Abstract
Poly(ADP-ribose) [pADPr] is a large, structurally complex polymer of repeating ADP-ribose units. It is biosynthesized from NAD+ by poly(ADP-ribose) polymerases [PARPs] and degraded to ADP-ribose by poly(ADP-ribose) glycohydrolase. PADPr is involved in many cellular processes and exerts biological function through covalent modification and non-covalent binding to specific proteins. Very little is known about molecular recognition and structure activity relationships for non-covalent interaction between pADPr and its binding proteins, in part because of lack of access to the polymer on large scale and to units of defined lengths. We prepared polydisperse pADPr from PARP1 and tankyrase 1 at hundreds of milligram scale by optimizing enzymatic synthesis and scaling up chromatographic purification methods. We developed and calibrated an anion exchange chromatography method to assign pADPr size and scaled it up to purify defined length polymers on the milligram scale. Furthermore, we present a pADPr profiling method to characterize the polydispersity of pADPr produced by PARPs under different reaction conditions and find that substrate proteins affect the pADPr size distribution. These methods will facilitate structural and biochemical studies of pADPr and its binding proteins.
Keywords: PARP, PARG, pADPr, PAR, ADPr, poly(adp-ribose), poly(ADP-ribose) polymerase, poly(ADP-ribose) glycohydrolase, polymer, NAD+, ADP ribose
Introduction
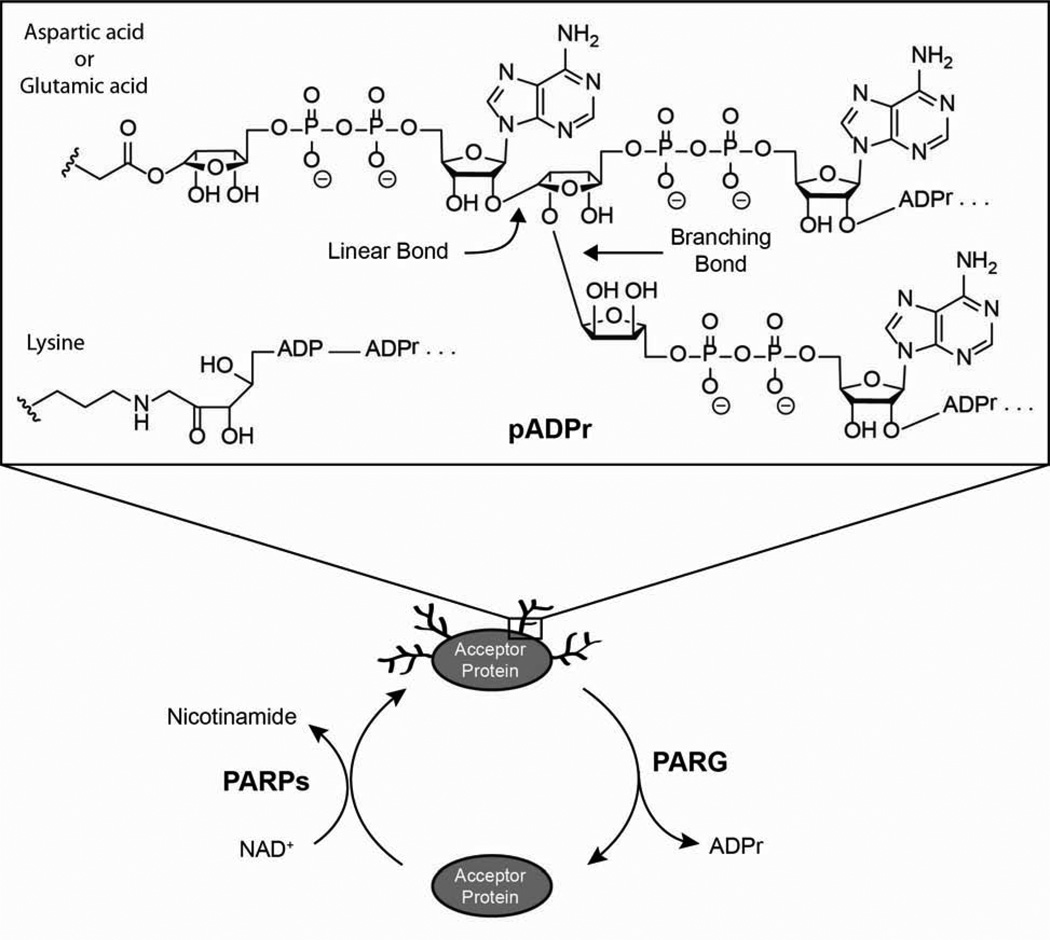
Poly(ADP-ribose) [pADPr] is a post-translational modification made up of ADPr (adenosine 5’-diphosphoribose) units connected linearly or in a branched fashion (Figure 1) [1]. It is covalently attached to acidic or basic residues of target proteins and can be quite heterogeneous in structure, varying in size (number of ADPr units) and shape (degree of branching) [2; 3]. PADPr is biosynthesized from NAD+ (β-nicotinamide adenine dinucleotide) by enzymes known as PARPs [poly (ADP ribose) polymerases] and degraded by poly (ADP ribose) glycohydrolase [PARG] to release ADPr molecules (Figure 1) [4].
Figure 1.
The metabolism of poly(ADP-ribose) [pADPr]. PARPs [poly(ADP-ribose) polymerases] biosynthesize pADPr from NAD+ while PARG [poly(ADP-ribose) glycohydrolase] degrades polymer to ADP-ribose [ADPr]. PADPr is covalently attached to aspartic acid, glutamic acid, or lysine residues of acceptor proteins and the ADPr units of the polymer are connected linearly or in a branched fashion.
There are 17 types of PARPs but only one PARG gene [5]. The members of the PARP superfamily are all related by the presence of a homologous PARP domain [6]. These enzymes can be subdivided into six groups based on domain architecture (PARP1 subgroup, vault PARP, tankyrases, CCCH-PARPs, macro PARPs, and other) and three categories based on enzymatic activity (poly, mono, and inactive) [7]. In this paper, we focused on human PARP1 (hPARP1) and human tankyrase 1 (hTNKS1) because both are known to synthesize polymers and are amongst the best characterized members of the PARP family. It should be noted that hPARP1 and hTNKS1 are also known as ADP-ribosyltransferase 1 and 5 (ARTD1 and ARTD5), respectively, under a recently proposed nomenclature [8].
PARPs and pADPr have been implicated in many cellular processes [1; 5; 9]. PARP1 is involved in DNA repair/maintenance and transcriptional regulation while TNKS1 has been implicated in telomere maintenance, assembly of the mitotic spindle, and regulation of protein stability [10; 11; 12; 13; 14; 15; 16; 17]. PADPr is thought to function through both covalent and non-covalent interactions. Covalent attachment of pADPr can affect the structure and function of the modified protein, in part due to the high density of negative charges on the polymer [18; 19; 20]. Two examples are automodification of PARPs, which tend to inactivate enzymatic activity, and heteromodification of histones, which relaxes chromosome packing and increases access to damaged DNA [21].
Non-covalent interactions between pADPr and specific binding proteins are thought to play a role in the DNA damage response and in regulation of protein stability [22; 23; 24; 25; 26; 27]. The chromatin remodeling enzyme ALC1 is rapidly recruited to DNA damage sites in a pADPr dependent manner. For the regulation of protein stability, the E3 ubiquitin ligase RNF146 interacts with tankyrase poly(ADP-ribosyl)ated axin and 3BP2, and ubiquitinylates them for degradation. Free pADPr has been reported to be a cytotoxic signaling molecule that causes caspase independent cell death by inducing the release of apoptosis inducing factor from the mitochondria [28; 29; 30; 31; 32].
Precisely how binding proteins recognize pADPr is unclear but structural studies of binding proteins interacting with fragments of the polymers have hinted alternative recognition modes. PADPr binding macro domains, which are present in several binding proteins including some PARPs, are thought to bind to ADPr units at the tips of pADPr while the pADPr-binding zinc finger and WWE domains are thought to recognize the PRAMP (phosphoribosyl AMP) units along the length of the polymer [24; 26; 33; 34]. The structure of pADPr itself in solution is also unknown. Spectral analysis on pADPr has suggested a helical structure but nuclear magnetic resonance studies on 13C and 15N labeled polymers showed an absence of any inherent regular structure [35; 36; 37]. By electron microscopy, bulk pADPr appeared as “root-like” structures about 100 nm long [38; 39]. Further characterization is required to fully understand the structure activity relationship of pADPr.
Detailed structural and biochemical characterization of pADPr has been limited in part because of lack of access to the polymer at large scale. Polydisperse pADPr is commercially available, at a current price of ~$175 for 1 nmol (~1 µg). Defined length pADPr is not commercially available. It has been prepared previously, but yields were meager and scale was insufficient for structural and biochemical studies [40; 41]. Herein we describe an improved procedure for preparing PARP1 and tankyrase 1 bulk and homogenous polymers in large scale. We also present convenient methods to assign pADPr size by anion exchange chromatography using high pressure liquid chromatography (HPLC).
Materials and Methods
HPARP1 (1–1014), hPARP1 (379–1014), and hTNKS1 (1093–1327) constructs were gifts from J. Pascal. HPARP1 (655–1014) and bovine PARG CF (residues 386–977) constructs were generously provided by P. Chang and H. Kleine, respectively [7]. The genes for hPARP1 (1–1014), hPARP1 (379–1014), and hPARP1 (655–1014) were cloned into the pET28 expression vector (Novagen) and hexahistidine tagged at the N-terminus [42]. A pET24(+) expression vector (Novagen) was used for hTNKS1 (1093–1327) to generate proteins with the hexahistidine tag at the C-terminus. Bovine PARG CF was cloned into a pGEX-2T expression vector (GE Healthcare) with an N-terminal GST tag.
All reagents were purchased from Sigma-Aldrich, Acros, Alfa Aesar, GE Healthcare, RPI Corp., Roche, Fisher, or VWR and were used without any further purification unless otherwise stated. Nickel nitriloacetic acid (Ni-NTA) agarose and glutathione agarose beads were obtained from Qiagen and Sigma-Aldrich, respectively. Gel filtration columns (superdex 200 and S75) and ECH Sepharose™ 4B media were purchased from GE Healthcare. DNA Pac PA100 (analytical, guard, and semi-preparative) and Kinetex™ C18 HPLC columns were obtained from Dionex and Phenomenex, respectively.
HPLC analysis and fractionation were performed on an Agilent 1100 series HPLC liquid chromatography system equipped with a diode array detector, thermostated automatic sample injector, and a time based fraction collector. For all HPLC analysis, the diode array detector monitored UV (ultraviolet) absorbance at 254 nm. Aqueous samples were concentrated on an EZ-2 series Genevac evaporator or a Labconco FreeZone 4.5 liter console freeze dry system (lyophilizer). All FPLC (fast pressure liquid chromatography) purifications were performed on a BioLogic DuoFlow™ FPLC system from Bio-Rad at 4°C. The cell disruptor and NanoDrop 8000 were manufactured by Avestin and Thermo Scientific, respectively.
Expression of hPARP1 (1–1014), hPARP1 (379–1014), hPARP1 (655–1014), bPARG CF and hTNKS1 (1093–1327)
All hPARP1 constructs and bovine PARG CF were expressed in Escherichia coli strain BL21 (DE3) Rosetta2 (Novagen). HTNKS1 (1093–1327) was expressed in strain BL21 (DE3) Rosetta pLysS (Novagen). The cells were grown in terrific broth medium containing 1% glucose, 100 µM ZnCl2, 50 µg/ml kanamycin, and 34 µg/ml chloramphenicol. Glucose and ZnCl2 were omitted for hTNKS1 (1093–1327) and bPARG CF, respectively. Ampicillin (100 µg/ml) was substituted for kanamycin in the bPARG CF growth media. Cells were grown at 37°C to OD600 = 0.5–1.0 and induced with isopropyl-β-D-thiogalactopyranoside (0.25 to 0.5 mM) at 16°C for 16–24 h. The cells were pelleted, medium discarded, flash frozen in liquid nitrogen, and stored at −80°C until purification.
Purification of hPARP1 (1–1014), hPARP1 (379–1014), and hPARP1 (655–1014)
Cells were resuspended in lysis buffer (20 mM Hepes, pH 7.5, 1 mM β-mercaptoethanol, 1 mM PMSF, 1 mM benzamidine, 0.2 % Nonidet™ P40 substitute, 0.2% Tween20, and 500 mM NaCl) and lysed with a cell disruptor. After pelleting cell debris for 90 min at 138,000 × g, the supernatant was incubated with Ni-NTA at 4°C for 1h. The Ni-NTA beads were washed with buffer 1 (20 mM Hepes, pH 7.5, 1 mM β-mercaptoethanol, 1 mM PMSF, 1 mM benzamidine, and 500 mM NaCl) containing 20 mM imidazole, and eluted with buffer 1 with 400 mM imidazole. After diluting with 3AB buffer (100 mM Tris, pH 7.5, 0.5 mM EDTA, and 14 mM β-mercaptoethanol) to decrease the NaCl concentration down to 400 mM, the proteins were slowly loaded (0.20 ml/min) via FPLC onto a 3-aminobenzamide ECH sepharose 4B column equilibrated with 4 CV (column volume) of 3AB buffer. The column was washed with 3AB buffer containing 400 mM NaCl (2 CV) and 800 mM NaCl before eluting with 3AB elution buffer (100 mM Tris, pH 7.5, 0.5 mM EDTA, 14 mM β-mercaptoethanol, 400 mM NaCl, and 1 mM 3-methoxybenzamide) [43]. Desired proteins were concentrated with an Amicon Ultra centrifugal filter (MWCO = 30 kDa, Millipore), and gel filtered on a superdex 200 column with GF (gel filtration) buffer 1 (100 mM Tris, pH 7.5, 0.1 mM EDTA, 1 mM β-mercaptoethanol, and 150 mM NaCl). Small aliquots of the proteins were flash frozen in liquid nitrogen and stored at −80°C.
Purification of hTNKS1 (1093–1327)
Cells were resuspended in lysis buffer (50 mM Hepes, pH 7.5, 1 mM β-mercaptoethanol, cOmplete EDTA-free protease inhibitor cocktail tablets (Roche), 10 % glycerol, 300 mM NaCl, and 10 mM imidazole) and lysed using a cell disruptor. Cell debris was pelleted for 45 min at 138,000 × g, and the supernatant was incubated with Ni-NTA at 4°C for 1h. The Ni-NTA beads were washed with buffer 2 (30 mM Hepes, pH 7.5, 1 mM β-mercaptoethanol, 10% glycerol, 500 mM NaCl, and 50 mM imidazole), and eluted with buffer 2 containing 500 mM imidazole. Desired proteins were gel filtered on a superdex 200 or S75 column with GF buffer 2 (30 mM Hepes, pH 7.5, 1 mM β-mercaptoethanol, 50% glycerol, and 300 mM NaCl). Small aliquots of the proteins were flash frozen in liquid nitrogen and stored at −80°C.
Purification of bPARG CF
Cells were resuspended in lysis buffer (20 mM Tris, pH 8.0, 5 mM β-mercaptoethanol, 1 mM EDTA, cOmplete protease inhibitor cocktail tablets [Roche], 10 % glycerol, 0.1% Tween20, and 500 mM NaCl) and lysed using a cell disruptor. The lysed sample was incubated at 4°C for 1 h to aid solubilization of bPARG CF proteins. Cell debris was pelleted for 30 min at 138,000 × g, and the supernatant was incubated with glutathione agarose at 4°C for 1h. The glutathione agarose beads were washed with buffer 3 (100 mM Tris, pH 8.0, 5 mM β-mercaptoethanol, and 500 mM NaCl), and eluted with buffer 3 containing 20 mM glutathione. Desired proteins were gel filtered on a superdex 200 column with GF buffer 3 (100 mM Tris, pH 8.0, 5 mM β-mercaptoethanol, and 120 mM NaCl). Small aliquots of the proteins were flash frozen in liquid nitrogen and stored at −80°C.
PADPr synthesis through automodification reaction: hPARP1 (1–1014), hPARP1 (379–1014), hPARP1 (655–1014), or hTNKS1 (1093–1327) alone
This method was adapted from Kiehlbauch et al. and Panzeter et al [41; 44]. A reaction mixture (500 µl) of 100 mM Tris, pH 8.0, 10 mM MgCl2, 1 mM DTT, activated DNA (25 µg/ml, Sigma Aldrich - #D4522), and PARP [10.8 µM, 23.2 µM, 45.5 µM, or 42.9 µM for hPARP1 (1–1014), hPARP1 (379–1014), hPARP1 (655–1014), or hTNKS1 (1093–1327), respectively] was pre-incubated at room temperature for 20 min. For hTNKS1 (1093–1327) pADPr biosynthesis, the activated DNA and pre-incubation step were both omitted. A solution of NAD+ (20 mM) was added and the reaction was incubated at 30°C [room temperature for hTNKS1 (1093–1327)] for 1h or more. A 50 µl aliquot was saved for ion pairing HPLC analysis.
PADPr synthesis through automodification and heteromodification reactions: hPARP1 (1–1014) + hPARP1 (379–1014), hTNKS1 (1093–1327) + hPARP1 (379–1014), or hTNKS1 (1093–1327) + histones
The same condition as automodification reaction was used except that two proteins were present in the reaction mixture. The protein concentrations were as follows: hPARP1 (1–1014) (1.88 µM) + hPARP1 (379–1014) (13.7 µM), hTNKS1 (1093–1327) (21.9 µM) + hPARP1 (379–1014) (21.9 µM), or hTNKS1 (1093–1327) (21.9 µM) + histones (0.43 mM, Sigma Aldrich - H9250). For hTNKS1 (1093–1327) containing reactions, the mixture was incubated at room temperature instead of 30°C and both activated DNA and pre-incubation step were omitted.
HPARP1 (379–1014) was originally added to hPARP1 (1–1014) to increase the protein concentration of the polymerization reaction to at least 1 mg/ml. When isolating bulk pADPr, it was essential for the protein concentration of the polymerization reaction to be at least 1 mg/ml. Below this level, it was difficult to obtain an acid insoluble protein precipitate when the polymerization reaction was treated with 20% trichloracetic acid to stop the reaction.
Purification of bulk pADPr
To stop the reaction, the mixture (450 µl) was treated with an equal volume of 20% trichloroacetic acid (450 µl), incubated at 0°C for 15 min, and centrifuged at 20,800 × g for 15 min at 4°C to pellet the precipitates. The pellet was redissolved in 1 M KOH/50 mM EDTA (100 µl), incubated at 60°C for 1 h with occasional vortexing to detach pADPr from proteins. AAGE9 buffer (900 µl; 250 mM ammonium acetate, pH 9.0, 6 M guanidine hydrochloride, and 10 mM EDTA) was added and the pH was adjusted to 9.0 with HCl. The alkali digested material was loaded onto a 1 ml DHBB (Dihydroxyboryl Bio-Rex 70) column sequentially equilibrated with 5–10 CV of AAGE9 buffer, 20 CV water, and 5–20 CV of AAGE9 buffer [43; 45]. Note, the binding capacity of DHBB column is around 3 mg of pADPr/ml of resin. The sample was washed with 10 CV of AAGE9 buffer and 20 CV of 1 M ammonium acetate (pH 9.0) before eluting the pADPr with 6 ml of 37°C water and collecting 1 ml fractions. Bulk pADPr eluted in the second and third fractions. Samples were concentrated to dryness with a Genevac evaporator or lyophilizer.
Determining pADPr concentration
PADPr concentration was determined using the equation: [pADPr] = [(A258)cm−1]/[13,500 cm−1M−1] (Trevigen®). UV absorbance at 258 nm was measured with a NanoDrop 8000.
Enzymatic degradation of pADPr
An aliquot (50 µl) of the pADPr formation reaction was filtered through a 3 kDa Nanosep® centrifugal device (VWR) to separate the small molecules (found in the filtrate) from proteins (found in the retentate). The filtrate was subjected to HPLC ion pairing analysis (see below). The retentate was reconstituted in water (50 µl), and digested with bPARG CF (0.34 nM) for 30 min at 37°C in a reaction mixture (150 µl final volume) of 100 mM Tris, pH 8.0, and 10 mM DTT. After filtering the reaction through a 3 kDa Nanosep® centrifugal device to remove proteins, the filtrate was subjected to ion pairing HPLC analysis.
Ion pairing HPLC analysis
Ion pairing analysis were performed using a Kinetex™ 2.6 µm C18, 100 Å, 50 × 2.1 mm column at a flow rate of 0.2 ml/min and monitoring UV absorbance at 254 nm. The method was adapted from Di Pierro et al. and uses tetrabutylammonium hydroxide as the counterion [46]. The column was initially equilibrated with ion pairing buffer A (10 mM tetrabutylammonium hydroxide, 10 mM KH2PO4, 0.25% methanol, pH 7.0) and a variable gradient of ion pairing buffer B (2.8 mM tetrabutylammonium hydroxide, 100 mM KH2PO4, 30% methanol, pH 5.5) was used for elution. The concentration of ion pairing buffer B in the 32 min method was set as follows: 0 min (0% B), 1 min (0% B), 8 min (60% B), 21 min (100% B), 23 min (100% B), 24 min (0% B), and 32 min (0% B).
HPLC pADPr profiling
HPLC profiling of pADPr was performed using a DNA Pac PA100 4 × 250 mm analytical column equipped with a 4 × 50 mm guard column at a flow rate of 1 ml/min and monitoring UV absorbance at 254 nm. The method was adapted from Kiehlbauch et al. and Fahrer et al [40; 41]. Bulk pADPr (0.72 µmoles) was loaded onto the column equilibrated with dionex buffer A (25 mM Tris, pH 9.0) and the polymers were eluted with a variable gradient of dionex buffer B (25 mM Tris, pH 9.0, and 1 M NaCl). The concentration of dionex buffer B in the 152 min method was set as follows: 0 min (0% B), 6 min (0% B), 12 min (30% B), 18 min (40% B), 48 min (47% B), 78 min (49% B), 138 min (55% B), 144 min (63% B), 145 min (100% B), 148 min (100% B), 149 min (0% B), and 152 min (0% B).
HPLC pADPr fractionation
A semi-preparative DNA Pac PA100 column (9 × 250 mm) was used to fractionate bulk pADPr into homogenous polymers. The flow rate was 2 ml/min and ~2.74 µmoles (~3 mg) of bulk pADPr was loaded onto the column. The column was pre-equilibrated with dionex buffer A and the concentration of dionex buffer B in the 186 min method was set as follows: 0 min (0% B), 6 min (0% B), 18 min (30% B), 30 min (40% B), 90 min (47% B), 150 min (49% B), 162 min (55% B), 174 min (63% B), 175 min (100% B), 179 min (100% B), 180 min (0% B), and 186 min (0% B). Two ml fractions were collected and evaporated to dryness at 35°C with a Genevac evaporator.
Desalting pADPr
Dried pADPr fractions from DNA Pac PA100 columns were reconstituted in water and desalted on a Sephadex G25 fine (23 ml) column connected to an HPLC system. A solution of polymers (loaded up to 2 ml max) were eluted isocratically with water at a flow rate of 1 ml/min for 45 min. Desired products, collected in 1 ml fractions, were evaporated to dryness at 35°C with a Genevac evaporator and a lyophilizer.
Characterization of pADPr with MALDI-TOF mass spectrometer
Desalted, dried, homogenous pADPr were submitted for MALDI-TOF mass spectrometry analysis at the Scripps Center for Metabolomics and Mass Spectrometry, Scripps Research Institute in San Diego. Samples were reconstituted in water and analyzed with THAP (2’, 4’, 6’-trihydroxyacetophenone monohydrate) as the matrix.
Results
Purification of PARP1, TNKS1 and PARG enzymes
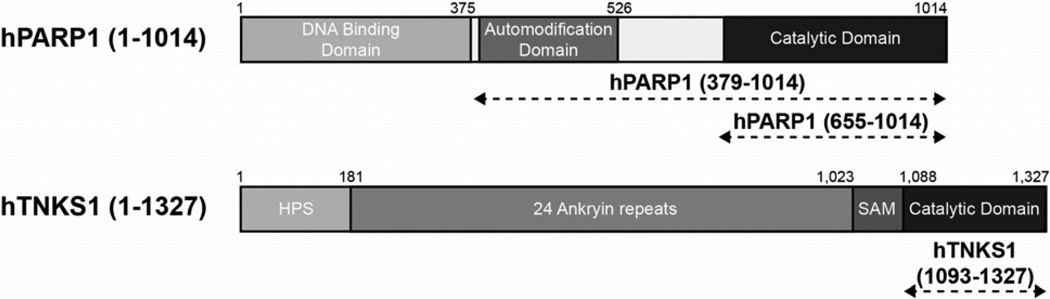
We expressed and purified three hPARP1 and one hTNKS1 constructs as candidate enzymes for scale up of pADPr enzymatic synthesis. The three hPARP1 constructs corresponded to full length protein [hPARP1 (1–1014)], protein lacking the DNA binding domain [hPARP1 (379–1014)], and protein missing both DNA binding and automodification domains [hPARP1 (655–1014)] (Figure 2). For hTNKS1, we used a construct that encompassed its C-terminal catalytic domain [hTNKS1 (1093–1327)] (Figure 2). These four constructs were chosen because they were all previously found to be capable of biosynthesizing pADPr. [42; 47; 48]
Figure 2.
Domain architecture of hPARP1 and hTNKS1 constructs. HPS, homopolymeric runs of histidines, prolines, and serines; SAM, sterile alpha motif.
All four constructs were expressed as hexahistidine tagged proteins, N-terminal for the hPARP1 constructs and C-terminal for hTNKS1 (1093–1327), in bacteria and induced with IPTG at 16°C for 16–24h. The three hPARP1 proteins were purified with a three step purification scheme consisting of immobilized metal affinity chromatography (IMAC), affinity chromatography with 3-aminobenzamide conjugated sepharose, and gel filtration. The affinity purification step was included to select for correctly folded proteins. The shortest construct, hPARP1 (655–1014), had the best yield (1247 nmoles of proteins/liter of culture) while full length hPARP1 (1–1014) had the lowest (14 nmoles of protein/liter of culture) (Table 1, Figure S1). For hTNKS1 (1093–1327), a two-step purification scheme of IMAC followed by gel filtration yielded 643 nmoles of proteins/liter of culture (Table 1).
Table 1.
PARPs and PARG protein purification summary.
| Protein | Yield | |
|---|---|---|
| mg/L of culture | nmoles/L of culture | |
| hPARP1 (1–1014) | 1.6 | 14 |
| hPARP1 (379–1014) | 39.2 | 537 |
| hPARP1 (655–1014) | 54.9 | 1247 |
| hTNKS1 (1093–1327) | 18.0 | 643 |
| bPARG CF | 1.1 | 13 |
Values are the average of at least two independent experiments.
In addition to the PARPs, we also expressed and purified a PARG protein. This protein was needed to digest pADPr to confirm the presence of any polymer. The PARG construct, bovine PARG catalytic fragment (bPARG CF), was also expressed in bacteria and induced with IPTG at 16°C for 16–24h [49]. Since it was a glutathione S-transferase tagged protein, purification was accomplished with glutathione agarose affinity chromatography followed by gel filtration. The bPARG CF yield was 13 nmoles of proteins/liter of culture (Table 1, Figure S1).
Purified PARP1, TNKS1, and PARG CF proteins are enzymatically active
Automodification reactions followed by PARG digestion assays were performed to determine if the purified enzymes were active. Each PARP protein was individually incubated with NAD+ to stimulate pADPr synthesis. After 1 h incubation, the reaction was filtered through a 3 kDa centrifugal filter to separate small molecules from proteins. The protein fraction was collected and treated with bPARG CF. After 30 min incubation, the reaction was filtered through a 3 kDa centrifugal filter and the filtrate was analyzed by ion pairing HPLC for the presence of ADPr [46]. If the enzymes were active, any pADPr made would be digested to ADPr by bPARG CF.
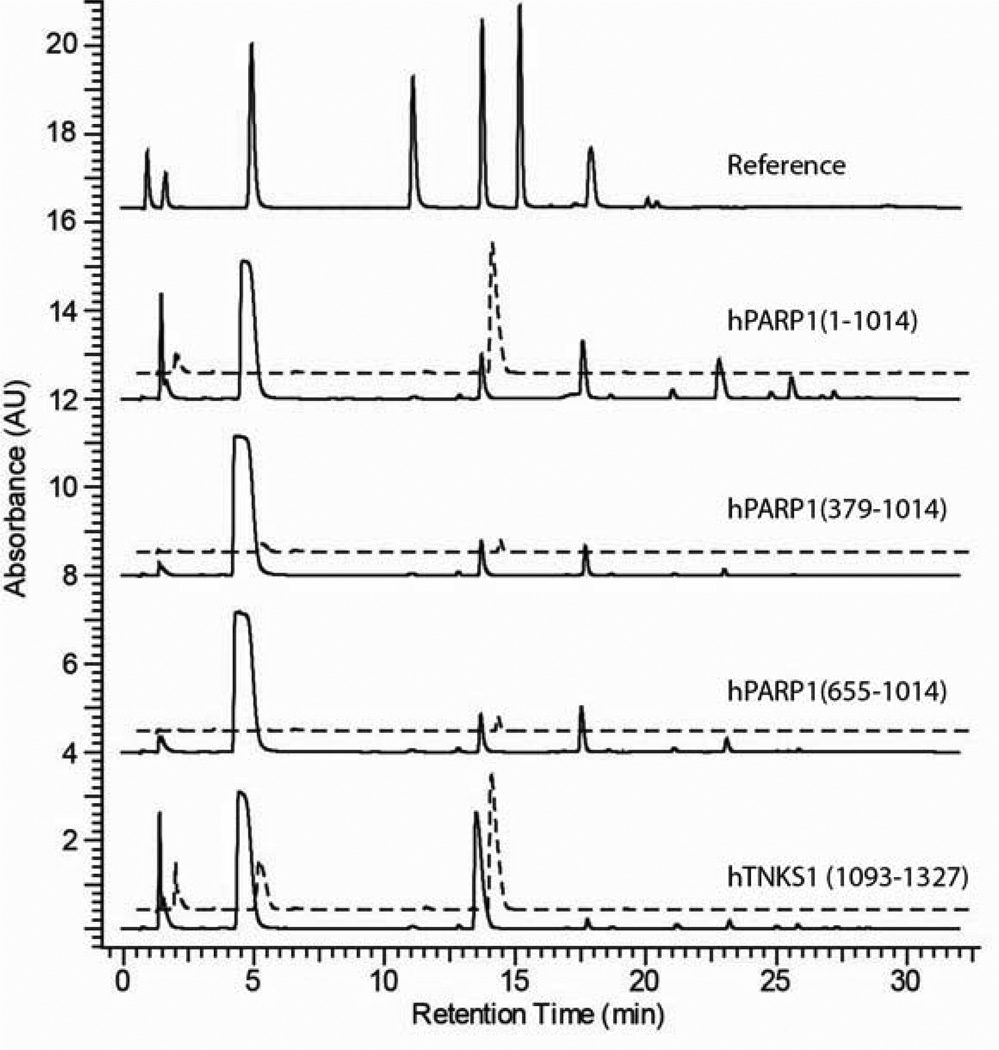
All purified PARP proteins were enzymatically active and capable of biosynthesizing pADPr as evidenced by the presence of an ADPr signal following bPARG CF digestion (Figure 3, dashed lines). Full length hPARP1 (1–1014) and hTNKS1 (1093–1327) were the most active proteins polymerizing 689 ± 202 and 109 ± 27 mol of ADPr/mol of PARP, respectively (Table 2). The polymerase activity of hPARP1 (1–1014) decreased as larger segments of its N-terminus were deleted. Removing the DNA binding domain [hPARP1 (379–1014)] reduced the polymerase activity ~40-fold (from 689 ± 202 to 16 ± 8 mol of ADPr/mol of PARP). An additional 2-fold decrease in polymerase activity (16 ± 8 to 7 ± 5 mol of ADPr/mol of PARP) was observed when the automodification domain [hPARP1 (655–1014)] was also deleted (Table 2). The polymerase activity of hPARP1 (1–1014), hPARP1 (379–1014), and hPARP1 (655–1014) were completely inhibited when treated with 100 µM of ABT-888, a potent hPARP1 inhibitor (Ki = 5.2 nM) (data not shown).
Figure 3.
Ion pairing HPLC analysis of purified PARP enzymes. Solid and dashed lines are traces during polymerization and PARG digestion reactions, respectively. Reference standards from left to right are nicotinamide mono-nucleotide, nicotinamide, NAD+, AMP, ADPr, ADP, and ATP.
Table 2.
Summary of PARP NADase and Polymerase Activity
| NADase Activity | PARP Activity | |
|---|---|---|
| mol of ADPr/ | mol of ADPr/ | |
| Protein | mol of PARP | mol of PARP |
| hPARP1 (1–1014) | 89 ± 23 | 689 ± 202 |
| hPARP1 (379–1014) | 41 ± 9 | 16 ± 8 |
| hPARP1 (655–1014) | 25 ± 7 | 7 ± 5 |
| hTNKS1 (1093–1327) | 141 ± 20 | 109 ± 27 |
Values are the average of at least two independent experiments. Reactions were conducted with excess amounts of NAD+.
Aside from making ADPr polymers, PARPs are also capable of hydrolyzing NAD+ to nicotinamide and ADPr. If the residual NADase activity is more efficient than the polymerase activity, it presents a problem for pADPr scale up because the enzyme would consume NAD+ without generating pADPr. To measure the NADase activity of PARPs, the small molecule fraction from the automodification reaction was analyzed for ADPr content by ion pairing HPLC. Any ADPr molecule generated at this stage originated from NAD+ and is indicative of the level of NADase activity.
HPARP1 (1–1014) and hTNKS1 (1093–1327) were also the most active in NADase activity (Figure 3, solid line). During the automodification reaction, hPARP1 (1–1014) and hTNKS1 (1093–1327) generated 89 ± 23 and 141 ± 20 mol of ADPr/mol of PARP, respectively, from NAD+ (Table 2). Similar to its polymerase activity, the NADase activity of hPARP1 (1–1014) decreased as more domains from its N-terminus were deleted. However, the reduction in NADase activity was less pronounced; decreasing only ~2 and ~3.5 fold when one [hPARP1 (379–1014), 41 ± 9 mol of ADPr/mol of PARP] and two domains [hPARP1 (655–1014), 25 ± 7 mol of ADPr/mol of PARP] were deleted, respectively (Table 2).
Chromatographic Profiling of pADPr
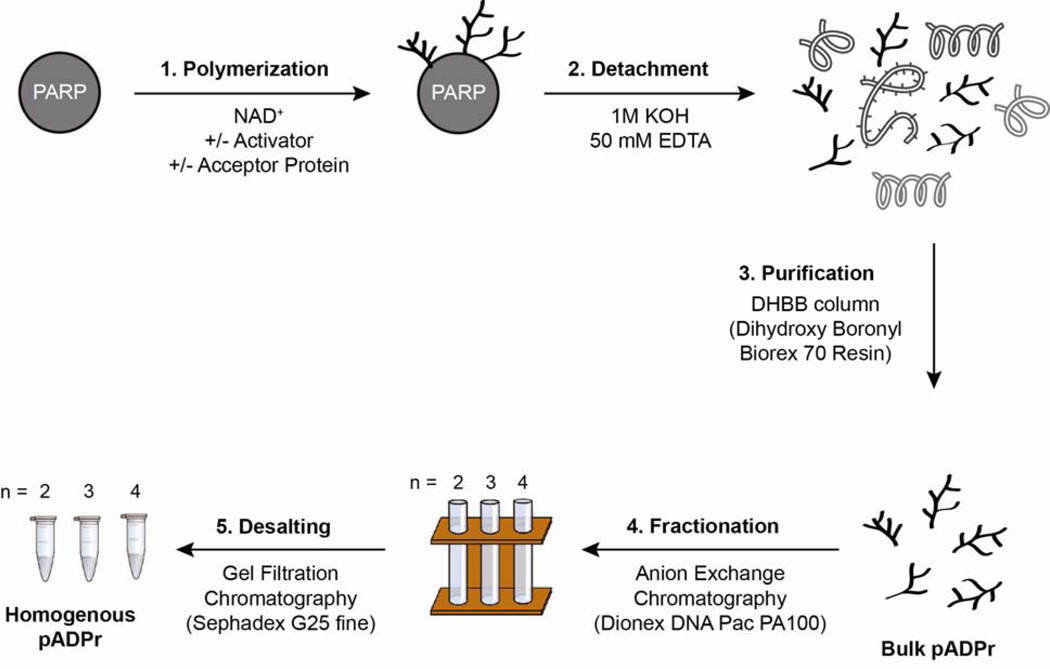
Based on polymerase activity, hPARP1 (1–1014) and hTNKS1 (1093–1327) were the most promising enzymes for pADPr scale up. We next profiled the bulk pADPr biosynthesis efficiency and polymer size distribution of these two proteins under different reaction conditions. The aim here was to prescreen for conditions that maximize pADPr yield and/or influence the size distribution of polymers being made. Starting from purified proteins, bulk and homogenous pADPr can be obtained in three and five steps, respectively (Figure 4). For each conditions tested, bulk pADPr were isolated by detachment with potassium hydroxide treatment followed by affinity chromatography purification with a DHBB column [43; 45]. Bulk polymers were then fractionated on an HPLC by anion exchange chromatography with an analytical Dionex DNA PAC PA100 column to profile pADPr size distribution [40].
Figure 4.
Schematic outline for the biosynthesis and isolation of bulk and homogenous pADPr. Starting from purified PARP proteins, bulk and homogenous pADPr can be obtained in three and five steps, respectively. The identities and concentrations of activator and acceptor protein can be found in the materials and methods.
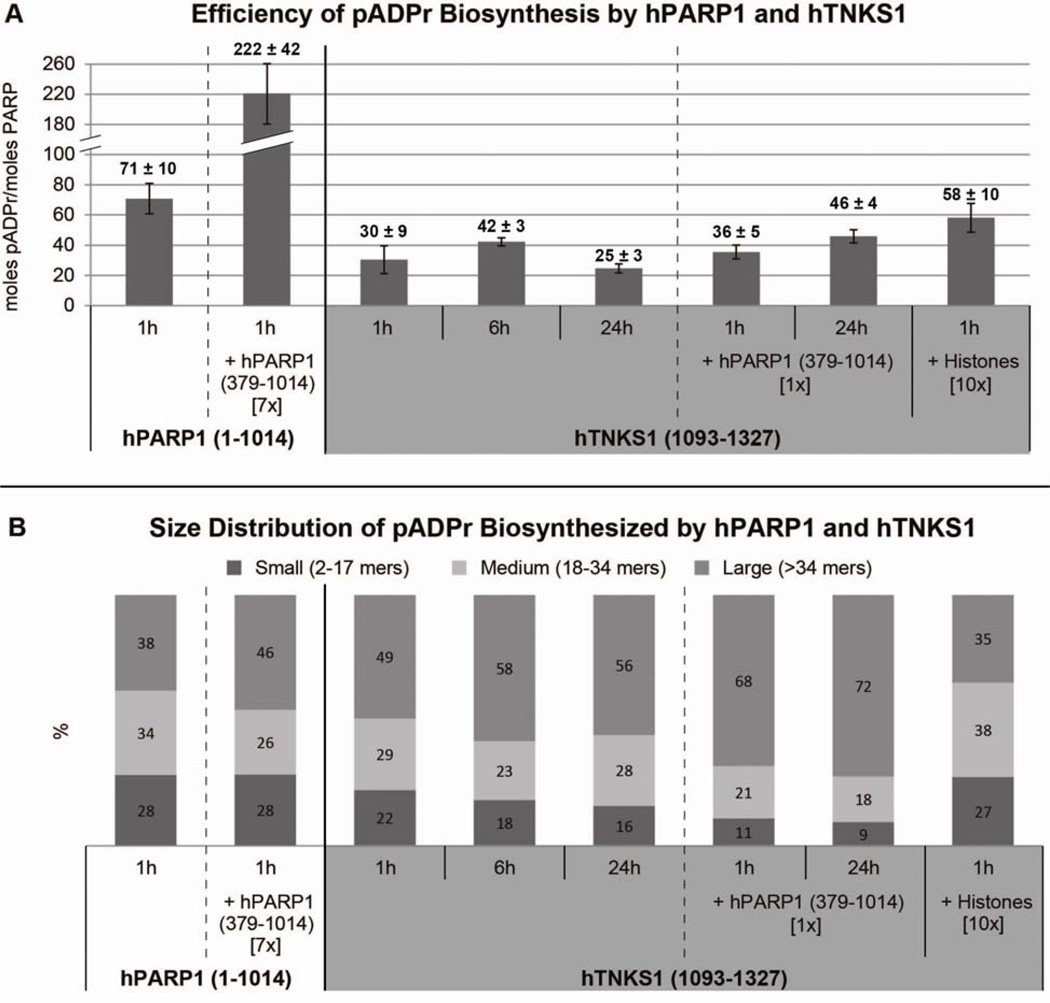
Under the reaction conditions screened, hPARP1 (1–1014) was more efficient at bulk pADPr biosynthesis than hTNKS1 (1093–1327). HPARP1 (1–1014) generated 71 ± 10 mol pADPr/mol PARP when incubated with activated DNA for 1 h (Figure 5a). When 7 mol equivalents of hPARP1 (379–1014) was added to the reaction, the efficiency of hPARP1 (1–1014) increased ~3 fold to 222 ± 42 mol of pADPr/mol PARP. For this measurement, we ignored the contribution from the polymerase activity of hPARP1 (379–1014) because it was ~40 fold less active than hPARP1 (1–1014) (Table 2). We essentially treated hPARP1 (379–1014) as an enzymatically inactive acceptor protein.
Figure 5.
PADPr profiling. (A) Efficiency of bulk pADPr biosynthesis by hPARP1 (1–1014) and hTNKS1 (1093–1327). (B) Size distribution of pADPr biosynthesized by hPARP1 (1–1014) and hTNKS1 (1093–1327). Hours represent reaction incubation time while numbers in [ ] represent molar equivalents. Measured values are the average of at least two independent experiments.
Compared to hPARP1 (1–1014), hTNKS1 (1093–1327) was about two fold less efficient (30 ± 9 mol pADPr/mol PARP) when incubated with NAD+ for 1 h (Figure 5a). The bulk pADPr yield of hTNKS1 (1093–1327) slightly increased at 6 h (42 ± 3 mol pADPr/mol PARP) and was practically the same when allowed to proceed for 24 h (24 ± 3 mol pADPr/mol PARP). Since hPARP1 (379–1014) increased pADPr production of hPARP1 (1–1014), we wondered if it would also enhance the efficiency of hTNKS1 (1093–1327). When one molar equivalent of hPARP1 (379–1014) was added, the pADPr production was the same at 1 h (36 ± 5 mol pADPr/mol PARP) and a slight increase was observed after 24 h (46 ± 4 mol pADPr/mol PARP) (Figure 5a). Interestingly, the addition of hPARP1 (379–1014) nearly doubled the bulk pADPr yield of hTNKS1 (1093–1327) at 24 h (46 ± 4 versus 25 ± 3 mol pADPr/mol PARP).
We next explored the effects of histones on the pADPr production of hTNKS1 (1093–1327). Histones are confirmed hPARP1 substrates that are known to influence the amount and size distribution of pADPr generated by hPARP1 [41; 50]. To our knowledge, it’s unknown if histones are also natural substrates of hTNKS1 inside the cell. Addition of 10 molar equivalents of whole histones doubled the pADPr yield of hTNKS1 (1093–1327) at 1 h incubation from 30 ± 9 to 58 ±10 mol pADPr/mol PARP (Figure 5a). This increase in pADPr yield can be attributed to heteromodification of histones (Figures S2).
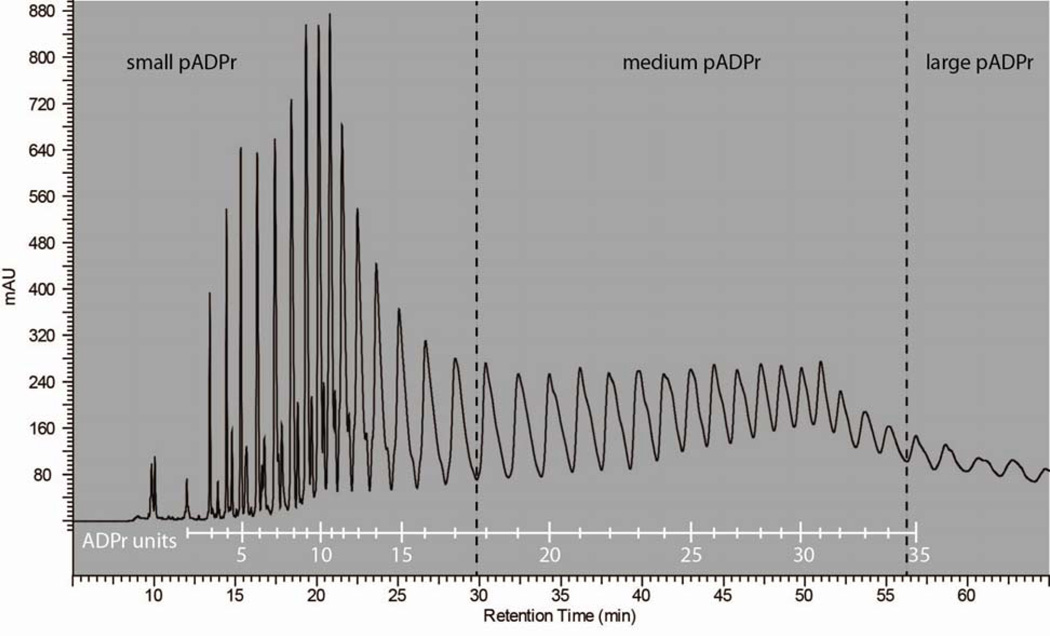
In understanding the biochemistry of PARPs, it is important to understand how pADPr size distribution is affected by substrates. To profile the pADPr size distribution, bulk polymers were fractionated by anion exchange chromatography and the area under the peaks were integrated to estimate the amount of distinct polymers (Figures S3 and S4). Peaks were arbitrarily grouped into three polymer sizes: small (2–17 mers), medium (18–34 mers), and large (>34 mers). Polymer size was determined by submitting a subset of desalted, homogenous pADPr for MALDI-TOF mass spectrometry analysis to determine the molecular weight (Table S1). Figure 6 is the resulting calibration trace correlating peaks to number of ADPr units. The measured molecular mass corresponded to polymers lacking a terminal ribose phosphate or a ribose bisphosphate group. This is consistent with literature findings that alkaline release of pADPr from proteins can result in three polymer isoforms: complete polymer, polymer lacking the terminal ribose phosphate, and polymer missing the terminal ribose bisphosphate group [51].
Figure 6.
Calibration trace correlating peaks to number of ADPr units.
For hPARP1 (1–1014) alone, the pADPr size distribution was 28% small, 34% medium, and 38% large (Figure 5b). In the presence of 7 molar equivalents hPARP1 (379–1014), the amount of large polymers increased 8% at the expense of medium polymers. For hTNKS1 (1093–1327) alone, the pADPr size distribution at 1 h incubation was 22% small, 29% medium, and 49% large (Figure 5b). Longer incubation times (6 and 24 h) increased the amount of large pADPr (+7–9%, from 49% to 56–58%) at the expense small (−4–6%, from 22% to 16–18%) and medium length polymers (−1–6%, from 29% to 23–28%). A shift towards large polymers were more pronounced when hPARP1 (379–1014) was added to the reaction. At 1 h incubation, there was a 19% increase in the amount of large polymers and a decrease of 11% and 8% for small and medium pADPr, respectively (Figure 5b). The same trend was also observed when comparing hTNKS1 (1093–1327) incubations at 24 h with or without hPARP1 (379–1014). The changes in small, medium, and large were −7% (from 16% to 9%), −10% (28% to 18%), and +16% (56% to 72%), respectively.
Addition of 10 molar equivalents of whole histones had the opposite effect from hPARP1 (379–1014) in that it preferentially shifted pADPr production towards small and medium sized polymers. Compared to hTNKS1 (1093–1327) alone, the amount of small and medium sized polymers increased 5% (from 22% to 27%) and 9% (29 % to 38%), respectively, at the expense of a 14% decrease (49% to 35%) in the amount of large pADPr (Figure 5b). A similar trend was previously observed when 10 molar equivalents of histones were added to hPARP1 (1–1014) [41].
Large scale preparation of bulk pADPr
In the large scale preparation of pADPr, we sought to (1) obtain polymers from different PARP enzymes, (2) produce tens to hundreds of milligram quantities of pADPr, and (3) make batches of pADPr enriched in small or large sized polymers. We carried out a total of four rounds of pADPr scale up, once with hPARP1 (1–1014) and three runs with hTNKS1 (1093–1327) as the active PARP enzyme (Table 3). In the only run with hPARP1 (1–1014), we were unable to perform an automodification reaction with hPARP1 (1–1014) alone because the low expression levels of this protein made it difficult to reach the necessary protein concentration of 1 mg/ml. We instead ran a reaction with hPARP1 (379–1014) mixed in to easily reach the 1 mg/ml protein concentration threshold. After lyophilization, we obtained 14.7 mg of hPARP1 (1–1014) bulk pADPr as a white powder.
Table 3.
Summary of bulk pADPr scale up
| Rxn | PARP | Acceptor | Rxn Condition |
Total pADPr Yield |
|---|---|---|---|---|
| A | hPARP1 (1–1014) (2.2 mg, 19 nmol) |
hPARP1 (379–1014) (10 mg, 137 nmol) |
1h, 30°C | 14.7 mg |
| B | hTNKS1 (1093–1327) (24.9 mg, 889 nmol) |
- | 6h, RT | 119.2 mg |
| C | hTNKS1 (1093–1327) (150 mg, 5.36 µmol) |
hPARP1 (379–1014) (150 mg, 2.05 µmol) |
21h, RT | 405 mg |
| D | hTNKS1 (1093–1327) (12 mg, 429 nmol) |
Histone (90 mg, 4.29 µmol) |
1h, RT | 24.8 mg |
In the next three runs, we switched over to hTNKS1 (1093–1327) as the active PARP because more hTNKS1 (1093–1327) was available and it was decently active at polymerizing ADPr (Table 1 and 2). Using only hTNKS1 (1093–1327), incubation with NAD+ for 6 hours at room temperature yielded 119.2 mg of bulk hTNKS1 pADPr after purification and lyophilization.
Having produced hPARP1 and hTNKS1 bulk pADPr, we next made a batch of pADPr enriched in large sized polymers. Compared to the previous run, we increased the scale of the reaction 6 fold and used 5.36 µmol of hTNKS1 (1093–1327) (Table 3). This reaction was carried out for 21 h at room temperature in the presence of hPARP1 (379–1014) because this condition was found to produce nearly 72% large polymers (Figure 5b). It should be noted that we used an equal amount of hPARP1 (379–1014) in terms of weight rather than molarity as in Figure 5b because an equimolar amount of hPARP1 (379–1014) would have required more protein than we had on hand. Although the reaction proceeded smoothly, we encountered column overloading issues during the DHBB purification step. The 30 ml DHBB column we had used for this preparation was saturated when all the bulk pADPr from 5.36 µmol of hTNKS1 (1093–1327) was loaded all at once. After purifying the crude pADPr sample in four batches, this reaction yielded 405 mg of bulk pADPr (Table 3).
In the last round of scale up, only 429 nmol of hTNKS1 (1093–1327) were used but 10 molar equivalents of whole histones were added. Histones were supplemented to bias polymer distribution towards shorter pADPr (Figure 5b). The reaction proceeded as planned and gave 24. 8 mg of bulk pADPr enriched in smaller sized pADPr.
Fractionation of bulk pADPr to homogenous pADPr
To obtain homogenous pADPr, we fractionated bulk pADPr into individual polymers using an anion exchange chromatography. This approach has been used to fractionate radioactive or biotin labeled bulk pADPr into distinct polymers [40; 41]. Since we were interested in obtaining homogenous small and medium sized polymers, we fractionated the 24.8 mg batch of bulk pADPr made in the presence of histones (Table 3). Using a semi-prep Dionex DNA PAC PA100 column (9 × 250 mm), the bulk pADPr were fractionated in 3 mg increments. In this method, the large polymers (>34 mer) were intentionally co-eluted together to reduce the run time. Resolution on the analytical and semi-prep anion exchange columns were comparable (Figure S5).
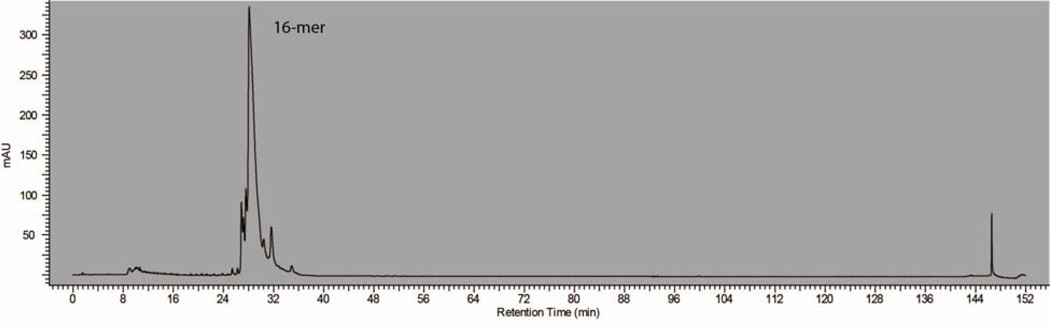
After fractionation, fractions of identical polymers were pooled together and concentrated to dryness using a Genevac evaporator. After reconstituting in a minimal amount of water, the samples were desalted in 2 ml batches by gel filtration chromatography on an HPLC system. Using an FPLC system gave poor results because the UV detector on the FPLC was significantly less sensitive than that of the HPLC. Samples were then concentrated to dryness with a Genevac evaporator and a lyophilizer to yield desalted, pADPr of distinct sizes as a white solid or foam in amounts of up to 2 mg. HPLC analysis of a purified 16-mer pADPr on an analytical anion exchange column showed a single peak and thus confirmed the successful isolation of polymer of a specified length (Figure 7).
Figure 7.
HPLC trace of an isolated 16-mer pADPr from hTNKS1 (1093–1327) with histones polymerization reaction.
Discussion
When activated, PARPs can add polymers to themselves via automodification. As the size of the tethered polymer increases, the buildup of negative charge is thought to kill the enzymatic activity of the PARP through protein denaturation [52; 53; 54]. Due to this property, a large supply of active PARP enzymes is needed to generate vast amounts of pADPr. Previous efforts to obtain bulk and homogenous pADPr have solely focused on using full length PARP1 [PARP1 (1–1014)] as the active enzyme. This is an excellent choice based on enzymatic activity but not in terms of protein expression levels in bacteria. HPARP1 (1–1014) was the most active of the three hPARP1 constructs tested but its bacterial protein yield of 1.57 mg/liter of culture was the poorest. It appears that hPARP1 (1–1014) is somewhat toxic to bacteria [55]. The smaller hPARP1 constructs, hPARP1 (379–1014) and hPARP1 (655–1014), expressed significantly better in bacteria but are unsuitable substitutes for hPARP1 (1–1014) in polymer scale up because their enzymatic activity are 43 to 98 fold lower. The N-terminal DNA binding domain of hPARP1, which are missing in both smaller constructs, was found to be important for catalytic activity [42; 56].
HTNKS1 is a viable alternative to hPARP1 (1–1014) for pADPr scale up. The catalytic domain of hTNKS1 [hTNKS1 (1093–1327)] is enzymatically active and expressed quite well in bacteria. Despite being ~6 fold less active than hPARP1 (1–1014) and having a roughly equal level of ADPr polymerization and NADase activity, we were able to obtain hundreds of milligram amounts of bulk pADPr from hTNKS1 (1093–1327) polymerization reactions. The most we could manage using hPARP1 (1–1014) as the active enzyme was 15 mg of bulk pADPr. The difference in yield can be readily attributed to having more hTNKS1 (1093–1327) than hPARP1 (1–1014) enzymes available for in vitro reactions. If one absolutely requires large amounts of polymers from hPARP1 (1–1014), we highly recommend switching to a baculovirus expression system to gain more access to hPARP1 (1–1014) enzymes. Expressing in Sf9 (Spodoptera frugiperada) cells have been shown to yield up to 40 mg of hPARP1 (1–1014) proteins per liter of culture [57].
This is the first instance of hTNKS1 polymers being prepared in large scale. Compared to the complex, branched pADPr from hPARP1 (1–1014), hTNKS1(1093–1327) is only thought to make linear, branchless polymers [7; 47]. Unfortunately, we are unable to confirm the presence or absence of branching in hTNKS1 (1093–1327) generated polymers because our ion pairing HPLC method was not sensitive enough. We could not detect the diagnostic branching degradation product when we digested bulk polymers from either hPARP1 (1–1014) or hTNKS1 (1093–1327) with phosphodiesterase (data not shown). Branching is a rare event that is known to occur randomly at a low frequency [41].
Fractionating bulk pADPr is the most feasible route to obtaining polymers of defined lengths. By biasing pADPr size distribution, the structural heterogeneity of bulk pADPr is decreased and our chance of accessing substantial amounts of homogenous polymers increases. Previous studies have shown high concentrations of histones influences hPARP1 (1–1014) to produce shorter polymers [41; 50]. The same effect was observed when 10 molar equivalents of histones were added to hTNKS1 (1093–1327) polymerization reactions. There was a 14% increase in the amount of small and medium sized polymers when histones were present. To our knowledge, histones have not been previously shown to be substrates of hTNKS1. We suspect histones shift pADPr distribution towards shorter polymers by capping polymer ends and blocking further polymer growth. The macrodomain present in histones are known to bind to the ADPr units at the tips of pADPr [26]. When ~25 mg of bulk pADPr enriched in shorter polymers were fractionated by anion exchange chromatography, we obtained up to two milligram amounts of homogenous pADPr as a dried, desalted powder. This is a significant milestone as it is the first instance of milligram quantities of homogenous polymers being prepared. With more bulk pADPr, it should be possible to resolve tens of milligram amounts of distinct pADPr. Access to these amounts of homogenous pADPr will greatly facilitate the further characterization of pADPr structure and biochemical properties.
If longer pADPr is desired, one should add hPARP1 (379–1014) to the reaction mixture. Adding seven molar equivalents of hPARP1 (379–1014) to hPARP1 (1–1014) increased the amount of large polymers by 12%. The same trend was also observed when hPARP1 (379–1014) was mixed with hTNKS1 (1093–1327). There was a 16–19% increase in large polymers after 1 and 24 hour incubation in the presence of one molar equivalent of hPARP1 (379–1014). It’s uncertain why the addition of hPARP (379–1014) would lead to longer polymers especially since hPARP1 (379–1014) was found to have poor polymerase activity in automodification reactions (Table 2). Perhaps hPARP1 (379–1014) is capable of adding ADPr units to preexisting polymers but struggles with initiating pADPr formation.
It’s fascinating to note that different acceptor proteins can have opposite effects on the polymer size distribution and the same acceptor protein can differentially affect hPARP1 (1–1014) and hTNKS1 (1093–1327). For both hPARP1 (1–1014) and hTNKS1 (1093–1327), histones shift the polymer distribution to smaller sizes, while hPARP1 (379–1014) shifts the profile to larger polymers [41; 50]. Interestingly hPARP1 (379–1014) affected the activity of hPARP1 (1–1014) and hTNKS1 (1093–1327) differently. For hPARP1 (1–1014), the increase in large pADPr comes at the expense of only medium size polymers (Figure 5b). In the case of hTNKS1 (1093–1327), however, both small and medium sized pADPr are converted into large polymers. Since small, medium, and large polymers are known to have different levels of biological activity, this highlights the importance of characterizing the size distribution of PARPs and how it is influenced by different reactions conditions as it can provide information on the regulation and biological activity of pADPr [28; 40].
The ion pairing HPLC method described above is a convenient assay for measuring the enzymatic activity of PARP and PARG enzymes. The original method from Di Pierro et al. was capable of resolving 38 nucleoside-like compounds in a single 100 minute run [46]. We have adapted this method into a 32 minute run that can readily detect small molecule starting material, byproducts, or degradation metabolites of pADPr. This single method was able to characterize the NADase and polymerase activity of PARP as well as the pADPr hydrolyzing activities of PARG and phosphodiesterase. This method is more sensitive than gel shift assays, more quantitative than western blotting, and does not require the use of radioactivity [42; 58; 59].
The prevailing method to identify pADPr size is to run a high resolution PAGE and visualize the resolved polymers by radioactivity, immunoblotting, or silver staining [51; 60; 61; 62; 63]. Bromophenol blue and xylene cyanol were used as size markers because they co-migrate with ~8 and 20 mers, respectively [59]. This method works well but it’s quite tedious requiring a large, custom sequencing gel to resolve long polymers and multiple steps in the visualization process.
An HPLC based method would be a faster and more convenient alternative. The anion exchange chromatography method to fractionate bulk pADPr was able to resolve at least 50 distinct polymer species. By determining the molecular weights of several peaks with MALDI-TOF mass spectrometry analysis, we created a reference trace correlating peaks to polymer size (Figure 6). In this non-isotopic HPLC method, polymer size can easily be determined by consulting our calibration trace and matching elution time to polymer size. This method also offers the advantage of not sacrificing precious polymer samples as one can collect the material when it elutes off the column.
In conclusion, the major challenges to accessing bulk and homogenous pADPr are the self-inactivating nature of PARPs and the structural heterogeneity of pADPr. Increasing the amount of PARP enzymes in polymerization reactions is the key to overcoming the first issue while adding size modulating proteins to reduce the structural heterogeneity of bulk pADPr can make fractionation into distinct polymers more manageable. Using hTNKS1 (1093–1327) as the active PARP enzyme, we successfully obtained bulk and homogenous pADPr in two to hundreds of milligram scale. By calibrating peaks to number of ADPr units on the anion exchange column trace, we established a convenient, non-isotopic HPLC method for assigning polymer size based on elution time. The same anion exchange chromatography method was readily applied towards profiling the polydispersity of pADPr of PARPs under different reaction conditions. With an improved, robust methods for polymer scale up and characterization, we hope to make pADPr more accessible for future structural and biochemical studies.
Supplementary Material
Acknowledgements
This work was supported by grants from the National Institutes of Health (NCI PO1 grant CA139980 to T.J.M.). E.S.T. was supported by postdoctoral fellowship #116420-PF-09-024-01-CCG from the American Cancer Society. K.A.K. is the Fayez Sarofim Fellow of the Damon Runyon Cancer Research. We thank John Pascal, Henning Kleine, and Paul Chang for generously providing constructs. We also thank Aaron Groen and Sujeong Kim for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hakme A, Wong HK, Dantzer F, Schreiber V. The expanding field of poly(ADP-ribosyl) ation reactions. 'Protein Modifications: Beyond the Usual Suspects' Review Series. EMBO Reports. 2008;9:1094–1100. doi: 10.1038/embor.2008.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altmeyer M, Messner S, Hassa PO, Fey M, Hottiger MO. Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic Acids Research. 2009;37:3723–3738. doi: 10.1093/nar/gkp229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tao Z, Gao P, Liu HW. Identification of the ADP-Ribosylation Sites in the PARP-1 Automodification Domain: Analysis and Implications. Journal of the American Chemical Society. 2009 doi: 10.1021/ja906135d. [DOI] [PubMed] [Google Scholar]
- 4.Diefenbach J, Burkle A. Introduction to poly(ADP-ribose) metabolism. Cellular and Molecular Life Sciences. 2005;62:721–730. doi: 10.1007/s00018-004-4503-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hassa PO, Hottiger MO. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Frontiers in Bioscience. 2008;13:3046–3082. doi: 10.2741/2909. [DOI] [PubMed] [Google Scholar]
- 6.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nature Reviews. Molecular Cell Biology. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 7.Kleine H, Poreba E, Lesniewicz K, Hassa PO, Hottiger MO, Litchfield DW, Shilton BH, Luscher B. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Molecular Cell. 2008;32:57–69. doi: 10.1016/j.molcel.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 8.Hottiger MO, Hassa PO, Luscher B, Schuler H, Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends in Biochemical Sciences. 2010 doi: 10.1016/j.tibs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 9.Heeres JT, Hergenrother PJ. Poly(ADP-ribose) makes a date with death. Current Opinion in Chemical Biology. 2007;11:644–653. doi: 10.1016/j.cbpa.2007.08.038. [DOI] [PubMed] [Google Scholar]
- 10.Chang P, Coughlin M, Mitchison TJ. Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nature Cell Biology. 2005;7:1133–1139. doi: 10.1038/ncb1322. [DOI] [PubMed] [Google Scholar]
- 11.Chang P, Coughlin M, Mitchison TJ. Interaction between Poly(ADP-ribose) and NuMA contributes to mitotic spindle pole assembly. Molecular Biology of the Cell. 2009;20:4575–4585. doi: 10.1091/mbc.E09-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang P, Jacobson MK, Mitchison TJ. Poly(ADP-ribose) is required for spindle assembly and structure. Nature. 2004;432:645–649. doi: 10.1038/nature03061. [DOI] [PubMed] [Google Scholar]
- 13.Huang SM, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, Charlat O, Wiellette E, Zhang Y, Wiessner S, Hild M, Shi X, Wilson CJ, Mickanin C, Myer V, Fazal A, Tomlinson R, Serluca F, Shao W, Cheng H, Shultz M, Rau C, Schirle M, Schlegl J, Ghidelli S, Fawell S, Lu C, Curtis D, Kirschner MW, Lengauer C, Finan PM, Tallarico JA, Bouwmeester T, Porter JA, Bauer A, Cong F. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009 doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 14.Kim MY, Zhang T, Kraus WL. Poly(ADP-ribosyl)ation by PARP-1: 'PAR-laying' NAD+ into a nuclear signal. Genes and Development. 2005;19:1951–1967. doi: 10.1101/gad.1331805. [DOI] [PubMed] [Google Scholar]
- 15.Levaot N, Voytyuk O, Dimitriou I, Sircoulomb F, Chandrakumar A, Deckert M, Krzyzanowski PM, Scotter A, Gu S, Janmohamed S, Cong F, Simoncic PD, Ueki Y, La Rose J, Rottapel R. Loss of Tankyrase-Mediated Destruction of 3BP2 Is the Underlying Pathogenic Mechanism of Cherubism. Cell. 2011;147:1324–1339. doi: 10.1016/j.cell.2011.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malanga M, Althaus FR. The role of poly(ADP-ribose) in the DNA damage signaling network. Biochemistry and Cell Biology. 2005;83:354–364. doi: 10.1139/o05-038. [DOI] [PubMed] [Google Scholar]
- 17.Smith S, Giriat I, Schmitt A, de Lange T. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science. 1998;282:1484–1487. doi: 10.1126/science.282.5393.1484. [DOI] [PubMed] [Google Scholar]
- 18.Ame JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 19.Burkle A. Poly(ADP-ribose). The most elaborate metabolite of NAD+ FEBS Journal. 2005;272:4576–4589. doi: 10.1111/j.1742-4658.2005.04864.x. [DOI] [PubMed] [Google Scholar]
- 20.Smith S. The world according to PARP. Trends in Biochemical Sciences. 2001;26:174–179. doi: 10.1016/s0968-0004(00)01780-1. [DOI] [PubMed] [Google Scholar]
- 21.Aubin RJ, Frechette A, de Murcia G, Mandel P, Lord A, Grondin G, Poirier GG. Correlation between endogenous nucleosomal hyper(ADP-ribosyl)ation of histone H1 and the induction of chromatin relaxation. EMBO Journal. 1983;2:1685–1693. doi: 10.1002/j.1460-2075.1983.tb01643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahel D, Horejsi Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, Flynn H, Skehel M, West SC, Jackson SP, Owen-Hughes T, Boulton SJ. Poly(ADP-ribose)-Dependent Regulation of DNA Repair by the Chromatin Remodeling Enzyme ALC1. Science. 2009 doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caiafa P, Guastafierro T, Zampieri M. Epigenetics: poly(ADP-ribosyl)ation of PARP-1 regulates genomic methylation patterns. FASEB Journal. 2008 doi: 10.1096/fj.08-123265. [DOI] [PubMed] [Google Scholar]
- 24.Eustermann S, Brockmann C, Mehrotra PV, Yang JC, Loakes D, West SC, Ahel I, Neuhaus D. Solution structures of the two PBZ domains from human APLF and their interaction with poly(ADP-ribose) Nature Structural and Molecular Biology. 2010 doi: 10.1038/nsmb.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guettler S, Larose J, Petsalaki E, Gish G, Scotter A, Pawson T, Rottapel R, Sicheri F. Structural basis and sequence rules for substrate recognition by tankyrase explain the basis for cherubism disease. Cell. 2011;147:1340–1354. doi: 10.1016/j.cell.2011.10.046. [DOI] [PubMed] [Google Scholar]
- 26.Timinszky G, Till S, Hassa PO, Hothorn M, Kustatscher G, Nijmeijer B, Colombelli J, Altmeyer M, Stelzer EH, Scheffzek K, Hottiger MO, Ladurner AG. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nature Structural and Molecular Biology. 2009 doi: 10.1038/nsmb.1664. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Liu S, Mickanin C, Feng Y, Charlat O, Michaud GA, Schirle M, Shi X, Hild M, Bauer A, Myer VE, Finan PM, Porter JA, Huang SM, Cong F. RNF146 is a poly(ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling. Nature Cell Biology. 2011;13:623–629. doi: 10.1038/ncb2222. [DOI] [PubMed] [Google Scholar]
- 28.Andrabi SA, Kim NS, Yu SW, Wang H, Koh DW, Sasaki M, Klaus JA, Otsuka T, Zhang Z, Koehler RC, Hurn PD, Poirier GG, Dawson VL, Dawson TM. Poly(ADP-ribose) (PAR) polymer is a death signal. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:18308–18313. doi: 10.1073/pnas.0606526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.David KK, Andrabi SA, Dawson TM, Dawson VL. Parthanatos, a messenger of death. Frontiers in Bioscience. 2009;14:1116–1128. doi: 10.2741/3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, Kim NS, Haince JF, Kang HC, David KK, Andrabi SA, Poirier GG, Dawson VL, Dawson TM. Poly(ADP-ribose) (PAR) binding to apoptosis-inducing factor is critical for PAR polymerase-1-dependent cell death (parthanatos) Science Signalling. 2011;4:ra20. doi: 10.1126/scisignal.2000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu SW, Andrabi SA, Wang H, Kim NS, Poirier GG, Dawson TM, Dawson VL. Apoptosis-inducing factor mediates poly(ADP-ribose) (PAR) polymer-induced cell death. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:18314–18319. doi: 10.1073/pnas.0606528103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu SW, Wang Y, Frydenlund DS, Ottersen OP, Dawson VL, Dawson TM. Outer mitochondrial membrane localization of apoptosis-inducing factor: mechanistic implications for release. ASN Neuro. 2009;1 doi: 10.1042/AN20090046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z, Michaud GA, Cheng Z, Zhang Y, Hinds TR, Fan E, Cong F, Xu W. Recognition of the iso-ADP-ribose moiety in poly(ADP-ribose) by WWE domains suggests a general mechanism for poly(ADP-ribosyl)ation-dependent ubiquitination. Genes and Development. 2012;26:235–240. doi: 10.1101/gad.182618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahel I, Ahel D, Matsusaka T, Clark AJ, Pines J, Boulton SJ, West SC. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–85. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- 35.Minaga T, Kun E. Probable helical conformation of poly(ADP-ribose). The effect of cations on spectral properties. Journal of Biological Chemistry. 1983;258:5726–5730. [PubMed] [Google Scholar]
- 36.Minaga T, Kun E. Spectral analysis of the conformation of polyadenosine diphosphoribose. Evidence indicating secondary structure. Journal of Biological Chemistry. 1983;258:725–730. [PubMed] [Google Scholar]
- 37.Schultheisz HL, Szymczyna BR, Williamson JR. Enzymatic Synthesis and Structural Characterization of (13)C, (15)N-Poly(ADP-ribose) Journal of the American Chemical Society. 2009 doi: 10.1021/ja903155s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Murcia G, Jongstra-Bilen J, Ittel ME, Mandel P, Delain E. Poly(ADP-ribose) polymerase auto-modification and interaction with DNA: electron microscopic visualization. EMBO Journal. 1983;2:543–548. doi: 10.1002/j.1460-2075.1983.tb01460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hayashi K, Tanaka M, Shimada T, Miwa M, Sugimura T. Size and shape of poly(ADP-ribose): examination by gel filtration, gel electrophoresis and electron microscopy. Biochemistry and Biophysical Research Communications. 1983;112:102–107. doi: 10.1016/0006-291x(83)91803-x. [DOI] [PubMed] [Google Scholar]
- 40.Fahrer J, Kranaster R, Altmeyer M, Marx A, Burkle A. Quantitative analysis of the binding affinity of poly(ADP-ribose) to specific binding proteins as a function of chain length. Nucleic Acids Research. 2007;35:e143. doi: 10.1093/nar/gkm944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kiehlbauch CC, Aboul-Ela N, Jacobson EL, Ringer DP, Jacobson MK. High resolution fractionation and characterization of ADP-ribose polymers. Analytical Biochemistry. 1993;208:26–34. doi: 10.1006/abio.1993.1004. [DOI] [PubMed] [Google Scholar]
- 42.Langelier MF, Servent KM, Rogers EE, Pascal JM. A third zinc-binding domain of human poly(ADP-ribose) polymerase-1 coordinates DNA-dependent enzyme activation. Journal of Biological Chemistry. 2008;283:4105–4114. doi: 10.1074/jbc.M708558200. [DOI] [PubMed] [Google Scholar]
- 43.Dantzer F, Ame JC, Schreiber V, Nakamura J, Menissier-de Murcia J, de Murcia G. Poly(ADP-ribose) polymerase-1 activation during DNA damage and repair. Methods in Enzymology. 2006;409:493–510. doi: 10.1016/S0076-6879(05)09029-4. [DOI] [PubMed] [Google Scholar]
- 44.Panzeter PL, Zweifel B, Althaus FR. Fast protein liquid chromatographic purification of poly(ADP-ribose) polymerase and separation of ADP-ribose polymers. Journal of Chromatography A. 1994;678:35–40. [Google Scholar]
- 45.Wielckens K, Bredehorst R, Adamietz P, Hilz H. Protein-bound polymeric and monomeric ADP-ribose residues in hepatic tissues. Comparative analyses using a new procedure for the quantification of poly(ADP-ribose) European Journal of Biochemistry. 1981;117:69–74. doi: 10.1111/j.1432-1033.1981.tb06303.x. [DOI] [PubMed] [Google Scholar]
- 46.Di Pierro D, Tavazzi B, Perno CF, Bartolini M, Balestra E, Calio R, Giardina B, Lazzarino G. An ion-pairing high-performance liquid chromatographic method for the direct simultaneous determination of nucleotides, deoxynucleotides, nicotinic coenzymes, oxypurines, nucleosides, and bases in perchloric acid cell extracts. Analytical Biochemistry. 1995;231:407–412. doi: 10.1006/abio.1995.0071. [DOI] [PubMed] [Google Scholar]
- 47.Rippmann JF, Damm K, Schnapp A. Functional characterization of the poly(ADP-ribose) polymerase activity of tankyrase 1, a potential regulator of telomere length. Journal of Molecular Biology. 2002;323:217–224. doi: 10.1016/s0022-2836(02)00946-4. [DOI] [PubMed] [Google Scholar]
- 48.Simonin F, Hofferer L, Panzeter PL, Muller S, de Murcia G, Althaus FR. The carboxyl-terminal domain of human poly(ADP-ribose) polymerase. Overproduction in Escherichia coli, large scale purification, and characterization. Journal of Biological Chemistry. 1993;268:13454–13461. [PubMed] [Google Scholar]
- 49.Lin W, Ame JC, Aboul-Ela N, Jacobson EL, Jacobson MK. Isolation and characterization of the cDNA encoding bovine poly(ADP-ribose) glycohydrolase. Journal of Biological Chemistry. 1997;272:11895–11901. doi: 10.1074/jbc.272.18.11895. [DOI] [PubMed] [Google Scholar]
- 50.Naegeli H, Althaus FR. Regulation of poly(ADP-ribose) polymerase. Histone-specific adaptations of reaction products. Journal of Biological Chemistry. 1991;266:10596–10601. [PubMed] [Google Scholar]
- 51.Alvarez-Gonzalez R, Jacobson MK. Quantification of Poly(ADP-Ribose) In Vitro: Determination of the ADP-Ribose Chain Length and Branching Pattern. Methods in Molecular Biology. 2011;780:35–46. doi: 10.1007/978-1-61779-270-0_2. [DOI] [PubMed] [Google Scholar]
- 52.D'Amours D, Desnoyers S, D'Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochemical Journal. 1999;342(Pt 2):249–268. [PMC free article] [PubMed] [Google Scholar]
- 53.Ferro AM, Olivera BM. Poly(ADP-ribosylation) in vitro. Reaction parameters and enzyme mechanism. Journal of Biological Chemistry. 1982;257:7808–7813. [PubMed] [Google Scholar]
- 54.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 2010 doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gagnon SN, Desnoyers S. Single amino acid substitution enhances bacterial expression of PARP-4D214A. Molecular and Cellular Biochemistry. 2003;243:15–22. doi: 10.1023/a:1021645327079. [DOI] [PubMed] [Google Scholar]
- 56.Ikejima M, Noguchi S, Yamashita R, Ogura T, Sugimura T, Gill DM, Miwa M. The zinc fingers of human poly(ADP-ribose) polymerase are differentially required for the recognition of DNA breaks and nicks and the consequent enzyme activation. Other structures recognize intact DNA. Journal of Biological Chemistry. 1990;265:21907–21913. [PubMed] [Google Scholar]
- 57.Giner H, Simonin F, de Murcia G, Menissier-de Murcia J. Overproduction and large-scale purification of the human poly(ADP-ribose) polymerase using a baculovirus expression system. Gene. 1992;114:279–283. doi: 10.1016/0378-1119(92)90588-g. [DOI] [PubMed] [Google Scholar]
- 58.Finch KE, Knezevic CE, Nottbohm AC, Partlow KC, Hergenrother PJ. Selective Small Molecule Inhibition of Poly(ADP-Ribose) Glycohydrolase (PARG) ACS Chemical Biology. 2012 doi: 10.1021/cb200506t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haince JF, Poirier GG, Kirkland JB. Nonisotopic Methods for Determination of Poly(ADP-Ribose) Levels and Detection of Poly(ADP-Ribose) Polymerase. Current Protocols in Cell Biology. 2003:18.7.1–18.7.26. doi: 10.1002/0471143030.cb1807s21. [DOI] [PubMed] [Google Scholar]
- 60.Affar EB, Duriez PJ, Shah RG, Winstall E, Germain M, Boucher C, Bourassa S, Kirkland JB, Poirier GG. Immunological determination and size characterization of poly(ADP-ribose) synthesized in vitro and in vivo. Biochimica et Biophysica Acta. 1999;1428:137–146. doi: 10.1016/s0304-4165(99)00054-9. [DOI] [PubMed] [Google Scholar]
- 61.Alvarez-Gonzalez R, Jacobson MK. Characterization of polymers of adenosine diphosphate ribose generated in vitro and in vivo. Biochemistry. 1987;26:3218–3224. doi: 10.1021/bi00385a042. [DOI] [PubMed] [Google Scholar]
- 62.Malanga M, Bachmann S, Panzeter PL, Zweifel B, Althaus FR. Poly(ADP-ribose) quantification at the femtomole level in mammalian cells. Analytical Biochemistry. 1995;228:245–251. doi: 10.1006/abio.1995.1346. [DOI] [PubMed] [Google Scholar]
- 63.Panzeter PL, Althaus FR. High resolution size analysis of ADP-ribose polymers using modified DNA sequencing gels. Nucleic Acids Research. 1990;18:2194. doi: 10.1093/nar/18.8.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.