Abstract
The protein tyrosine kinase Jak2 mediates signaling through numerous cytokine receptors. Jak2 possesses a pseudokinase domain (JH2) and a tyrosine kinase domain (JH1). Through unknown mechanisms, JH2 regulates the catalytic activity of JH1, and hyperactivating mutations in the JH2 region of human Jak2 are causative for myeloproliferative neoplasms (MPNs). We showed previously that Jak2 JH2 is in fact catalytically active. Here, we present crystal structures of human Jak2 JH2, both wild-type and the most prevalent MPN mutant, V617F. The structures reveal that JH2 adopts the fold of a prototypical protein kinase but binds Mg-ATP non-canonically. The structural and biochemical data indicate that the V617F mutation rigidifies α-helix C in the N lobe of JH2, which facilitates trans-phosphorylation of JH1. The crystal structures of JH2 afford new opportunities for the design of novel Jak2 therapeutics targeting MPNs.
Jak2, a member of the Janus family of protein tyrosine kinases (Jak1–3, Tyk2), associates with the cytoplasmic regions of various cytokine receptors, including those for growth hormone, erythropoietin, leptin, interferon-γ and interleukins IL-3 and IL-5 (ref. 1). Jak2 is activated through cytokine-mediated receptor dimerization or rearrangement and signals through the Jak-Stat (signal transducer and activators of transcription) pathway2, which is essential for myeloid cell development, proliferation and survival, as well as for the initial stages of the immune response.
Jaks possess an N-terminal FERM (band 4.1, ezrin, radixin, moesin) domain, a Src homology-2 (SH2)-like domain, a pseudokinase domain (Jak homology-2, JH2), and a C-terminal tyrosine kinase domain (JH1). Extensive biochemical studies on Jaks have established that1: (i) the FERM domain is primarily responsible for the association of Jaks with cytokine receptors, (ii) the SH2-like domain does not function as a conventional phosphotyrosine-binding domain and its precise role is unclear, (iii) JH1 is activated via trans-phosphorylation of tandem tyrosines in the activation loop (Tyr1007 and Tyr1008 in Jak2) and (iv) JH2 regulates the activity of JH1. At present, structural information for Jaks is limited to JH1 (refs. 3–6).
Mutations in Jaks are causative for myeloproliferative neoplasms (MPNs) in humans, which are clonal proliferative disorders affecting different myeloid lineages7. The molecular etiologies for the more common MPNs, namely, polycythemia vera, essential thrombocythemia and primary myelofibrosis, have been identified and shown to be caused in most cases by mutations in the JH2 region of Jak2 (refs. 7,8). Mutations in JH2 of Jak1–3 have also been linked to leukemias, such as acute lymphoblastic leukemia and acute myeloid leukemia8. All of these mutations result in constitutive tyrosine kinase activity of Jak2. V617F in JH2 of Jak2 is the most commonly identified mutation in MPNs and is responsible for >95% of cases of polycythemia vera and ~50% of cases of essential thrombocythemia and primary myelofibrosis9–12. This mutation has also recently been implicated in non-small-cell lung cancer13.
The molecular mechanisms underlying JH2 regulation of JH1 in Jaks are poorly understood. The MPN pathogenic data imply an inhibitory function for JH2, but some mutations in JH2 of Jak3 are loss-of-function, causing severe combined immunodeficiency (SCID)14, suggestive of a positive regulatory function for JH2. The dual regulatory nature of JH2 is supported by biochemical studies of JH2 deletion mutants, which showed that basal activity is elevated, but cannot be further stimulated by cytokine and is well below that of cytokine-stimulated full-length Jak2 (refs. 15–17). A molecular model for an autoinhibitory interaction between JH2 and JH1, which would hinder trans-phosphorylation of the JH1 activation loop, has been proposed18 but never verified experimentally. The molecular nature of the positive (stimulatory) interaction mediated by JH2, which would facilitate trans-phosphorylation of JH1, has received far less attention.
We recently showed that, despite several amino-acid substitutions thought to render it inactive, Jak2 JH2 is catalytically active and phosphorylates two negative regulatory sites in Jak2, Ser523 and Tyr570 (ref. 19). To gain insights into the molecular basis for this catalytic activity and for the constitutive activity of the pathogenic mutant V617F, we undertook crystallographic and biochemical studies of Jak2 JH2. The results show that Jak2 JH2 adopts the fold of a eukaryotic protein kinase, but binds Mg-ATP via a novel mode. Molecular dynamics simulations in conjunction with the crystallographic studies indicate that the mutation V617F rigidifies an α helix (αC) in the N lobe of JH2, which stabilizes the JH2 stimulatory interaction necessary for Jak2 activation.
RESULTS
Crystal structure of Jak2 JH2
We initially engineered two baculoviruses to encode human Jak2 JH2, residues 536–827, both wild-type and V617F. We expressed these two proteins in soluble form in insect cells, but the proteins showed signs of aggregation during purification and did not yield crystals. Based on our homology model of Jak2 JH2, we identified three putative solvent-exposed hydrophobic residues (all in the C lobe) for possible substitution: Trp659, Trp777 and Phe794 (each of which is not conserved in Jaks). Full-length Jak2 bearing the mutations W659A, W777A and F794H was phosphorylated in cells to the same extent as wild-type (data not shown). The mutated versions of wild-type and V617F JH2 (see Methods for details), also with a new C-terminus at residue 812, expressed at higher levels, were better behaved during purification and readily yielded crystals. These proteins will hereafter be referred to simply as JH2-WT (wild-type) and JH2-VF (V617F).
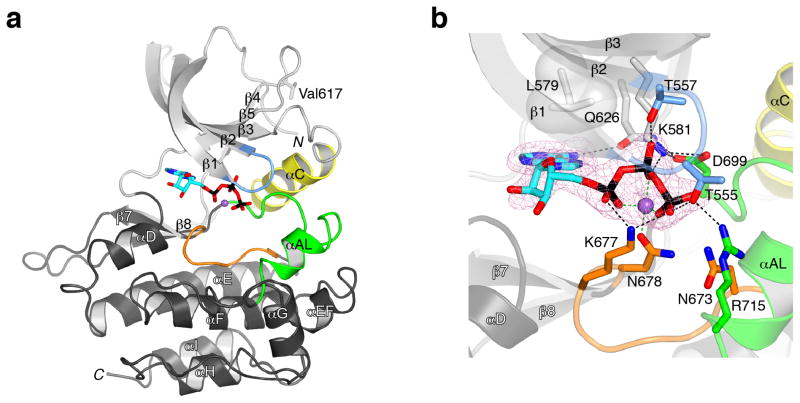
We obtained crystal structures of JH2-WT without nucleotide (apo; 2.0-Å resolution) and with Mg-ATP bound (co-crystallization; 1.75-Å resolution), each with one JH2 molecule per asymmetric unit. Data collection and refinement statistics appear in Table 1. Jak2 JH2 adopts the prototypical serine-threonine and tyrosine kinase fold, with an N lobe comprising a five-stranded β sheet and one α helix (αC), and a C lobe that is mainly α-helical (Fig. 1a). The domain structure of JH2 begins at Phe537 and ends at Leu808, residues that are conserved in JH2 of Jaks. Notable overall features of the JH2 structure include a relatively short (non-phosphorylatable) activation loop (seven residues shorter than in Jak2 JH1), which terminates in an α helix, and an extended loop between β-strand 7 (β7) and β8 (eight residues longer than the corresponding loop in JH1).
Table 1.
X-ray data collection and refinement statistics
| JH2-WT (apo) | JH2-WT (Mg-ATP) | JH2-VF (Mg-ATP) | |
|---|---|---|---|
| Data collection | |||
| Space group | P21 | P21 | P21 |
| Cell dimensions | |||
| a, b, c (Å) | 44.4, 57.1, 61.0 | 44.6, 57.5, 60.9 | 46.9, 57.3, 60.5 |
| α β γ(°) | 90.0, 110.4, 90.0 | 90.0, 110.8, 90.0 | 90.0, 111.8, 90.0 |
| Resolution (Å) | 50.0–2.0 | 50.0–1.75 | 50.0–2.0 |
| Rsym or Rmerge | 3.9 (12.1) | 6.6 (43.1) | 5.5 (21.5) |
| I/σI | 37.3 (12.1) | 25.5 (2.6) | 19.1 (2.8) |
| Completeness (%) | 99.6 (99.4) | 99.2 (94.1) | 95.3 (66.4) |
| Redundancy | 3.7 | 6.4 | 3.5 |
| Refinement | |||
| Resolution (Å) | 50.0–2.0 | 50.0–1.75 | 50.0–2.0 |
| No. reflections | 18,109 | 27,543 | 18,524 |
| Rwork/Rfree | 18.1/21.7 | 17.8/20.6 | 18.1/22.8 |
| No. atoms | |||
| Protein | 2,145 | 2,133 | 2,144 |
| Ligand/ion | 12 | 42 | 34 |
| Water | 152 | 147 | 111 |
| B-factors | |||
| Protein | 31.9 | 32.7 | 39.6 |
| Solvent | 39.6 | 39.3 | 41.8 |
| r.m.s deviations | |||
| Bond lengths (Å) | 0.007 | 0.009 | 0.008 |
| Bond angles (°) | 1.2 | 1.4 | 1.3 |
One crystal was used per data set. Values in parentheses are for highest-resolution shell.
Figure 1.
Crystal structure of Jak2 JH2. (a) Ribbon diagram of the structure of JH2. The N lobe of JH2 is colored light gray expect for the nucleotide-binding loop (blue) and αC (yellow). The C lobe is colored dark gray except for the catalytic loop (orange) and the activation loop (green). The α helices and β strands are labeled (shown semi-transparent), as are the N- and C-termini (N and C). ATP is shown in stick representation and colored cyan (carbon), red (oxygen), blue (nitrogen) or black (phosphorus), and the Mg2+ ion is colored purple. The side chain of Val617, the site of the pathogenic mutation V617F (in the β4-β5 loop), is shown in stick representation. (b) Mode of ATP binding in JH2. The viewing angle is approximately the same as in a. Select side chains are shown, with carbon atoms colored according to the residue’s location, as in a (e.g., orange for catalytic loop). Superimposed is an electron density map (Fo-Fc, pink mesh, contoured at 3σ) computed without Mg-ATP in the model (but present during refinement). Hydrogen bonds and salt bridges are represented by black dashed lines, and Mg2+ coordination (≤2.1 Å) is represented by green dashed lines.
JH2s of Jaks lacks several residues that, in canonical protein kinases, are important for catalysis (Supplementary Fig. 1). The catalytic loop in canonical protein kinases contains a conserved aspartate that plays a key role in the phosphoryl transfer reaction. In Jak JH2s, an asparagine (Asn673 in Jak2) replaces the aspartate. The activation loop of canonical protein kinases begins with a DFG sequence motif, whereas Jak JH2s contain DPG. Finally, in canonical protein kinases, a conserved lysine in β3 (Lys581 in Jak2) is salt-bridged to a conserved glutamate in αC, the latter of which is replaced by alanine or threonine in Jak JH2s (Ala597 in Jak2).
ATP binding mode and comparison with other protein kinases
As in canonical protein kinases, Mg-ATP binds in the cleft between the N and C lobes of Jak2 JH2 (Fig. 1a,b). The structures of JH2-WT, with and without bound Mg-ATP, are very similar, with a root-mean-square deviation (r.m.s.d.) in Cα positions (residues 537–808) of only 0.44 Å. The apo structure exhibits a closed lobe configuration, which is not appreciably altered upon Mg-ATP binding. This trait is probably due in part to a C lobe-N lobe contact mediated by conserved (Jak family) Arg715 at the end of the activation loop, which in both the apo and ATP-bound forms of JH2 makes a hydrogen bond with Thr555 in the nucleotide-binding loop (Fig. 1b). One salient difference between the apo and Mg-ATP-bound structures is found in αC. In the apo structure, αC is disrupted midway by an intercalating water molecule that bridges backbone atoms Phe594(O) and Ala598(N) in the helix. Binding of Mg-ATP displaces this water molecule, but the backbone hydrogen-bonding in αC remains irregular (Supplementary Fig. 2).
There are several features that distinguish the ATP-binding mode in JH2 from that in canonical protein kinases (Fig. 1b). In JH2, the residue from the N lobe that flanks the adenine base is Leu579 (β3), whereas alanine is highly conserved at this position (VA(V/I)K) motif) in canonical protein kinases. Two threonine residues in the nucleotide-binding loop of JH2 are hydrogen-bonded to ATP phosphate groups: Thr555 with the γ phosphate and Thr557 with the β phosphate. Thr557 is typically a glycine in canonical protein kinases (GXGXXG motif, where X is any amino acid). The so-called gatekeeper residue in the back of the ATP-binding cleft is typically hydrophobic, but in JH2 is a glutamine (Gln626), which is hydrogen-bonded to the adenine base and to Asp699 (DPG) in the activation loop.
A single Mg2+ ion is present in the structure, which is coordinated by Asn678 (catalytic loop), an oxygen atom from each of the three ATP phosphate groups and one water molecule (Fig. 1b). In canonical protein kinases such as (serine-threonine) protein kinase A (PKA)20 or the insulin receptor tyrosine kinase21, two Mg2+ ions are present, and the aspartate of the DFG motif coordinates both ions (Supplementary Fig. 3a). Rather than coordinating a Mg2+ ion, Asp699 in JH2 (DPG) is salt-bridged to Lys581, the conserved β3 lysine. This interaction evidently substitutes for the canonical β3 lysine-αC glutamate salt bridge. A superposition of the active sites of JH2 and PKA reveals that the γ phosphate of ATP in the JH2 structure is positioned for phosphoryl transfer (Supplementary Fig. 3a).
JH2s of Jaks shares several sequence characteristics with Her3 (ErbB3), a member of the epidermal growth factor receptor family. Her3 was also characterized as a pseudokinase, but was recently shown to possess weak catalytic activity22. In both kinases, the conserved glutamate inαC is absent, and asparagine replaces aspartate in the catalytic loop. In the Her3 structure22,23, Asp833 from the DFG motif is salt-bridged to the β3 lysine (Lys723), similar to Asp699 in Jak2 JH2, but it also coordinates the lone Mg2+ ion (Supplementary Fig. 3b). The γ phosphate of the nucleotide is in a different position in the two structures, which could be due to co-crystallization with AMPPNP (Her3) versus ATP (Jak2).
Cis- versus trans-autophosphorylation of Ser523 and Tyr570
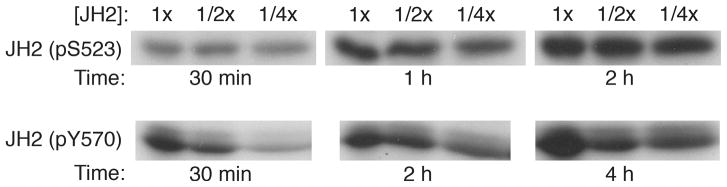
We determined whether the two sites we had identified previously19, Ser523 and Tyr570, are autophosphorylated via a cis- or trans-mechanism. Simple modeling based on the JH2 crystal structure suggests that Ser523, in the SH2-JH2 linker, could possibly reach the JH2 active site to be autophosphorylated in cis, whereas Tyr570 in the β2-β3 loop of JH2, far removed from the active site (Supplementary Fig. 4), would necessarily be autophosphorylated in trans. As described previously19, we purified a longer form of JH2 (residues 513–827, which includes Ser523) by anion-exchange chromatography, which yielded two JH2 peaks representing unphosphorylated and Ser523-phosphorylated JH2. We incubated unphosphorylated JH2 with Mn-[γ-32P]ATP at several different protein concentrations and for several time points. The data show that the phosphorylation level of Ser523 is independent of JH2 concentration, consistent with autophosphorylation in cis (Fig. 2, top). Moreover, these data provide further proof, beyond expression of a catalytically inactive JH2 mutant (K581A)19, that Ser523 is phosphorylated by JH2 and not by a contaminating protein kinase, which would necessarily be concentration-dependent. Similarly, we performed an autophosphorylation reaction with Ser523-phosphorylated JH2 (from the second ion-exchange peak). In this case, the phosphorylation level of Tyr570 was concentration-dependent, consistent with autophosphorylation in trans (Fig. 2, bottom).
Figure 2.
Cis- versus trans-autophosphorylation of Jak2 JH2. Autophosphorylation reactions were performed at three different JH2 concentrations (1/4x, most dilute), and aliquots were taken at the indicated time points. Equal amounts of protein were loaded in each lane for SDS-PAGE and autoradiography. Top: the starting sample is unphosphorylated JH2, which becomes phosphorylated at Ser523 (see ref. 19). Bottom: the starting sample is Ser523-phosphorylated JH2, which becomes phosphorylated at Tyr570.
Crystal structure of Jak2 JH2 V617F
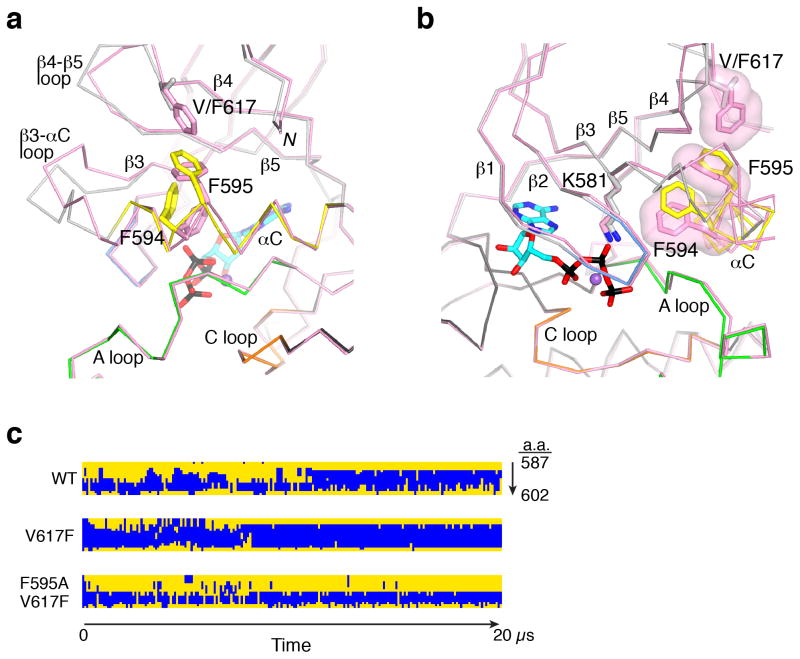
We obtained a crystal structure at 2.0-Å resolution of the pathogenic JH2 mutant, V617F (JH2-VF), with Mg-ATP bound, in the same monoclinic lattice as JH2-WT (Table 1). Val617 is situated in the β4-β5 loop in the N lobe of JH2 (Fig. 1a). Overall, the structures of JH2-VF and JH2-WT are highly similar, with an r.m.s.d. in Cα positions (residues 537–808) of 0.76 Å (Fig. 3a). The mode of nucleotide binding to JH2-VF is indistinguishable from that of JH2-WT. Substantive structural deviations between JH2-VF and JH2-WT occur in αC and in the β3-αC and β4-β5 loops. Most notably, in contrast to its distorted structure in JH2-WT, αC in JH2-VF exhibits continuous backbone hydrogen-bonding and is extended by an additional turn on the N-terminal end (Supplementary Fig. 2). Phe617 (replacing valine) causes a rotation in the phenyl ring of Phe595 (αC) and induces a major shift in the side-chain position of neighboring Phe594 (Fig. 3a). Phe617, Phe595 and Phe594 form π-stacking interactions (T-shaped) (Fig. 3b), with a closest inter-ring carbon-carbon distance of 3.8 Å for both Phe617-Phe595 and Phe595-Phe594. The Phe594 perturbation alters slightly the side-chain position of Lys581 (Fig. 3b), which might explain why the catalytic activity of JH2 is impaired in V617F19.
Figure 3.
Comparison of JH2 V617F and wild-type structures. (a) Cα trace comparing JH2-VF to JH2-WT in the N lobe. The view is approximately 90° from that in Fig. 1a, with the same coloring of secondary-structure elements for JH2-WT (e.g., yellow for αC). JH2-VF is colored pink. The side chains of Phe594, Phe595 and Val or Phe617 are shown in stick representation. Mg-ATP is represented as in Fig. 1a. The N-terminus (residue 536) is labeled N. The catalytic loop (C loop), the activation loop (A loop), αC and other select segments are labeled. (b) The view is rotated by 90° from that in a, and the molecular surfaces of Phe594, Phe595 and Phe617 are included. (c) Results of molecular dynamic simulations for wild-type (WT), V617F and F595A V617F JH2. The secondary-structure assignment for each residue in the αC region (amino acids (a.a.) 587–602), as given by DSSP47, is plotted top-to-bottom as a function of simulation time (20 μs total). Residues in α-helical conformation are colored blue, and residues in all other conformations (e.g., coil, turn, 310 helix, etc.) are colored yellow.
Molecular dynamics simulations
To explore further the structural differences between JH2-WT and JH2-VF and the relative stability of αC, we performed long-scale (~20 μs) Molecular dynamics simulations of the two proteins in their apo forms. The simulations show that αC in wild-type JH2 is prone to melting and that Phe617 stabilizes the helix (Fig. 3c), corroborating the crystallographic data. Stabilization of αC is likely due in part to the π-stacking interactions between Phe617, Phe595 and Phe594. However, during the 20-μs simulation, these three residues are in contact only transiently, suggesting that Phe617 stabilizes αC through indirect mechanisms as well. The simulations also indicate that, overall, JH2 is intrinsically flexible in comparison with other protein kinases. The catalytic loop is one of the more stable polypeptide segments in protein kinases and, over the simulation, the r.m.s.d. in Cα positions for the catalytic loop of JH2-WT (residues 671–678) was 1.99 Å, versus 0.76 Å for the same eight residues in the catalytic loop of the Src tyrosine kinase (residues 384–391). This is probably due, at least in part, to a glycine in the JH2 catalytic loop (Gly672 in Jak2) in place of the canonical arginine (HRD motif; Supplementary Fig. 1). A small amino acid is required at this position to accommodate the shorter activation loop of JH2 and its divergent conformation.
Effects of JH2 mutations in full-length Jak2
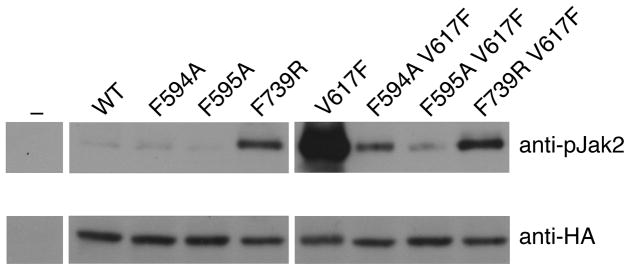
To probe the importance of Phe594 and Phe595 in αC for the constitutive activity of V617F, we introduced the point mutations F594A or F595A into either HA-tagged wild-type Jak2 or V617F (double mutants) and expressed the proteins in γ2A mammalian cells that lack endogenous Jak2. As observed previously, the basal (non-cytokine-stimulated) activation state of Jak2 V617F, as measured by JH1 activation-loop phosphorylation (pY1007, pY1008), is markedly enhanced compared to wild-type Jak2 (Fig. 4). The single point mutations in αC (F594A or F595A) did not substantially change the basal activation state of Jak2, but they caused a dramatic loss of constitutive activity of V617F (Fig. 4 and refs. 24,25). To investigate whether destabilization ofαC by F595A could account for the loss of V617F activity, we performed molecular dynamic simulations on the double mutant, F595A V617F. Indeed, F595A caused a reversion of αC stability back to the wild-type level, if not below (Fig. 3c). Because F595A suppresses not only the hyperactivity of (proximal) V617F, but also of R683G (β7-β8 loop; Supplementary Fig. 4), and even of T875N in JH1 (ref. 24), it suggests that F595A intrinsically destabilizes αC and that the structural integrity of αC is critical for the stimulatory interaction mediated by JH2.
Figure 4.
Basal activation state of Jak2 mutants in mammalian cells. Lysates from transiently transfected (or not, ‘−’) γ2A cells were immunoprecipitated with anti-HA antibodies and immunoblotted with either anti-pJak2 (pY1007-1008) antibodies (top) or anti-HA antibodies (bottom; Jak2 expression control). These data are all from the same two blots.
Because destabilization of αC in the N lobe was found to suppress the constitutive activity of V617F, we asked what effect destabilization of the C lobe would have on Jak2 activity. For this purpose, we introduced the mutation F739R. Phe739 in αF is buried in the hydrophobic core of the C lobe (Supplementary Fig. 4), and mutation to arginine should severely destabilize the C lobe. In contrast to F594A and F595A, F739R showed a marked increase in basal phosphorylation of Jak2 in γ2A cells (Fig. 4). Similar to F594A and F595A, but to a lesser extent, F739R suppressed the activity of V617F. The behavior of F739R largely mimics that of the Jak2 JH2 deletion mutant15 —increased but sub-maximal Jak2 activity—and, from the comparison with F595A (no increase in basal activity, major suppression of V617F activity), suggests that the C lobe plays a more important role in the JH2-mediated inhibitory interaction than it does in the stimulatory interaction.
DISCUSSION
The crystal structure of Jak2 JH2, a domain that was originally characterized as a pseudokinase but shown by us to possess catalytic activity19, reveals that it adopts a prototypical eukaryotic protein kinase fold, but engages Mg-ATP via a novel binding mode (Fig. 1b). Previously, we measured a Kd for (mant-)ATP binding of 1 μM19, which is substantially lower than for typical protein kinases (Kd = 36 μM for PKA26). The higher affinity is evidently due to the additional, non-canonical interactions with Mg-ATP described above (Fig. 1b). The functional role of the high-affinity binding is not known, but it may be important for the structural stability of JH2, which is required for the JH2-mediated stimulatory interaction.
Previously, we mapped Ser523 and Tyr570 as JH2 phosphorylation sites19, which had been shown earlier to be important for maintaining low basal activity of Jak2 (refs. 27–30). Here, we show that Ser523 is phosphorylated by JH2 in cis and Tyr570 in trans. It is conceivable that phosphorylation of these sites fortifies the inhibitory interaction mediated by JH2, through electrostatic interactions between pSer523 and pTyr570 and basic residues in Jak2 (presumably in JH1 or JH2). These phosphorylation sites are not conserved in the other Jaks, which suggests that Jak2 may be the only Jak to possess this additional negative regulatory mechanism. Of note, Jak2 is the only Jak that trans-activates via homodimeric cytokine receptors (e.g., erythropoietin receptor and growth hormone receptor), which juxtapose two Jak2 molecules. Other Jaks (including Jak2) trans-activate via heterodimeric receptors, which juxtapose, for example, Jak1 and Jak3.
Her3, like JH2 of Jaks, contains an asparagine in the catalytic loop in place of the canonical aspartate (Asn815 in Her3, Asn673 in Jak2 JH2) (Supplementary Fig. 3b). Quantum mechanics-molecular mechanics (QM-MM) simulations on Her3 provided theoretical support for phosphoryl transfer (via an associative mechanism) with asparagine at this key position22. Interestingly, when introduced into full-length Jak2, the mutation N673D (restoring the canonical aspartate) resulted in suppression of V617F hyperactivity (data not shown), suggesting that the non-canonical asparagine plays a positive role in Jak2 activation. We speculate that, because of the non-canonical mode of Mg-ATP binding in JH2, aspartate at this position (proximal to the γ phosphate) might destabilize ATP binding as a consequence of (negative) charge repulsion.
The exact nature of the JH2-mediated inhibitory and stimulatory interactions that regulate Jak2 tyrosine kinase (JH1) activity are not known. The majority of MPN-causative mutations in Jak2 map to either the N lobe of JH2 (in exons 12 or 14) or the β7-β8 region (in exon 16) (Supplementary Fig. 4), with relatively few mutations mapped to JH1 (ref. 8). In addition, in a random mutagenesis study of Jak2 (encompassing the entire molecule)31, gain-of-function mutations mapped either to JH2 (the same regions: N lobe, β7-β8) or to residues just N-terminal to JH2, with no mutations in JH1. These considerations suggest that, rather than an autoinhibitory interaction between JH2 and JH1 (ref. 18), the prevailing model in the field, the domain interactions may predominantly involve JH2 itself, i.e., JH2 dimerization, both in the basal state on pre-dimerized cytokine receptors32 (inhibitory dimer) and after cytokine binding and receptor rearrangement (stimulatory dimer). Further structural and biochemical studies will be required to characterize the requisite molecular interactions.
MPN mutations in JH2 evidently drive formation of the stimulatory interaction in the basal state, either by destabilizing the inhibitory interaction (without compromising the ability of JH2 to form the stimulatory interaction) or by hyperstabilizing the stimulatory interaction. Our structural and mutagenesis data indicate that V617F is in the latter category. In addition, V617F and other MPN mutations tested19 impair JH2 catalytic activity, which further enhances Jak2 (JH1) activity due to loss of Ser523 and Tyr570 phosphorylation (negative regulatory). Therefore, MPN mutations in Jak2 JH2 appear to achieve hyperactivity through gain-of-function steric (on JH1) and loss-of function catalytic (JH2) mechanisms.
Elucidation of the central role of Jak2 in the pathogenesis of MPNs and other human diseases has spurred development of small-molecule inhibitors of Jak2. Numerous clinical trials with these inhibitors are ongoing, and the first Jak2 inhibitor (ruxolitinib) was recently approved by the U.S. Food and Drug Administration for treatment of myelofibrosis. All Jak inhibitors in clinical trials target JH1, which is identical in wild-type Jak2 and in most (JH2-based) MPN mutants, and side effects such as anemia and thrombocytopenia inevitably arise33. Because JH2 is a key regulatory domain and a hot spot for disease mutations, this domain represents an alternative small-molecule target. Our high-resolution structures of Jak2 JH2 should enable the design of such compounds.
ONLINE METHODS
Protein expression, purification and crystallization
We expressed human Jak2 JH2 (residues 536–812), wild-type (JH2-WT) and V617F (JH2-VF), with a C-terminal thrombin-cleavable His6-tag in baculovirus-infected Spodoptera frugiperda-9 cells. JH2-WT harbored the designed mutations W659A, W777A and F794H (described in Results). JH2-VF harbored W777A and F794H, but due to a cloning oversight contained native Trp659. We resuspended cell pellets in lysis buffer containing 20 mM Tris-HCl (pH 8.5), 500 mM NaCl, 10% (v/v) glycerol, 0.5 mM TCEP, 1 mM sodium orthovanadate and 20 mM imidazole, supplemented with protease inhibitors (Roche Diagnostics). We lysed the cells using a cell disruptor (Avestin), clarified the lysate by centrifugation and purified the protein by nickel-affinity chromatography (Ni-NTA, QIAGEN) and anion-exchange chromatography (Source-Q, GE Healthcare). JH2 eluted as a single peak on the Source-Q column, and fractions corresponding to the peak were concentrated to to ~6 mg ml−1 using Amicon centrifugal filters (Millipore). Crystals of JH2-WT and JH2-VF, apo or with Mg-ATP (3 mM MgCl2, 1 mM ATP), grew in hanging drops at 4 °C in 200 mM sodium acetate, 100 mM Tris-HCl (pH 8.0 or 8.5) and 18–21% (w/v) polyethylene glycol (PEG) 4000. Macroseeding was required to grow single crystals of sufficient size.
X-ray data collection, structure determination and refinement
We collected diffraction data for apo JH2-WT (see Table 1 for unit-cell information) on a MicroMax-007 x-ray generator (Rigaku) (λ= 1.5418 Å) and an RAXIS IV++ image plate detector (Rigaku). We collected data for ATP-bound JH2-WT and JH2-VF at beam line X25 (λ=0.9789 Å) at the National Synchrotron Light Source, Brookhaven National Laboratory on a Pilatus 6M CCD detector (DECTRIS). We processed the data using HKL-2000 (ref. 34). We determined the structure of apo JH2-WT by molecular replacement (one molecule in the asymmetric unit), trying many tyrosine kinase structures (separate N and C lobes or both lobes) as search models. Ultimately, we were successful in placing the C lobe of JH2-WT with MOLREP35, using as a search model the C lobe (poly-alanine) of the epidermal growth factor receptor (EGFR) kinase domain (PDB code 1M14). We were unable to obtain a solution for the N lobe of JH2-WT (with the C lobe fixed) by molecular replacement, and manually placed the N lobe (poly-alanine) of the EGFR kinase domain in the (weak) 2Fo-Fc electron-density map generated from the JH2-WT C-lobe model, followed by rigid-body refinement in REFMAC36. Iterative rounds of model-building in Coot37 and positional and B-factor refinement (with TLS38) with REFMAC led to an atomic model of apo JH2-WT at 2.0-Å resolution that includes all residues from 536 to 810. We determined the crystal structures of JH2-WT and JH2-VF with bound Mg-ATP by molecular replacement using the JH2-WT apo structure as the search model.
Molecular dynamics simulations
For the starting structures in the simulations, the mutations introduced to increase protein solubility (see above) were reverted back to their wild-type residues. Simulation systems were set up by placing the protein at the center of a cubic simulation box (with periodic boundary conditions) of at least 70 Å per side. Explicitly represented water molecules were added to fill the system, and Na+ and Cl ions were added to maintain physiological salinity (150 mM) and to obtain a neutral total charge for the system. The systems were parameterized using the Amber ff99SB-ILDN force field with TIP3P water39–41. Equilibrium molecular dynamic simulations were performed on the special-purpose molecular dynamics machine Anton42 in the NVT ensemble at 310 K using the Nose-Hoover thermostat43 with a relaxation time of 1.0 ps and a time step of 2.5 fs. All bond lengths to hydrogen atoms were constrained using a recently developed implementation44 of M-SHAKE45. The Lennard-Jones and the Coulomb interactions in the simulations were calculated using a force-shifted cutoff of 12 Å (ref. 46).
In vitro autophosphorylation of JH2
We produced and purified human Jak2 JH2, residues 513–824, as described previously19, isolating two peaks from the Mono-Q (GE Healthcare) column: non-phosphorylated and Ser523-phosphorylated JH2. The autophosphorylation reaction buffer contained 10 mM ATP, 10 μCi [γ-32P]ATP (PerkinElmer), 20 mM Tris-HCl (pH 8.0), 20mM MnCl2, 300 mM NaCl, 10% (v/v) glycerol and 0.5 mM TCEP. We performed the reactions at room temperature and stopped them by adding 2x SDS-PAGE sample buffer.
Cell transfection and Western blotting
Transfection of HA-tagged human Jak2, wild-type or mutants (obtained by QuikChange (Stratagene)), into Jak2-deficient γ2A cells has been described19. We used the following antibodies for immunoprecipitation or Western blotting: anti-HA (Covance), anti-Jak2 pY1007–1008 (Cell Signaling Technology) and anti-Jak2 pSer523 (ref. 28).
Supplementary Material
Acknowledgments
This work was supported in part by NIAID grant R21 AI095808 (S.R.H.), the Medical Research Council of Academy of Finland (O.S.), the Sigrid Juselius Foundation (O.S.), the Finnish Cancer Foundation (O.S.), Competitive Research Funding and Centre of Laboratory Medicine of the Tampere University Hospital (O.S.), the Maire Lisko Foundation (O.S.) and the Tampere Tuberculosis Foundation (O.S.). NCI training grant T32 CA009161 supported R.M.B. Financial support for beam line X25 at the National Synchrotron Light Source, Brookhaven National Laboratory, comes principally from the U.S. Department of Energy, from the National Center for Research Resources (P41 RR012408) and from NIGMS (P41 GM103473). We thank E. Koskenalho for technical assistance, V. DiGiacomo for peptide modeling, A. Heroux for x-ray data collection, W.T. Miller, M. Mohammadi and K. Gnanasambandan for critical reading of the manuscript and M. Eck (Dana-Farber Cancer Institute) for discussions and for providing coordinates prior to publication.
Footnotes
ACCESSION CODES
Atomic coordinates and structure factors for the JAK2 JH2 structures have been deposited in the Protein Data Bank with ID codes 4FVP (JH2-WT, apo), 4FVQ, (JH2-WT, Mg-ATP) and 4FVR (JH2-VF, Mg-ATP).
AUTHOR CONTRIBUTIONS
R.M.B., crystallographic studies and manuscript preparation; D.U., in-cell biochemical studies and manuscript preparation; Y.S. and D.E.S., molecular dynamics simulations and manuscript preparation; O.S., project supervisor and manuscript preparation; S.R.H., project supervisor and principal manuscript author.
References
- 1.Ghoreschi K, Laurence A, O’Shea JJ. Janus kinases in immune cell signaling. Immunol Rev. 2009;228:273–287. doi: 10.1111/j.1600-065X.2008.00754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 3.Lucet IS, et al. The structural basis of Janus kinase 2 inhibition by a potent and specific pan-Janus kinase inhibitor. Blood. 2006;107:176–183. doi: 10.1182/blood-2005-06-2413. [DOI] [PubMed] [Google Scholar]
- 4.Boggon TJ, Li Y, Manley PW, Eck MJ. Crystal structure of the Jak3 kinase domain in complex with a staurosporine analog. Blood. 2005;106:996–1002. doi: 10.1182/blood-2005-02-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams NK, et al. Dissecting specificity in the Janus kinases: the structures of JAK-specific inhibitors complexed to the JAK1 and JAK2 protein tyrosine kinase domains. J Mol Biol. 2009;387:219–232. doi: 10.1016/j.jmb.2009.01.041. [DOI] [PubMed] [Google Scholar]
- 6.Chrencik JE, et al. Structural and thermodynamic characterization of the TYK2 and JAK3 kinase domains in complex with CP-690550 and CMP-6. J Mol Biol. 2010;400:413–433. doi: 10.1016/j.jmb.2010.05.020. [DOI] [PubMed] [Google Scholar]
- 7.Vainchenker W, Delhommeau F, Constantinescu SN, Bernard OA. New mutations and pathogenesis of myeloproliferative neoplasms. Blood. 2011;118:1723–1735. doi: 10.1182/blood-2011-02-292102. [DOI] [PubMed] [Google Scholar]
- 8.Haan C, Behrmann I, Haan S. Perspectives for the use of structural information and chemical genetics to develop inhibitors of Janus kinases. J Cell Mol Med. 2010;14:504–527. doi: 10.1111/j.1582-4934.2010.01018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kralovics R, et al. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352:1779–1790. doi: 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- 10.James C, et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434:1144–1148. doi: 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- 11.Baxter EJ, et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365:1054–1061. doi: 10.1016/S0140-6736(05)71142-9. [DOI] [PubMed] [Google Scholar]
- 12.Levine RL, et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387–397. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 13.Lipson D. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Medicine AOP. 2012 doi: 10.1038/nm.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Russell SM, et al. Mutation of Jak3 in a patient with SCID: essential role of Jak3 in lymphoid development. Science. 1995;270:797–800. doi: 10.1126/science.270.5237.797. [DOI] [PubMed] [Google Scholar]
- 15.Saharinen P, Silvennoinen O. The pseudokinase domain is required for suppression of basal activity of Jak2 and Jak3 tyrosine kinases and for cytokine-inducible activation of signal transduction. J Biol Chem. 2002;277:47954–47963. doi: 10.1074/jbc.M205156200. [DOI] [PubMed] [Google Scholar]
- 16.Saharinen P, Takaluoma K, Silvennoinen O. Regulation of the Jak2 tyrosine kinase by its pseudokinase domain. Mol Cell Biol. 2000;20:3387–3395. doi: 10.1128/mcb.20.10.3387-3395.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen M, et al. Complex effects of naturally occurring mutations in the JAK3 pseudokinase domain: evidence for interactions between the kinase and pseudokinase domains. Mol Cell Biol. 2000;20:947–956. doi: 10.1128/mcb.20.3.947-956.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindauer K, Loerting T, Liedl KR, Kroemer RT. Prediction of the structure of human Janus kinase 2 (JAK2) comprising the two carboxy-terminal domains reveals a mechanism for autoregulation. Protein Eng. 2001;14:27–37. doi: 10.1093/protein/14.1.27. [DOI] [PubMed] [Google Scholar]
- 19.Ungureanu D, et al. The pseudokinase domain of JAK2 is a dual-specificity protein kinase that negatively regulates cytokine signaling. Nat Struct Mol Biol. 2011;18:971–976. doi: 10.1038/nsmb.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng J, et al. 2.2 A refined crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MnATP and a peptide inhibitor. Acta Cryst. 1993;D49:362–365. doi: 10.1107/S0907444993000423. [DOI] [PubMed] [Google Scholar]
- 21.Hubbard SR. Crystal structure of the activated insulin receptor tyrosine kinase in complex with peptide substrate and ATP analog. EMBO J. 1997;16:5572–5581. doi: 10.1093/emboj/16.18.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi F, Telesco SE, Liu Y, Radhakrishnan R, Lemmon MA. ErbB3/HER3 intracellular domain is competent to bind ATP and catalyze autophosphorylation. Proc Natl Acad Sci USA. 2010;107:7692–7697. doi: 10.1073/pnas.1002753107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jura N, Shan Y, Cao X, Shaw DE, Kuriyan J. Structural analysis of the catalytically inactive kinase domain of the human EGF receptor 3. Proc Natl Acad Sci USA. 2009;106:21608–21613. doi: 10.1073/pnas.0912101106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dusa A, Mouton C, Pecquet C, Herman M, Constantinescu SN. JAK2 V617F constitutive activation requires JH2 residue F595: a pseudokinase domain target for specific inhibitors. PLoS ONE. 2010;5:e11157. doi: 10.1371/journal.pone.0011157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gnanasambandan K, Magis A, Sayeski PP. The constitutive activation of Jak2-V617F is mediated by a pi stacking mechanism involving phenylalanines 595 and 617. Biochemistry. 2010;49:9972–9984. doi: 10.1021/bi1014858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ni Q, Shaffer J, Adams JA. Insights into nucleotide binding in protein kinase A using fluorescent adenosine derivatives. Protein Sci. 2000;9:1818–1827. doi: 10.1110/ps.9.9.1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Argetsinger LS, et al. Autophosphorylation of JAK2 on tyrosines 221 and 570 regulates its activity. Mol Cell Biol. 2004;24:4955–4967. doi: 10.1128/MCB.24.11.4955-4967.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishida-Takahashi R, et al. Phosphorylation of Jak2 on Ser(523) inhibits Jak2-dependent leptin receptor signaling. Mol Cell Biol. 2006;26:4063–4073. doi: 10.1128/MCB.01589-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feener EP, Rosario F, Dunn SL, Stancheva Z, Myers MG. Tyrosine phosphorylation of Jak2 in the JH2 domain inhibits cytokine signaling. Mol Cell Biol. 2004;24:4968–4978. doi: 10.1128/MCB.24.11.4968-4978.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mazurkiewicz-Munoz AM, et al. Phosphorylation of JAK2 at serine 523: a negative regulator of JAK2 that is stimulated by growth hormone and epidermal growth factor. Mol Cell Biol. 2006;26:4052–4062. doi: 10.1128/MCB.01591-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao L, et al. A JAK2 interdomain linker relays Epo receptor engagement signals to kinase activation. J Biol Chem. 2009;284:26988–26998. doi: 10.1074/jbc.M109.011387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livnah O, et al. Crystallographic evidence for preformed dimers of erythropoietin receptor before ligand activation. Science. 1999;283:987–990. doi: 10.1126/science.283.5404.987. [DOI] [PubMed] [Google Scholar]
- 33.Tefferi A. JAK inhibitors for myeloproliferative neoplasms: clarifying facts from myths. Blood. 2012 doi: 10.1182/blood-2011-11-395228. [DOI] [PubMed] [Google Scholar]
- 34.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 35.Vagin AA, Teplyakov A. MOLREP: an automated program for molecular replacement. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 36.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 37.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 38.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 39.Hornak V, et al. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lindorff-Larsen K, et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 42.Shaw DE, et al. Millisecond-scale molecular dynamics simulations on Anton. In ACM/IEEE Conference on Supercomputing. ACAM/IEEE Conference on Supercomputing; New York, NY, ACM Press. 2009. [Google Scholar]
- 43.Hoover WG. Canonical dynamics: Equilibrium phase-space distributions. Phys Rev A. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 44.Lippert RA, et al. A common, avoidable source of error in molecular dynamics integrators. J Chem Phys. 2007;126:046101. doi: 10.1063/1.2431176. [DOI] [PubMed] [Google Scholar]
- 45.Krautler V, Van Gunsteren WF, Hunenberger PH. A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J Comput Chem. 2001;22:501–508. [Google Scholar]
- 46.Fennell CJ, Gezelter JD. Is the Ewald summation still necessary? Pairwise alternatives to the accepted standard for long-range electrostatics. J Chem Phys. 2006;124:234104. doi: 10.1063/1.2206581. [DOI] [PubMed] [Google Scholar]
- 47.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.