of specific cleavages by thrombin, factor Xa, and activated protein C with activation and inactivation of factor VIII coagulant activity. Biochemistry. 1986;25:505–512. doi: 10.1021/bi00350a035. [DOI] [PubMed] [Google Scholar]
- 6.Parker ET, Pohl J, Blackburn MN, Lollar P. Subunit structure and function of porcine factor Xa-activated factor VIII. Biochemistry. 1997;36:9365–9373. doi: 10.1021/bi970599o. [DOI] [PubMed] [Google Scholar]
- 7.Pieters J, Lindhout T, Hemker HC. In situ-generated thrombin is the only enzyme that effectively activates factor VIII and factor V in thromboplastin-activated plasma. Blood. 1989;74:1021–1024. [PubMed] [Google Scholar]
- 8.Fay PJ, Mastri M, Koszelak ME, Wakabayashi H. Cleavage of factor VIII heavy chain is required for the functional interaction of A2 subunit with factor IXa. J. Biol. Chem. 2001;276:12434–12439. doi: 10.1074/jbc.M009539200. [DOI] [PubMed] [Google Scholar]
- 9.Donath MJ, Lenting PJ, Van Mourik JA, Mertens K. Kinetics of factor VIII light-chain cleavage by thrombin and factor Xa. A regulatory role of the factor VIII heavy-chain region Lys713-Arg740. Eur. J. of Biochem. 1996;240:365–372. doi: 10.1111/j.1432-1033.1996.0365h.x. [DOI] [PubMed] [Google Scholar]
- 10.Lollar P, Hill-Eubanks DC, Parker CG. Association of the factor VIII light chain with von Willebrand factor. J. Biol. Chem. 1988;263:10451–10455. [PubMed] [Google Scholar]
- 11.Regan LM, Fay PJ. Cleavage of factor VIII light chain is required for maximal generation of factor VIIIa activity. J. Biol. Chem. 1995;270:8546–8552. doi: 10.1074/jbc.270.15.8546. [DOI] [PubMed] [Google Scholar]
- 12.Fay PJ, Haidaris PJ, Smudzin TM. Human factor VIIIa subunit structure. Reconstruction of factor VIIIa from the isolated A1/A3-C1-C2 dimer and A2 subunit. J. Biol. Chem. 1991;266:8957–8962. [PubMed] [Google Scholar]
- 13.Lollar P, Parker ET. Structural basis for the decreased procoagulant activity of human factor VIII compared to the porcine homolog. J. Biol. Chem. 1991;266:12481–12486. [PubMed] [Google Scholar]
- 14.Fay PJ, Smudzin TM, Walker FJ. Activated protein C-catalyzed inactivation of human factor VIII and factor VIIIa. Identification of cleavage sites and correlation of proteolysis with cofactor activity. J. Biol. Chem. 1991;266:20139–20145. [PubMed] [Google Scholar]
- 15.Plantier JL, Rolli V, Ducasse C, Dargaud Y, Enjolras N, Boukerche H, Negrier C. Activated factor X cleaves factor VIII at arginine 562, limiting its cofactor efficiency. J. Thromb. Haemost. 2010;2:286–293. doi: 10.1111/j.1538-7836.2009.03675.x. [DOI] [PubMed] [Google Scholar]
- 16.Di Cera E. Thrombin interactions. Chest. 2003;124:11S–17S. doi: 10.1378/chest.124.3_suppl.11s. [DOI] [PubMed] [Google Scholar]
- 17.Krishnaswamy S. Exosite-driven substrate specificity and function in coagulation. J. Thromb. Haemost. 2005;3:54–67. doi: 10.1111/j.1538-7836.2004.01021.x. [DOI] [PubMed] [Google Scholar]
- 18.Orcutt SJ, Pietropaolo C, Krishnaswamy S. Extended interactions with prothrombinase enforce affinity and specificity for its macromolecular substrate. J. Biol. Chem. 2002;277:46191–46196. doi: 10.1074/jbc.M208677200. [DOI] [PubMed] [Google Scholar]
- 19.Varfaj F, Wakabayashi H, Fay PJ. Residues surrounding Arg336 and Arg562 contribute to the disparate rates of proteolysis of factor VIIIa catalyzed by activated protein C. J. Biol. Chem. 2007;282:20264–20272. doi: 10.1074/jbc.M701327200. [DOI] [PubMed] [Google Scholar]
- 20.Nogami K, Zhou Q, Myles T, Leung LL, Wakabayashi H, Fay PJ. Exosite-interactive regions in the A1 and A2 domains of factor VIII facilitate thrombin-catalyzed cleavage of heavy chain. J. Biol. Chem. 2005;280:18476–18487. doi: 10.1074/jbc.M412778200. [DOI] [PubMed] [Google Scholar]
- 21.Newell JL, Fay PJ. Cleavage at Arg-1689 influences heavy chain cleavages during thrombin-catalyzed activation of factor VIII. J. Biol. Chem. 2009;284:11080–11089. doi: 10.1074/jbc.M900234200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bode W, Brandstetter H, Mather T, Stubbs MT. Comparative analysis of haemostatic proteinases: structural aspects of thrombin, factor Xa factor IXa and protein C. Thromb. Haemost. 1997;78:501–511. [PubMed] [Google Scholar]
- 23.Hill-Eubanks DC, Lollar P. von Willebrand factor is a cofactor for thrombin-catalyzed cleavage of the factor VIII light chain. J. Biol. Chem. 1990;265:17854–17858. [PubMed] [Google Scholar]
- 24.Mimms LT, Zampighi G, Nozaki Y, Tanford C, Reynolds JA. Phospholipid vesicle formation and transmembrane protein incorporation using octyl glucoside. Biochemistry. 1981;20:833–840. doi: 10.1021/bi00507a028. [DOI] [PubMed] [Google Scholar]
- 25.Casillas G, Simonetti C, Pavolvsky A. Artificial substrate for the assay of factors V and VIII. Coagulation. 1971;4:107–111. [Google Scholar]
- 26.Jenkins PV, Freas J, Schmidt KM, Zhou Q, Fay PJ. Mutations associated with hemophilia A in the 558–565 loop of the factor VIIIa A2 subunit alter the catalytic activity of the factor Xase complex. Blood. 2002;100:501–508. doi: 10.1182/blood-2001-12-0361. [DOI] [PubMed] [Google Scholar]
- 27.Wakabayashi H, Freas J, Zhou Q, Fay PJ. Residues 110–126 in the A1 domain of factor VIII contain a Ca2+ binding site required for cofactor activity. J. Biol. Chem. 2004;279:12677–12684. doi: 10.1074/jbc.M311042200. [DOI] [PubMed] [Google Scholar]
- 28.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 29.Lollar P, Fay PJ, Fass DN. Factor VIII and factor VIIIa. Methods Enzymol. 1993;222:128–143. doi: 10.1016/0076-6879(93)22010-d. [DOI] [PubMed] [Google Scholar]
- 30.Newell JL, Fay PJ. Proteolysis at Arg740 facilitates subsequent bond cleavages during thrombin-catalyzed activation of factor VIII. J. Biol. Chem. 2007;282:25367–25375. doi: 10.1074/jbc.M703433200. [DOI] [PubMed] [Google Scholar]
- 31.Fay PJ. Activation of factor VIII and mechanisms of cofactor action. Blood Rev. 2004;18:1–15. doi: 10.1016/s0268-960x(03)00025-0. [DOI] [PubMed] [Google Scholar]
- 32.Edwards PD, Mauger RC, Cottrell KM, Morris FX, Pine KK, Sylvester MA, Scott CW, Furlong ST. Synthesis and enzymatic evaluation of a P1 arginine aminocoumarin substrate library for trypsin-like serine proteases. Bioorg. Med. Chem. 2000;10:2291–2294. doi: 10.1016/s0960-894x(00)00460-1. [DOI] [PubMed] [Google Scholar]
- 33.Kemball-Cook G, Tuddenham EG, Wacey AI. The factor VIII Structure and Mutation Resource Site: HAMSTeRS version 4. Nucleic Acids Res. 1998;26:216–219. doi: 10.1093/nar/26.1.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rezaie AR. Role of residue 99 at the S2 subsite of factor Xa and activated protein C in enzyme specificity. J. Biol. Chem. 1996;271:23807–23814. doi: 10.1074/jbc.271.39.23807. [DOI] [PubMed] [Google Scholar]
- 35.Schechter I, Berger A. On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]