Summary
Yeast RNA polymerase (Pol) II general factor TFIIE and the TFIIH subunit Ssl2/XPB function in transition of the preinitiation complex (PIC) to the open complex. We find that the three TFIIE winged helix (WH) domains form a heterodimer, with the Tfa1/TFIIEα WH binding the Pol II clamp and the Tfa2/TFIIEβ tandem WH domain encircling promoter DNA that becomes single stranded in the open complex. Ssl2 lies adjacent to TFIIE, enclosing downstream promoter DNA. In contrast to previous proposals, comparison of the PIC and open complex models strongly suggests that Ssl2 promotes DNA opening by functioning as a double stranded DNA translocase, feeding 15 bp of double stranded DNA into the Pol II cleft. Right-handed threading of DNA through the Ssl2 binding groove, combined with the fixed position of upstream promoter DNA, will lead to DNA unwinding and the open state.
Introduction
Protein coding genes are transcribed by RNA polymerase (Pol) II in conjunction with a set of six general transcription factors (GTFs): TFIIA, TBP, TFIIB, TFIIF, TFIIE, and TFIIH 1,2. A key intermediate in transcription initiation is the preinitiation complex (PIC). In this state, several DNA-GTF interactions anchor Pol II to double stranded promoter DNA. The PIC then transitions to the open complex state where, in an ATP-dependent reaction, ~10 bp of DNA surrounding the transcription start site are unwound and the DNA template strand is positioned within the Pol II cleft 3,4. Pol II then locates the transcription start site, RNA synthesis is initiated, and Pol II enters a processive elongation state.
Biochemical data combined with x-ray structures have led to models for the partially assembled PIC and open complex 5–10. In the PIC models, binding of TFIIB to the Pol II surface sets the path of promoter DNA across the cleft directly above the enzyme active site, while two structured domains of TFIIF are positioned on the outside surface of Pol II, connected by a flexible linker located near the Pol II protrusion domain 6,7. Two flexible TFIIB segments termed reader and linker are positioned within the cleft and, in the open complex state, the reader is proposed to recognize template strand DNA in the enzyme active site 9,10. A key unanswered question is how the PIC transitions to the open complex. TFIIE and TFIIH function in this step, since the requirement for these factors is eliminated by promoter DNA containing a preformed heteroduplex bubble 8,11–13. However, detailed molecular information on the location of these factors in the PIC and how they function is lacking, leading to a major gap in our understanding of the initiation mechanism.
TFIIE binds the PIC intermediate containing Pol II, TBP, TFIIB, and TFIIF, which is then bound by TFIIH to complete PIC assembly. Mammalian TFIIE is a conserved heterodimer consisting of two subunits, TFIIEα and TFIIEβ, corresponding to the S. cerevisiae subunits Tfa1 and Tfa2 14,15. Tfa1 contains an N-terminal winged helix (WH) domain followed by a zinc ribbon (ZR) domain and both are essential for TFIIE function 16–19 (Figure 1). Although WH domains typically bind DNA 20, the predicted DNA binding groove of the Tfa1 WH is negatively charged and may not bind DNA in the PIC 21. The TFIIEβ/Tfa2 subunit contains a central tandem WH domain, an architecture conserved in the Pol I and Pol III subunits Rpa49 and Rpc34 22. The TFIIEβ/Tfa2 N-terminal WH (WH1) binds double stranded DNA in vitro 23,24, while WH2 resembles the WH fold with an additional C-terminal α-helix found in the archaeal transcription factor Sto12a 25 that is otherwise unrelated to TFIIE. Previous findings suggest that the Tfa1 N-terminus and the Tfa2 C-terminus are involved in dimerization 16,19,26 and residues flanking the Tfa1 ZR crosslink to the C-terminus of Tfa2 27. Protein-protein and protein-DNA crosslinking studies have all indicated that TFIIE binds Pol near the clamp domain 5,28,29. The WH of TFE, the archaeal ortholog of Tfa1 21, binds the coiled-coil element in the Pol clamp where it competes for binding with the elongation factor Spt4/5 30 and also crosslinks to the non-template DNA strand in the open complex 31.
Figure 1.
Functionally important regions in TFIIE. Summary of TFIIE subunit domains showing intramolecular FeBABE cleavage and in vivo function. Structured domains in either subunit are labeled (WH = winged helix, ZR = zinc ribbon), as are positions that were linked to FeBABE in the course of this work. Shaded bars summarize functional assays shown in Supplementary Fig. 1a, b (black = essential for yeast growth, grey = partially redundant WH domains (lethal when both deleted), white = no phenotype upon mutation). Lines connecting Tfa1 and Tfa2 represent intramolecular FeBABE cleavages when FeBABE was conjugated to the indicated positions in Tfa1 (Supplementary Fig. 1c; Supplementary Table 1).
ATPase activity of the helicase-related TFIIH subunit XPB (yeast Ssl2/Rad25) is essential for promoting DNA strand separation 32. Photocrosslinking and biochemical cleavage data showed that Ssl2/XPB interacts with DNA downstream from the site of melting 28,29, ruling out a mechanism whereby XPB directly unzips the DNA strands. Two models have been suggested for the mechanism of XPB. First, it was proposed that XPB binds to a specific downstream location on promoter DNA and, while remaining at that location, rotates DNA to generate torque that results in DNA melting 28. Alternatively, it was proposed that the XPB ATPase but not helicase activity is required, possibly by directing a conformational change in the PIC that drives open complex formation 33. However, the lack of information on the precise location of Ssl2/XPB in the PIC has made it difficult to distinguish between these and other mechanisms.
Here, we used biochemical probes, genetic assays, and molecular modeling to locate both TFIIE and Ssl2 in the PIC. We find that the Tfa1 WH anchors TFIIE to the Pol II clamp and dimerizes with the Tfa2 tandem WH domain, positioning it to encircle upstream promoter DNA in the PIC and in position to interact with single stranded DNA in the open complex. Furthermore, probes on TFIIE and Ssl2 fix the location of Ssl2 as directly adjacent to TFIIE over downstream promoter DNA. Combined with previous work, our results strongly suggest that Ssl2/XPB acts as a double stranded DNA translocase, inserting 15 bp of double stranded DNA into the Pol II cleft, thereby promoting DNA melting. Finally, crosslinking probes on Ssl2 show an unexpected interaction with TFIIB and suggest that the PIC exists in at least two conformations, differing in the location of the TFIIB reader/linker domains.
Results
Identification of functionally important regions in TFIIE
To determine the ideal locations for placement of biochemical probes within TFIIE, we first identified important functional regions using a yeast genetic assay. A series of small internal deletions were generated in Tfa1 and Tfa2 and a plasmid shuffle assay used to test in vivo function (summarized in Figure 1; Supplementary Fig. 1). Western blot analysis showed that nearly all of the deletion variants were expressed at wild type levels. We found that disruption of either the Tfa1 WH or ZR domains is lethal (Supplementary Fig. 1a). Strains containing small deletions throughout the Tfa1 C-terminal region had normal growth rates, showing that the Tfa1 C-terminus contains no single essential element, however, strains with large C-terminal deletions grew slowly (e.g., Tfa1 Δ213-482), consistent with previous results 18,19. Within the Tfa2 tandem WH domain, deletion of WH1 had no growth phenotype, deletion of WH2 showed slow growth, and deletions removing both WH1 and WH2 were lethal (Supplementary Fig. 1b). This suggests that the individual Tfa2 WHs have partially redundant function. Within Tfa2, the only lethal small deletions were those associated with the TFIIE dimerization region: The lethal mutation Tfa2 Δ249-268 removes a helix in WH2 that associates with Tfa1 (see below) and deletion Δ282-328 removes residues of the probable dimerization region that crosslink to Tfa1 34.
Architecture of the TFIIE dimerization domain
Based on the above analysis, structure models were generated for the functionally important TFIIE domains (Online Methods). The individual Tfa2 WH domain structures were oriented relative to each other by alignment with the tandem WH domains in the TFIIE/F-related Pol I factor Rpa49 22. When this Tfa2 model was docked within the PIC, the Rpa49-based alignment of the WH domains agreed exceptionally well with results from the biochemical probes (see below).
The TFIIE models were then analyzed, and predicted surface exposed residues were selected within the TFIIE WH and ZR domains as sites for linkage of the hydroxyl radical-generating probe FeBABE 35,36 (Figure 1, Supplementary Table 1). These residues were substituted with cysteine in Tfa1 and Tfa2 derivatives that lacked endogenous cysteines apart from those in the ZR domain. The recombinant TFIIE derivatives were purified and conjugated to FeBABE 5 and all the TFIIE-FeBABE derivatives rescued transcription from yeast nuclear extracts depleted for TFIIE function (see below).
PICs were formed on an immobilized TATA-containing yeast HIS4 promoter 37 using yeast nuclear extracts supplemented with the TFIIE-FeBABE derivatives. FeBABE-induced protein cleavage was activated with H2O2 and cleavage monitored on Western blots, using a ladder of in vitro translated polypeptides as size markers 36. FeBABE typically induces protein cleavage within ~30 Å from the probe 5,36. To investigate the nature of the TFIIE dimerization domain, we first measured intramolecular cleavage within TFIIE using FeBABE on Tfa1, detecting cleavages on C-terminal Flag-tagged Tfa2. In the PIC, FeBABE at Tfa1 WH residues Glu53, Pro56, and Arg62 generated cleavage within Tfa2 WH2, whereas FeBABE at Tfa1 His93 strongly cleaved Tfa2 at residues C-terminal to WH2 (summarized in Figure 1; Supplementary Fig. 1c; supplementary Table 1). In contrast, probes positioned within the Tfa1 ZR did not cleave Tfa2 in the PIC, but did generate multiple cleavages within WH2 and the C-terminal region of Tfa2 in the presence of only Pol II. This result suggests that the Tfa1 ZR changes its orientation with respect to the remainder of TFIIE when incorporated into the PIC, possibly due to TFIIE interactions with other PIC components. Our combined biochemical and genetic data suggest that in the PIC, the TFIIE dimerization domain contains the Tfa1 WH domain, and the C-terminal region of Tfa2, including the predicted helix 4 of the Tfa2 WH2 domain. As described above, both of these regions are essential for viability.
TFIIE interacts with Pol II and spans the active site cleft
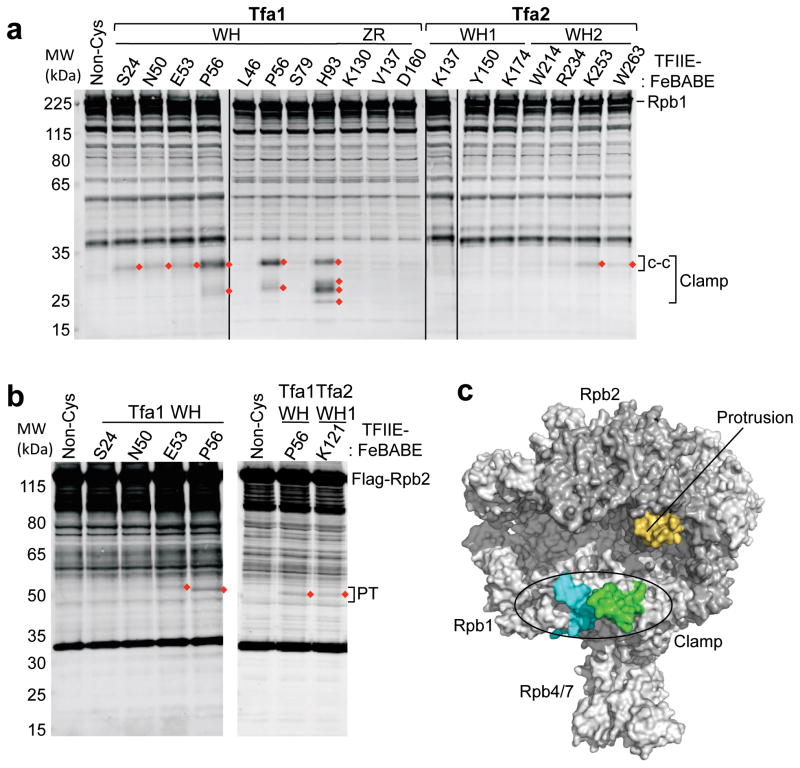
To locate the position of TFIIE in PICs, the cleavage experiments were repeated using Pol II containing epitope tags on either Rpb2, Rpb4, or Rpb7. Rpb1 cleavage was monitored using either an antibody recognizing the Rpb1 N-terminus or a C-terminal Flag tag where indicated. Protein cleavage from the TFIIE-FeBABE derivatives was observed exclusively in Rpb1 and Rpb2 (Supplementary Table 1; representative data in Figure 2a,b). The locations of these cleavages are shown on the Pol II structure in Figure 2c, with the calculated cleavage sites ± 10 residues on either side highlighted to account for uncertainty in the cleavage position 6. We first examined Pol II cleavage generated by probes within the Tfa1 WH, which cleaved Rpb1 exclusively in the clamp domain. FeBABE at the tightly clustered residues Ser24, Asn50, Glu53, and Pro56 all cleaved within the coiled-coil region of the Pol II clamp (Figure 2c, green). FeBABE at Tfa1 His93 and, to a lesser extent, Pro56 cleaved a segment of the Rpb1 clamp head domain (residues 179–222) adjacent to the coiled-coil motif (Figure 2c, cyan). FeBABE positioned at the C-terminus of Tfa2 WH2 (residues Lys253 and Trp263) also cleaved the clamp coiled-coil, consistent with the above findings that this region of Tfa2 and Tfa1 WH comprise part of the TFIIE dimerization region. Two positions in the Tfa1 WH and one position in Tfa2 WH1 all generated weak cleavage in the Rpb2 protrusion domain (Figure 2b, c, orange). Combined, these results show that TFIIE is situated so that it spans the Pol II cleft in the PIC.
Figure 2.
Mapping of TFIIE-FeBABE cleavage in Pol II. (a) Hydroxyl radical-mediated cleavage in Pol II subunit Rpb1 by the indicated TFIIE-FeBABE variants. PICs were formed at the HIS4 promoter using yeast nuclear extracts that were supplemented with recombinant TFIIE-FeBABE. Rpb1 cleavage products were visualized by Western blot using an antibody against the N-terminus of Rpb1 6. Red diamonds indicate reproducibly observed cleavage products compared to a reaction with a non cysteine-containing TFIIE (Non-Cys; lane 1). Brackets on the right indicate cleavage in the clamp and the coiled-coil domain (c-c). Bands in the Non-Cys control (lane 1) correspond to full length Rpb1 and non-specific reactivity of the primary/secondary antibodies with other proteins in the reaction. (b) Rpb2 cleavage using the indicated TFIIE-FeBABE derivatives. The right bracket indicates cleavage in the protrusion domain (PT). (c) Summary of the TFIIE-FeBABE mediated cleavages on Pol II. All cleavage events that are summarized in Table S1 were mapped on the Pol II surface and 10 residues on either side of the calculated cleavage sites are colored (green = cleavage in the Rpb1 clamp coiled-coil domain generated by FeBABE linked to residues in the TFIIE dimerization domain; cyan = cleavage in the Rpb1 clamp head by FeBABE conjugated to residue Tfa1 His93; orange = cleavage in the Rpb2 protrusion domain by FeBABE at Tfa1 Glu53, Pro56, and Tfa2 Lys121).
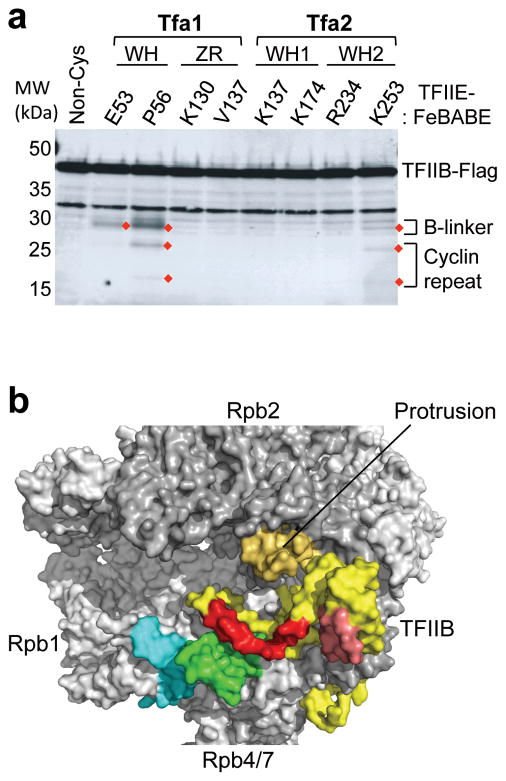
To confirm and extend the above findings, we used the TFIIE-FeBABE derivatives to assay for cleavage in TFIIB, since part of TFIIB is positioned within the Pol II cleft 9,10. Cleavage of a C-terminal Flag tagged TFIIB was visualized by Western blot. We found one strong and two weaker cleavage sites in TFIIB that were generated by FeBABE conjugated to residues in the TFIIE dimerization domain (Figure 3a). When mapped in the PIC model, the major cleavage site is located in the TFIIB linker (Figure 3b, red), as predicted by the position of the TFIIE dimerization domain at the Rpb1 clamp. The weaker cleavage sites are localized in the first TFIIB cyclin repeat (Figure 3b, salmon).
Figure 3.
Cleavage of TFIIB by TFIIE-FeBABE in the PIC. (a) TFIIB cleavage products generated by TFIIE-FeBABE. PICs containing the indicated TFIIE-FeBABE derivatives and TFIIB-Flag were treated with H2O2 and analyzed by Western blot. Cleavage products are highlighted with red diamonds, and the brackets on the right indicate cleavage in either the B-linker or the first TFIIB cyclin repeat domain. (b) Cleavage results from a were mapped to TFIIB (yellow) where 10 residues on either side of the calculated cleavage site are highlighted (red = cleavage in the TFIIB linker region; salmon = cleavage in the first TFIIB cyclin repeat). Cleavages in Pol II are colored as described in Figure 2c.
Position of TFIIE in the PIC
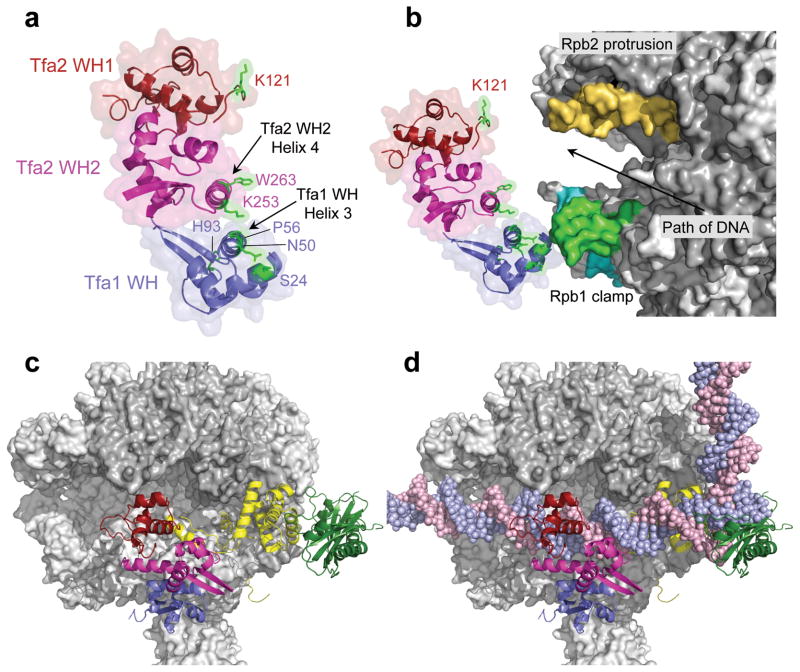
To map the molecular position of TFIIE in the PIC, we first modeled the relative alignment of the three TFIIE WH domains based on the above intramolecular FeBABE cleavage results, which strongly suggests that helix 3 in the Tfa1 WH dimerizes with helix 4 of the Tfa2 WH2 domain (Figure 4a; Supplementary Fig. 2a). This organization of the TFIIE dimerization domain is further supported by genetic assays discussed below. In Figure 4, TFIIE residues where FeBABE attachment cleaves Pol II and TFIIB are colored green. This arrangement of the three WH domains is consistent with previous intramolecular TFIIE crosslinking/mass spectrometry data 34 as well as our finding that helix 4 in Tfa2 WH2 is essential for function.
Figure 4.
Position of TFIIE in the PIC. (a) The TFIIE dimerization domain is formed in part by helix 3 in the Tfa1 WH domain and helix 4 in the Tfa2 WH2 domain. FeBABE-positions that cleave Pol II are highlighted in green. The Tfa1 WH domain is colored blue; the Tfa2 WH1 and WH2 domains are colored red and pink, respectively. (b) Position of the three TFIIE WH domains in the PIC based on the combined FeBABE cleavage data. Residues in TFIIE that cleave Pol II when linked to FeBABE are highlighted in green, as is the corresponding cleavage site in the coiled-coil region of the Pol II clamp. The cleavage site in the clamp head by Tfa1 residue His93 is colored in cyan, and cleavage in the Rpb2 protrusion domain is colored orange. An arrow indicates the path of the double stranded DNA in the PIC. (c, d) Top view of the Pol II PIC model containing Pol II (grey), TFIIB (yellow), TBP (dark green), and TFIIE without (c) and with (d) promoter DNA.
Next, TFIIE was docked to the surface of Pol II, based on the cleavage data from individual TFIIE-FeBABE variants (Figure 4b). In this model, the Tfa1 WH directly interacts with the clamp coiled-coil region while the Tfa2 tandem WH domain spans the Pol II cleft. FeBABE at Tfa1 His93 mainly cleaves the Rpb1 clamp in a region facing downstream DNA, while FeBABE at residues near Tfa1 Pro56 and helix 4 in Tfa2 WH2 strongly cleave the Rpb1 clamp coiled-coil, a region facing upstream DNA (Supplementary Fig. 2b). Residues in TFIIE that point away from either Rpb1 or Rpb2 do not cleave Pol II (Supplementary Fig. 2c). Consistent with this positioning, residue Lys121 in Tfa2 WH1, which points towards Rpb2, cleaves in the protrusion domain (Figure 4b). Finally, cleavage of the TFIIB linker and cyclin repeat domains by the TFIIE probes is in excellent agreement with the structure model.
To extend these findings, we assayed for cleavage within Tfg2, the small TFIIF subunit. Tfg2 contains two structured domains, binding to separate positions on Rpb2, that are connected by an unstructured loop near the Pol II protrusion 6,7. Consistent with our TFIIE modeling, we found that FeBABE at Tfa2 WH1 residues Lys137, Tyr150, Lys154 and Tfa1 Asn50 all cleaved the Tfg2 loop (Supplementary Fig. 2d). Based on the cleavage pattern in the protrusion domain and the space occupied by TFIIE, this new data, combined with our previous results, suggests that the Tfg2 loop lies on top of the protrusion. From this location, the Tfg2 loop likely interacts with Tfa2 WH1, perhaps explaining why TFIIE joins the PIC after TFIIF incorporation. The Tfg2 loop may assist in ordering the flexible protrusion domain and act to modulate the interaction of Pol II with other GTFs that bind nearby.
The arrangement of TFIIE on Pol II fits well with the proposed path of promoter DNA in the PIC 9,29,38. First, there is sufficient space in the model for promoter DNA between the tandem WH domain and Pol II (Figure 4). In the PIC model, TFIIE closely approaches promoter DNA bases −18 to −8 (using the mammalian numbering system defining the first T of TATA as −31), the segment where FeBABE linked to promoter DNA gives TFIIE cleavage 29. This also agrees with previous TFIIE-DNA promoter crosslinking in the human system 28,39. From this location in the PIC model, TFIIE encircles upstream promoter DNA near the site of initial DNA opening, where it is in position to potentially assist with melting, as well as to interact with single stranded non template DNA in the open complex. Coordinates of the PIC and open complex model discussed below are available at the Hahn laboratory web site: http://labs.fhcrc.org/hahn/Research/structure_coord/
Mutations disrupt TFIIE dimerization and Pol II-TFIIE interaction
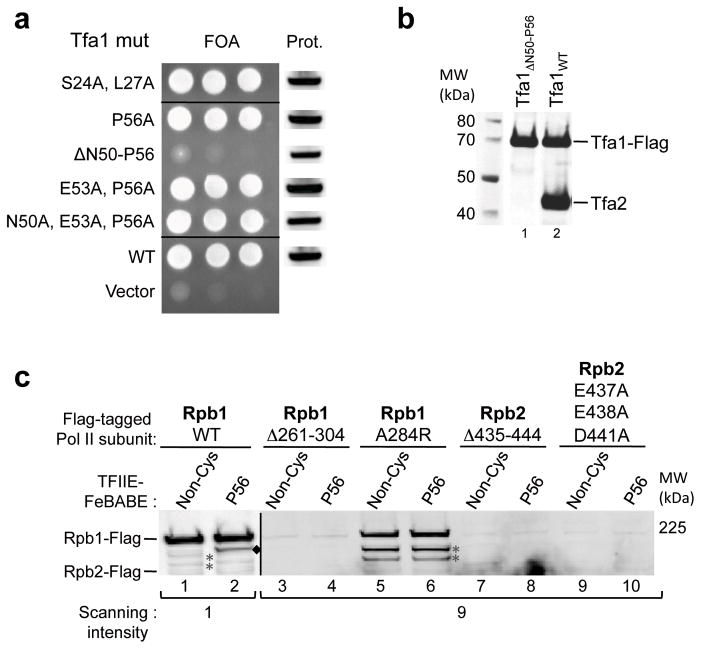
To confirm that the Tfa1 residues used to attach FeBABE were not critical for TFIIE function, we first supplemented nuclear extracts depleted for TFIIE with the FeBABE conjugated TFIIE variants. All of these variants rescued transcription to wild type levels (Supplementary Fig. 3a). Furthermore, yeast containing single or multiple alanine substitutions at Tfa1 residues Ser24, Leu27, Asn50, Glu53, and Pro56 showed wild type growth (Figure 5a; Supplementary Fig. 3b). In contrast, a small deletion within helix 3 of the Tfa1 WH domain, suggested above to be part of the TFIIE dimerization interface, was lethal (Tfa1 Δ50-56). To address the effect of this mutation on TFIIE dimerization, Flag-tagged mutant and wild type Tfa1 were purified from yeast whole-cell extracts via anti-Flag affinity beads and analyzed by Western blot. Whereas Tfa2 efficiently co-purified with wild type Tfa1, the Tfa1 Δ50-56 variant did not bind Tfa2 (Figure 5b), confirming the key role for Tfa1 helix 3 in TFIIE dimerization. This was further confirmed by bacterial co-expression and purification of Tfa2 with either wild type Tfa1 or a Tfa1 mutant in which helix 3 of the WH domain was deleted (Tfa1 Δ51-64). As predicted, this Tfa1 deletion abolished co-purification of Tfa1 and Tfa2 (Supplementary Fig. 3c).
Figure 5.
Effect of mutations in the TFIIE dimerization, Pol II clamp and protrusion domains. (a) Wild type TFIIE was selected against by plasmid shuffle assay and growth of TFA1 mutants measured on 5-fluoroorotic acid plates. On the left is growth of serial yeast cell dilutions and the right shows Western blot analysis of Flag- Tfa1 expression level in cells before plasmid shuffle. (b) A small deletion in helix 3 of the Tfa1 WH domain abolishes TFIIE dimerization. Tfa1 in which residues 50–56 of helix 3 were deleted (lane 1), and wild type Tfa1 (lane 2) were purified via anti-Flag affinity beads from yeast whole cell extracts and analyzed for CoIP of Tfa2. (c) The effect of Pol II mutations on PIC formation and TFIIE binding. PIC formation was attempted with yeast nuclear extracts containing Flag-tagged Pol II mutants (Rpb1 Δ261-304: deletion of the clamp coiled-coil domain; Rpb1 A284R: substitution of conserved A284 at the clamp coiled-coil; Rpb2 Δ435-444: deletion of the unstructured Rpb2 protrusion; Rpb2 E437A E438A D441A: substitution of acidic residues in the Rpb2 protrusion) supplemented with TFIIE −/+ FeBABE linked to residue Pro56. Purified PICs and cleavage products were analyzed by Western Blot, probing for Flag-tagged Rpb1 or Rpb2. Cleavage of wild type Rpb1 (WT) is labeled with black diamonds. Due to different Pol II expression levels, lanes containing wild type Pol II were scanned at intensity 1 on the Odyssey LI-COR system, reactions containing mutant Pol II were scanned at intensity 9. Asterisks indicate non specific Western blot signals, darker in lanes 5,6 because of the higher scanning intensity.
Mutations were also generated in the two domains of Pol II that are cleaved by TFIIE-FeBABE derivatives to test if these domains affect TFIIE-Pol II binding and PIC assembly. Two mutations were made in the Rpb1 clamp coiled-coil element: A complete deletion of the coiled-coil (Rpb1 Δ261-304), and a point mutation at the tip of the coiled coil (A284R) analogous to the A260R mutation made in archaeal Rpb1 that retains the ability to form PICs 9. Two mutations were also made in the Rpb2 protrusion domain: a deletion of the unstructured region (Rpb2 Δ435-444), and a triple mutation that eliminates three closely spaced acidic residues in the unstructured region (Rpb2 E437A E438A D441A). The mutant Pol II subunits all contained a 3X Flag tag at the C-terminus and were co-expressed in yeast with the corresponding untagged wild type subunit, since the mutants were expected to be detrimental to yeast growth. The mutant Pols were purified via anti-Flag affinity beads and analyzed by SDS PAGE and silver stain (Supplementary Fig. 3d), showing that the mutant subunits were all incorporated into Pol II.
To analyze whether the mutant Pols had reduced interaction with TFIIE, we first attempted immune precipitation using the purified Pol II derivatives and wild type recombinant TFIIE. However, this method was not effective, due to the low affinity of TFIIE for wild type Pol II. As an alternative, we prepared nuclear extracts from strains that co-expressed both the Flag-tagged Pol II variants and untagged wild type Pol II. These extracts, along with the recombinant TFIIE derivative Tfa1 Pro56-FeBABE, were used to form PICs at the immobilized HIS4 promoter. The TFIIE-FeBABE derivative allowed us to assay cleavage of the Flag-tagged Pol II subunit as a measure of TFIIE and mutant Pol II binding within the same PIC (Figure 5c). Western analysis of PICs formed using the various Pol II mutants surprisingly showed that all except Rpb1 A284R were strongly impaired for PIC formation (Figure 5c, lanes 1,3,5,7,9). The ability of the Rpb1 A284R derivative to bind HIS4 allowed us to test if mutation of the coiled-coil domain affects TFIIE binding. While Tfa1 Pro56-FeBABE cleaved within the coiled-coil domain of wild type Pol II, no FeBABE-specific cleavage was detected with the Rpb1 A284R derivative (Figure 5c, lanes 2,6). This finding suggests that in the PIC, TFIIE-Pol II binding is impaired or altered by a mutation in the Rpb1 clamp coiled-coil.
Position of the TFIIH subunit Ssl2/XPB in the PIC
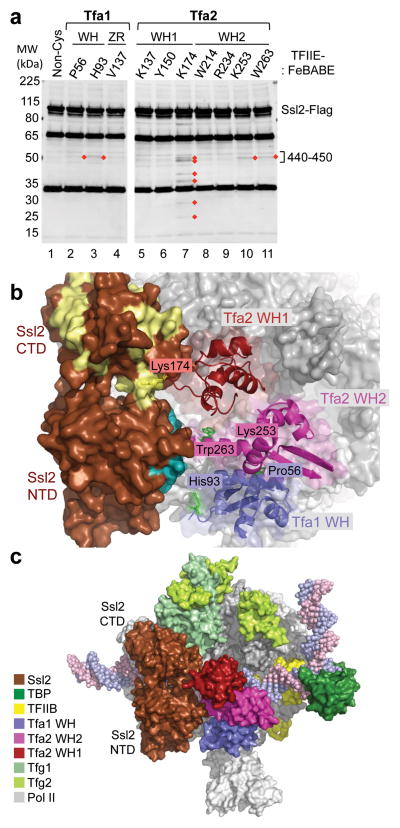
Determining the precise position of the TFIIH subunit Ssl2/XPB within the PIC is essential for understanding how it acts to promote DNA strand separation. Previous DNA-protein crosslinking and DNA-FeBABE cleavage results showed that Ssl2/XPB is located within a ~30 bp window downstream of TFIIE in our new PIC model 28,29. To more precisely localize the RecA-like domains of Ssl2 in the PIC, we assayed for cleavage of Flag-tagged Ssl2 by TFIIE-FeBABE derivatives (Figure 6a). FeBABE linked to Tfa2 WH2 at the residues Lys253 and Trp263 both cleave Ssl2 within the N-terminal RecA repeat. Nearly identical cleavage sites are observed using FeBABE linked to the Tfa1 WH residues Pro56 and His93. In contrast, FeBABE at Tfa2 Lys174, in a flexible loop in WH2, cleaves at 6 sites spread over both the N- and C-terminal RecA-like domains (Figure 6a, lane 7).
Figure 6.
Orientation of the TFIIH subunit Ssl2/XPB in the PIC. (a) Cleavage in Ssl2 by TFIIE-FeBABE variants. PICs were formed with nuclear extracts containing Ssl2-Flag and supplemented with TFIIE-FeBABE. Cleavage was induced by addition of H2O2 and products were visualized by Western blot using an anti-Flag antibody. Reproducibly observed cleavage products are highlighted with red diamonds. The majority of TFIIE-FeBABE variants that cut Ssl2 cleaved in a region spanning residues 440–450 on the Ssl2 N-terminal domain. (b) PIC model containing TFIIE and Ssl2. Ssl2 was positioned in the PIC based on hydroxyl-radical cleavage by FeBABE linked to various positions in TFIIE (shown as green, or yellow for Lys174). Ssl2 cleavage by residues in the TFIIE dimerization domain is colored teal, cleavage by Tfa2 residue Lys174 is colored pale yellow. (c) Model of the PIC containing Ssl2. Different components of the PIC are colored according to the color-legend.
To interpret these results, we mapped the cleavage sites on the Ssl2 structure model, based on sequence alignment with archaeal XPB, where the two RecA domains have been arranged in the closed ATP-bound conformation 29,40. We have also analyzed the cleavage data using an Ssl2 model with the two RecA-like domains arranged in the open form, based on an alignment with the double stranded DNA translocase Rad54 41. As shown below, the closed conformation of Ssl2 in the PIC is most consistent with all our biochemical data.
The Ssl2 cleavage results indicate that the RecA-like domains are directly adjacent to TFIIE with the N-terminal RecA domain facing Rpb1 and the C-terminal RecA domain facing up toward Rpb2 (Figure 6b,c). In this orientation, the Ssl2 ATP binding site is facing TFIIE and the expected DNA binding surface is near promoter DNA. This position is in excellent agreement with all the TFIIE-FeBABE cleavage results (Supplementary Fig. 4, summarized in Supplementary Fig. 5a). First, TFIIE residues where FeBABE attachment generates cleavage in Ssl2 (Tfa1 Pro56, His93 and Tfa2 Lys253, Trp263) are all near the sites of Ssl2 cleavage at residues 442–446 on the N-terminal RecA domain. Second, FeBABE at Tfa2 Lys174, in a flexible loop near Ssl2, generates cleavage on both faces of the C-terminal RecA domain as well as cleavage at 442–446 in the N-terminal domain. This can be explained by two alternate positions of FeBABE due to the flexible position of Lys174, one over the top surface of Ssl2 and the other near the Ssl2 ATP binding site. Alternatively, the Lys174 FeBABE results could be explained if the C-terminal RecA domain was rotated to the open orientation. However, results described below using Ssl2 crosslinking probes seem incompatible with the open conformation. Finally, the TFIIE probes that do not cleave Ssl2 are all pointed away from Ssl2 in the PIC model (Supplementary Fig. 4a).
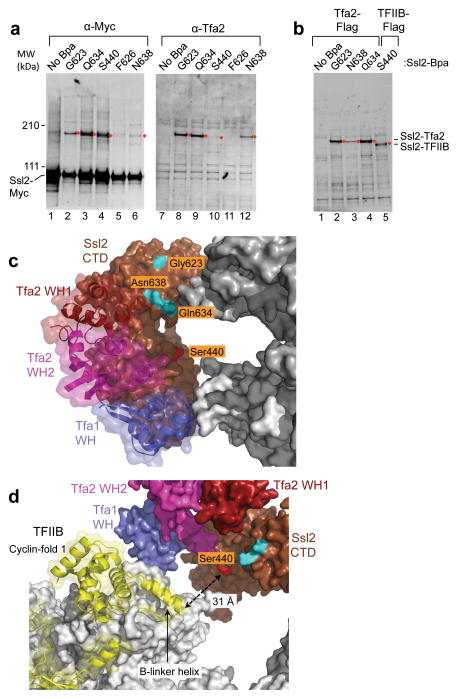
To extend these findings and to distinguish between the closed and open forms of Ssl2, we inserted the photoreactive non-natural amino acid BPA at surface exposed residues in the Ssl2 RecA-like domains 5,42. This was accomplished using a nonsense suppressor tRNA to insert Bpa at specific positions in Ssl2 (Supplementary Table 2). For these experiments, Ssl2 was Myc-tagged at the C-terminus. Nuclear extracts were made from strains with the Ssl2-BPA derivatives, used to form PICs, and then treated with UV light to induce protein crosslinking. Consistent with our PIC model, Ssl2 residues Gly623, Gln634, and Asn638 all strongly crosslinked to Tfa2 (Figure 7a, b). In this experiment, the mobility of specific Ssl2-Bpa derivatives is retarded upon UV crosslinking (Figure 7a, lanes 1–6). Probing for an identical shift in the mobility of Tfa2 showed that BPA positions Gly623, Gln634, and Asn638 gave a strong corresponding UV-dependent mobility shift for Tfa2 (Figure 7a, lanes 8,9,12). These findings are in excellent agreement with our positioning of Ssl2 in the PIC, as all Ssl2 residues that crosslinked to Tfa2 are facing in the direction of Tfa2 (Figure 7c). Thus, the Ssl2 crosslinks likely occur within either the N- or C-terminal Tfa2 residues flanking the tandem WH domain that may lie between Pol II and Ssl2. Rotation of the C-terminal Ssl2 RecA domain to the open conformation rotates these three crosslinking positions away from Tfa2. Therefore, our crosslinking data is most consistent with Ssl2 in the PIC being in the closed conformation.
Figure 7.
Protein crosslinking reveals Ssl2 interactions with TFIIE and TFIIB. (a) Nuclear extracts from strains with the crosslinker Bpa inserted at the indicated positions of Ssl2-13xMyc were used to form PICs and UV crosslinked. Proteins were separated on a 3–8% tris-acetate gel for 3 hrs, then visualized by probing with anti-Myc or anti-Tfa2. Anti-Myc blots contain 1X PIC per lane and anti-Tfa2 blots contain 4X PIC per lane. Crosslinking products are indicated by a red diamond. (b) Same as a except that strains used for the nuclear extract also contained either a 3x Flag epitope tagged Tfa2 or TFIIB as indicated. Anti-Flag blots contain 3X PIC per lane. (c) Residues in Ssl2 that crosslink to Tfa2 and TFIIB are shown on the Ssl2 surface in the PIC model. Residues that strongly and exclusively crosslinked to Tfa2 are colored in cyan. Colored in red is residue Ser440 which crosslinks strongest to TFIIB. (d) Same as c except that the model has been rotated and Rpb2 subunit removed to reveal TFIIB. Ssl2 Ser440 is colored red and is pointed toward the TFIIB linker helix, which is ~31Å distant.
An unexpected interaction between Ssl2 and TFIIB in the PIC
Surprisingly, Bpa at Ssl2 residues Ser440, Gln422, and Asn434 gave a UV-dependent mobility shift nearly identical to that for Tfa2 crosslinking, but the majority of this crosslinked product was not Tfa2 (Figure 7a, compare lanes 4 and 10; Supplementary Fig. 5). Instead, we found that these positions all crosslinked much more efficiently to TFIIB. This was demonstrated by repeating these experiments using an extract containing Flag-tagged TFIIB, where a UV-dependent shift in TFIIB mobility was specifically observed only with these Ssl2 derivatives (Figure 7b, lane 5; Supplementary Fig. 5c).
Since BPA crosslinks to proteins within a range of ~7Å, this result was unexpected because the TFIIB linker region, the nearest segment of TFIIB to Ssl2 in the PIC model, is over 30Å distant (Figure 7d). Interestingly, Ssl2 Ser440-BPA, which is the site of the strongest Ssl2-TFIIB crosslinking, is pointed directly at the cleft and the TFIIB linker. The simplest model to explain our results is that in the PIC, the TFIIB reader/linker exists in at least two conformations, one loosely bound in the cleft as indicated by the x-ray structures, and another where the linker/reader is flipped out and positioned near Ssl2. Since positioning of the reader/linker in the cleft is thought to be required for initiation, presumably the flipped out state is not poised for initiation.
Mechanism of DNA melting
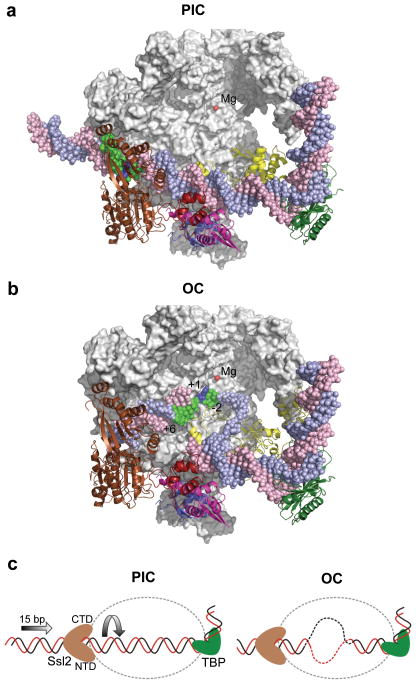
Our positioning of the Ssl2 RecA-like domains allowed us to investigate the implications of this location on the mechanism of promoter opening. We first modeled the architecture of the open complex by replacing double stranded DNA in the PIC with the DNA from Cramer and colleagues’ open complex model 9, which is derived from the DNA structure in the Pol II elongation complex 43. Figure 8 compares top views of the PIC and open complex models with the Rpb2 subunit removed for clarity. Promoter DNA bases −2 to +6, which are in contact with Ssl2 in the PIC model, are colored green in both models. In the open complex model, downstream double stranded DNA enters the cleft, making close contacts with the Pol II clamp. The DNA strands then separate so that only the template strand is in the active center of the enzyme. Comparison of the position occupied by bases −2 to +6 in the PIC and open complex shows that 15 bases of double stranded promoter DNA are inserted into the Pol II cleft (Figure 8, green; Supplemental movie). This finding is consistent with Ssl2 acting as a double stranded DNA translocase, using the energy of ATP hydrolysis to insert DNA into the cleft. DNA insertion by right handed threading of downstream DNA through the Ssl2 binding groove, combined with the fixed position of upstream DNA, leads to DNA unwinding –exactly what is necessary to promote separation of DNA strands in the open complex (Figure 8c). Our findings also suggest it is unlikely that DNA opening is being driven by only a conformational change in Ssl2. From our model, rotation of the C-terminal RecA-like domain to the open conformation is not predicted to cause a shift in the conformation of Pol II.
Figure 8.
Comparison of the PIC and open complex (OC) models suggests the role of Ssl2 in DNA strand opening. (a) The PIC model containing Pol II (grey), TBP (dark green), TFIIB (yellow), TFIIE (Tfa1 WH (blue), Tfa2 WH1 (red), and Tfa2 WH2 (pink)), Ssl2 (brown) and the active site Mg2+ (red). Rpb2 is removed to allow viewing into the Pol cleft. The blue template strand DNA is colored green from position −2 to +6 relative to the mammalian transcription start site (blue). (b) The OC model where double stranded DNA in the PIC was replaced with the DNA from the OC model 9. Compared to the PIC, 15 bases of upstream DNA is moved into the Pol II cleft and template strand DNA spanning positions −2 to +6 (green) relative to the transcription start site +1 (blue) is positioned close to the active center. (c) Diagram of Ssl2 function in OC formation. In the left panel, upstream promoter DNA is in a fixed position due to binding by TBP/TFIIB/Pol II. Ssl2 lies on downstream DNA at a fixed position due to interaction with TFIIE. Ssl2 feeds DNA into the cleft by a right handed threading mechanism, rotating and unwinding DNA.
Discussion
Understanding how the PIC transitions to the open complex state is a key question for understanding the mechanism of transcription initiation. Three general factors play central roles in this process: TFIIB, TFIIE and TFIIH. The TFIIB reader/linker regions are loosely bound to the Pol II active site cleft, and mutations that abolish their function are bypassed by pre opening the promoter template 8,9, which also eliminates the requirement for TFIIE and TFIIH in initiation 8,11–13. Here, we map the molecular location of TFIIE and the RecA-like domains of the TFIIH subunit Ssl2/XPB within the PIC. Their locations suggest roles for each of the three TFIIE WH domains and for the mechanism of Ssl2/XPB in promoting DNA strand separation. Our studies also suggest that the position of the TFIIB reader/linker is dynamic, flipping between at least two alternate conformations.
Function of the three TFIIE WH domains
Our results strongly suggest that the two TFIIE subunits form a heterodimer with a triple WH structure. Within this structure, the Tfa1 WH functions to anchor TFIIE on the Rpb1 clamp coiled-coil domain while the Tfa2 tandem WH domain spans the Pol II cleft, encircling promoter DNA in the PIC. This positions the tandem WH domain close to the site of DNA melting. From this location, the tandem WH domain likely captures the non template strand promoter DNA generated by the action of Ssl2/XPB, thereby promoting stable DNA opening. Our genetic experiments show that each Tfa2 WH is partially redundant, so either WH can likely fulfill these roles.
TFIIE ZR-FeBABE probes as well as PEAS photocrosslinker linked to the Tfa1 ZR domain (data not shown) gave no evidence for ZR-protein interactions in the PIC. Since the ZR domain is essential for initiation 16 we speculate that it makes important interactions with factors in the open complex state. FRET experiments using archaeal Pol and the general factors showed that the ZR of archaeal TFE is near the Pol stalk, (RpoE′/F, analogous to Pol II Rpb4/7) 30. However, the archaeal system spontaneously forms open complexes that are stabilized by TFE 44 so presumably these FRET measurements were done in the open complex state.
Our results on positioning of the Tfa1 WH agree with studies of archaeal TFE. These FRET experiments showed the TFE WH domain binding to the Pol clamp coiled-coil element 30. Analogous to the archaeal results, we find that a missense mutation at the tip of the coiled-coil alters or abolishes TFIIE binding in the PIC. However, archaeal TFE lacks the tandem WH domain found in Tfa2 and its counterparts in the Pol I and III systems 45, possibly because the TFE WH fulfills two roles, binding to Pol II as well as binding single stranded DNA in the open complex 31.
The Pol I subunit Rpa49, the Pol III subunit Rpc34, and the TFIIE subunit Tfa2 all contain tandem WH domains 22,46. Importantly, for all three Pol systems, tandem WH domains are located close to or above the cleft 22,47–49. Rpa49 was shown to bind double stranded and single stranded DNA through the tandem WH domain in vitro 22, a shared property with the TFIIE tandem WH domain 50. It was previously shown that the tandem WH domain of Rpc34 plays an important role in Pol III open complex formation 51. Based on low-resolution cryo-EM data, Cramer and colleagues 52 modeled Rpc34 on the surface of Pol III spanning the cleft in a similar location to the TFIIE tandem WH. It is very likely that these tandem WH domains exhibit similar functions during PIC formation and promoter opening for all the eukaryotic Pols.
Mechanism of Ssl2/XPB in DNA strand unwinding
The TFIIH subunit Ssl2/XPB is required for DNA opening and transition to the open complex, an ATP-dependent activity unique to the Pol II system. Recombinant XPB is a weak 3′ → 5′ ATP-dependent DNA helicase, however, this activity is not detectable when incorporated into TFIIH 53. Two incompatible models were proposed to explain the role of Ssl2/XPB in DNA opening suggesting that either DNA rotation or an ATP-dependent conformational change is the key activity of Ssl2/XPB 28,33. Until now, it has not been possible to distinguish between these or other models, in part because the Ssl2/XPB RecA-like domains have not been precisely localized within the PIC.
Our findings locate Ssl2 as adjacent to TFIIE, enclosing downstream promoter DNA at positions −2 to +6. This positioning is consistent with all other lower resolution biochemical mapping studies 28,29,39. The interaction between TFIIE and Ssl2 also agrees with the finding that TFIIE can bind and modulate the ATPase activity of XPB 54. Comparing the position of DNA bound by Ssl2 in the PIC with its position in the open complex model shows that 15 bp of double stranded DNA is inserted into the Pol II cleft during promoter opening. This strongly suggests that, in contrast to the previously proposed models, Ssl2/XPB acts as a double stranded DNA translocase to promote DNA opening. Since the position of upstream DNA is fixed by interaction of promoter DNA with TBP/TFIIB/Pol II and Ssl2 is fixed by interaction with TFIIE, right handed rotation of duplex DNA through Ssl2 as it tracks along one DNA strand will feed DNA into the Pol II cleft leading to DNA unwinding and the open state (Figure 8c). This model is distinct from the torque-generating model which suggested that Ssl2/XPB remained bound to a downstream position at the promoter while rotating DNA without feeding it into the Pol II cleft 28. In contrast to this model, it is not possible for Ssl2 to be in physical contact with positions −2 to +6 in the open complex, since it is buried deep within the Pol II. Finally, modeling the position of Ssl2 in the open form by reorientation of the C-terminal RecA-like domain, suggests that this change will not cause a major Pol II conformational shift leading to DNA strand opening.
Alternate PIC conformations and proposed functional significance
A surprising finding was the crosslinking of TFIIB to the Ssl2 N-terminal RecA domain. In our PIC model, based in part on the TFIIB-Pol II structure 9,10, Ssl2 is positioned too far away from TFIIB to crosslink via the short range BPA crosslinker. The Ssl2 residues which crosslink to TFIIB are all pointed at the TFIIB linker in the PIC model, located >30Å distant in the Pol II cleft. However, since the TFIIB linker/reader loops seem loosely associated with Pol II, given the fuzziness in the x-ray structure 9,10, the simplest model is that the reader/linker exists in two alternate conformations in the PIC, one similar to the x-ray structure and the other with the TFIIB loops flipped out and adjacent to Ssl2. This model is supported by recent findings showing that a TFIIB mutant that is defective for start site selection can be suppressed by a mutation in the Ssl2 RecA domains 41. An alternative explanation is that the reader/linker flips out as a result of partial PIC dissociation where the TFIIB core domain releases from Pol II as is observed in minimal open complexes 8,55. We think this second model unlikely, since it would require TFIIE dissociation and has only been observed in minimal complexes formed on heteroduplex DNA, not in the PICs analyzed here. The TFIIB-Ssl2 interaction may partly explain the inefficiency in Pol II initiation 37 and the alternate TFIIB conformations may be involved in the switch between transcription start site scanning and productive initiation.
Online Methods
Yeast strains and plasmids
Strains used for Ssl2-Bpa incorporation were derived from SHY706: matα Δade2::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 ssl2Δ::KanMx/pSH620 (ars cen URA3 SSL2), pLH157 (pADH1-EcTyrRS TRP1 2μ) 5. All SSL2-amber containing variants were generated in plasmid pLH173: pADH1-SSL2-13xMyc, 2μ LEU2, which uses the strong yeast ADH1 promoter to drive expression of SSL2. pLH173 or amber-containing variants were used to replace wild type SSL2 in strain SHY706 by plasmid shuffle assay in the presence of Bpa-containing media 5. 3XFlag-tagged variants of these strains were epitope tagged at the C-terminus of the indicated genes using the Hygromycin marker.
Strains used for TFA1 and TFA2 genetic assays were respectively: SHY734: matα Δade2::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 tfa1Δ::KanMx/pSH633 (ars cen URA3 TFA1) and SHY735: matα Δade2::hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63 ura3Δ0 tfa2Δ::KanMx/pSH634 (ars cen URA3 TFA2). TFA1 and TFA2 mutations were generated in plasmids pSH810 (ars cen LEU2 3xFlag-TFA1) and plasmid pSH811 (ars cen LEU2 3xFlag-TFA2), respectively. Internal deletions were designed to insert the flexible amino acid sequence GSGSGS at the deletion junction. Expression of the mutant derivatives was quantitated by anti-Flag Western blot in strains before the plasmid shuffle so that even lethal variants could be tested for protein expression.
Antisera
Anti-Flag M2 monoclonal antibody was from Sigma and anti-Myc monoclonal antibody (9E10) was from Covance. Anti-Tfa2 polyclonal antibody was described previously 37.
Yeast nuclear extracts
Yeast nuclear extracts were prepared as described 37. Yeast were grown in YPD medium containing Bpa (Bachem), 2.5 ml Bpa (100 mM in 1 M HCl) and an equal volume of 1 M NaOH was added per liter of YPD media before inoculation with a saturated culture to grow cells for nuclear extract preparation.
Pol II immobilized template assay for PIC formation and photo-crosslinking
The immobilized template assay for PIC formation was done as described 37. Gal4-VP16 and nuclear extracts prepared from the SSL2 amber strains grown in Bpa media were used to assemble PICs on immobilized DNA templates containing the yeast HIS4 promoter and a single upstream Gal4-binding site. After washing to remove nonspecifically bound proteins, UV irradiation was applied to the immobilized PICs in a Spectrolinker XL-1500 UV oven (Spectronics) with a total energy of 2.55 J cm−2 (~5 min). The UV irradiated PICs were analyzed by SDS-PAGE and western blotting. Identification of the crosslinking target using Western analysis was done as described 5.
Purification of recombinant TFIIE and linking to FeBABE
TFIIE subunits Tfa1 and Tfa2 were co expressed using a pET DUET vector (EMD4Biosciences) containing either no endogenous cysteines or single cysteine substitutions. Tfa2 was N-terminally 6xHis-SUMO-tagged (Invitrogen) for purification purposes. The expression plasmid was transformed into E. coli strain BL21(DE3)RIL (Stratagene), and cells were grown at 37 °C in 3 l YT Ampicillin +chloramphenicol-containing medium to an OD600 of 0.7. Expression was induced by the addition of IPTG to 1 mM, and TFIIE was expressed for 3.5 h at 30 °C. Cells were harvested by centrifugation, resuspended in Lysis Buffer (LB; 20 mM Tris/Cl pH 7.8, 250 mM KCl, 10 % (v/v) Glycerol, 1 mM PMSF), and stored at −80 °C. EDTA and DTT were added to 1 mM and 0.1 mM, respectively, prior to cell-lysis by lysozyme treatment (1 mg/ml) and sonication in 10 ml LB. The extracts were clarified by centrifugation and Imidazole (20 mM) was added to the supernatant, before incubation with 2 ml pre-equilibrated Nickel-Sepharose beads (GE Healthcare) for 90 min at 4 °C. The beads were washed three times with 20 ml LB containing 20 mM Imidazole, 1 mM PMSF, 2.5 mM β-Mercaptoethanol (BME), followed by five elution steps, each with 4 ml Elution Buffer (20 mM Tris/Cl pH 7.8, 250 mM KCl, 10 % (v/v) Glycerol, 250 mM Imidazole, 1 mM PMSF, 5 mM BME). Eluted TFIIE was then dialyzed against SUMO digestion buffer 5, followed by digestion with SUMO protease to a final concentration of 1.5 μg/ml for 15 min at RT. Finally, the digested protein sample was loaded onto a HiTrap® Heparin HP 5 ml column (GE Healthcare), pre-equilibrated with TFIIE storage buffer (20 mM Tris/Cl pH 7.8, 100 mM KCl, 10% Glycerol, 1 mM DTT, 0.5 mM PMSF), and eluted from the column with a gradient of 100–600 mM KCl. Fractions containing TFIIE were pooled, frozen in liquid nitrogen and stored at −80 °C.
FeBABE was conjugated to TFIIE essentially as described for TFIIB 36 with the following changes: A NAP-5 desalting column (GE Healthcare) was used to remove DTT and to change the buffer to Conjugation Buffer II (CBII), containing 30 mM MOPS pH 8.0, 300 mM KOAc, 0.5 mM EDTA, 10 % (v/v) Glycerol, and 5 mM TCEP. Oxygen was removed from CBII by sparging the buffer with nitrogen. The usage of CBII significantly increased FeBABE linking to TFIIE. Unbound FeBABE was removed using a second NAP-5 column, and the labeled TFIIE samples were frozen in liquid nitrogen until use in hydroxyl-radical cleavage assays (see below). All the TFIIE-FeBABE derivatives were tested for activity in complementing in vitro transcription assays, using yeast nuclear extracts depleted for TFIIE function 37.
FeBABE cleavage assay
Immobilized PIC formation and the hydroxyl-radical cleavage assays were performed essentially as described 5,6,36, but the protocol was modified to accommodate the weak binding of TFIIE to the PIC. 150 μl reactions contained 720 μg yeast nuclear extract and 100 nM recombinant TFIIE-FeBABE. After PIC formation for 40 min at room temp (RT) on a Dynal Roller at 30 rpm, samples were gently centrifuged (<2500 rpm, slow acceleration), washed once in TWB (transcription wash buffer; 20 mM HEPES pH 7.8, 100 mM KOAc, 1 mM EDTA, 5 mM MgOAc, 0.05 % (v/v) NP-40), and resuspended in 7.5 μl of TBW. 2.5 μl 50 % (v/v) Glycerol and 1.25 μl 50 mM sodium ascorbate were added, and the reactions were vortexed at low speed for 1–2 sec. Cleavage was induced by the addition of 1.25 μl H2O2/EDTA mix (0.17 % (v/v) H2O2, 10 mM EDTA). The reactions were vortexed at low speed for 1–2 sec and incubated at 30 °C for 2 min. Reactions were stopped with 5.5 μl of Stop/Loading Buffer (3.3x NuPAGE® LDS sample buffer, 100 mM DTT, 10 % (v/v) Glycerol). Proteins were separated on a 4–12% Bis-Tris gradient gel (Invitrogen) with MOPS running buffer and analyzed via Western blot. The size of the cleavage products was calculated by comparison with in vitro translated protein standards of epitope tagged TFIIB, Tfa2, and the Pol II subunit Rpb2. Cleavage products of untagged Rpb1 were calculated by an in vitro translated Rpb1 protein standard detected with an Rpb1N200 antibody 6.
Structure Modeling
To model the structured yeast TFIIE domains, HHpred 56 was used to identify and align the most related proteins of known structure with Tfa1 and Tfa2. These alignments and the structures of the related proteins were used with the program Modeller 9v5 57 using very thorough MD optimization with simulated annealing repeated three times for each model. 30 models of each domain were generated and examined using ERRAT 58, verify3D 59, and molprobity 60 and the top scoring models were used for analysis of protein cleavage results. The models scored well in these standard quality evaluations due to the similarity between yeast TFIIE domains and the known structures. Manual docking of TFIIE domains with the PIC model was used to find the positions of TFIIE consistent with the observed FeBABE cleavage results. Coordinates of the final model are available in PyMOL format at: http://labs.fhcrc.org/hahn/Research/structure_coord/index.html
Sequences and templates used for structure modeling (alignments are available upon request):
Tfa1 WH residues 12–95 aligned with Sulfolobus solfataricus TFIIE WH residues 7–88, PDB ID 1Q1H 21.
Tfa1 ZR residues 108–169 aligned with human TFIIEα WH residues 113–175, PDB ID 1VD4 17.
Tfa2 WH1 residues 118–187 aligned with human TFIIEβ WH1 residues 66–146, PDB ID 1D8J 23.
Tfa2 WH2 residues 191–268 aligned with Sulfolobus tokodaii factor STO12A residues 25–106, PDB ID 2D1H 25.
Analysis of Ssl2 binding in the PIC used both a previously published structure model for Ssl2/Rad25 29 and a model based on a new HHpred alignment of yeast Ssl2 residues 290–537 and 548–714 aligned with Archaeoglobus fulgidis XPB residues 4–209 and 238–434. Structure modeling was done as described above for TFIIE using Modeller 9v9 and top models were selected as described above. While slightly different, the new and old Ssl2 models gave identical results for Ssl2 positioning when FeBABE cleavage and Bpa crosslinking results were analyzed.
Supplementary Material
Acknowledgments
We thank Jie Luo and Jeff Ranish for sharing their unpublished TFIIE crosslinking data, James Fishburn for help with TFIIE purification, transcription assays and advice, Alessandro Vannini for coordinates of the Rpc34 WH model, Gerry Smith for discussions, and Bruce Knutson, Ivanka Kamenova and Mike Bartlett for comments on the manuscript. This work was supported by grant RO1GM053451 to SH, and Forschungsstipendium GR 3776/2-1:1 of the Deutsche Forschungsgemeinschaft to SG.
Footnotes
Author contributions
S.G. designed and performed all biochemical experiments except for the Bpa crosslinking which was designed and performed by L.W. S.G. and S.H. designed and performed Tfa1,2 genetic assays, performed structure modeling, and prepared the manuscript.
References
- 1.Hahn S, Young ET. Transcriptional regulation in Saccharomyces cerevisiae: transcription factor regulation and function, mechanisms of initiation, and roles of activators and coactivators. Genetics. 2011;189:705–36. doi: 10.1534/genetics.111.127019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41:105–78. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 3.Wang W, Carey M, Gralla JD. Polymerase II promoter activation: closed complex formation and ATP-driven start site opening. Science. 1992;255:450–3. doi: 10.1126/science.1310361. [DOI] [PubMed] [Google Scholar]
- 4.Holstege FCP, Fiedler U, Timmers HTM. Three transitions in the RNA polymerase II transcription complex during initiation. EMBO J. 1997;16:7468–7480. doi: 10.1093/emboj/16.24.7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen HT, Warfield L, Hahn S. The positions of TFIIF and TFIIE in the RNA polymerase II transcription preinitiation complex. Nat Struct Mol Biol. 2007;14:696–703. doi: 10.1038/nsmb1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eichner J, Chen HT, Warfield L, Hahn S. Position of the general transcription factor TFIIF within the RNA polymerase II transcription preinitiation complex. EMBO J. 2010;29:706–16. doi: 10.1038/emboj.2009.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen ZA, et al. Architecture of the RNA polymerase II-TFIIF complex revealed by cross-linking and mass spectrometry. EMBO J. 2010;29:717–26. doi: 10.1038/emboj.2009.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fishburn J, Hahn S. Architecture of the yeast RNA polymerase II open complex and regulation of activity by TFIIF. Mol Cell Biol. 2012;32:12–25. doi: 10.1128/MCB.06242-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kostrewa D, et al. RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature. 2009;462:323–30. doi: 10.1038/nature08548. [DOI] [PubMed] [Google Scholar]
- 10.Liu X, Bushnell DA, Wang D, Calero G, Kornberg RD. Structure of an RNA polymerase II-TFIIB complex and the transcription initiation mechanism. Science. 2010;327:206–9. doi: 10.1126/science.1182015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan G, Greenblatt J. Initiation of transcription by RNA Polymerase II is limited by melting of the promoter DNA in the region immediately upstream of the initiation site. J Biol Chem. 1994;269:30101–30104. [PubMed] [Google Scholar]
- 12.Tantin D, Carey M. A heteroduplex template circumvents the energetic requirement for ATP during activated transcription by RNA Pol II. J Biol Chem. 1994;269:17937–17400. [PubMed] [Google Scholar]
- 13.Holstege FC, van der Vliet PC, Timmers HT. Opening of an RNA polymerase II promoter occurs in two distinct steps and requires the basal transcription factors IIE and IIH. Embo J. 1996;15:1666–77. [PMC free article] [PubMed] [Google Scholar]
- 14.Ohkuma Y. Multiple functions of general transcription factors TFIIE and TFIIH in transcription: possible points of regulation by trans-acting factors. J Biochem (Tokyo) 1997;122:481–9. doi: 10.1093/oxfordjournals.jbchem.a021777. [DOI] [PubMed] [Google Scholar]
- 15.Feaver WJ, et al. Yeast TFIIE. Cloning, expression, and homology to vertebrate proteins. J Biol Chem. 1994;269:27549–53. [PubMed] [Google Scholar]
- 16.Ohkuma Y, Hashimoto S, Wang CK, Horikoshi M, Roeder RG. Analysis of the role of TFIIE in basal transcription and TFIIH-mediated carboxy-terminal domain phosphorylation through structure-function studies of TFIIE-alpha. Mol Cell Biol. 1995;15:4856–66. doi: 10.1128/mcb.15.9.4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okuda M, et al. A novel zinc finger structure in the large subunit of human general transcription factor TFIIE. J Biol Chem. 2004;279:51395–403. doi: 10.1074/jbc.M404722200. [DOI] [PubMed] [Google Scholar]
- 18.Kuldell NH, Buratowski S. Genetic analysis of the large subunit of yeast transcription factor IIE reveals two regions with distinct functions. Mol Cell Biol. 1997;17:5288–98. doi: 10.1128/mcb.17.9.5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakurai H, Ohishi T, Fukasawa T. Promoter structure-dependent functioning of the general transcription factor IIE in Saccharomyces cerevisiae. J Biol Chem. 1997;272:15936–42. doi: 10.1074/jbc.272.25.15936. [DOI] [PubMed] [Google Scholar]
- 20.Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol Rev. 2005;29:231–62. doi: 10.1016/j.femsre.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 21.Meinhart A, Blobel J, Cramer P. An extended winged helix domain in general transcription factor E/IIE alpha. J Biol Chem. 2003;278:48267–74. doi: 10.1074/jbc.M307874200. [DOI] [PubMed] [Google Scholar]
- 22.Geiger SR, et al. RNA polymerase I contains a TFIIF-related DNA-binding subcomplex. Mol Cell. 2010;39:583–94. doi: 10.1016/j.molcel.2010.07.028. [DOI] [PubMed] [Google Scholar]
- 23.Okuda M, et al. Structure of the central core domain of TFIIE beta with a novel double- stranded DNA-binding surface. Embo J. 2000;19:1346–56. doi: 10.1093/emboj/19.6.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanaka A, Watanabe T, Iida Y, Hanaoka F, Ohkuma Y. Central forkhead domain of human TFIIE beta plays a primary role in binding double-stranded DNA at transcription initiation. Genes Cells. 2009;14:395–405. doi: 10.1111/j.1365-2443.2008.01278.x. [DOI] [PubMed] [Google Scholar]
- 25.Shinkai A, et al. The putative DNA-binding protein Sto12a from the thermoacidophilic archaeon Sulfolobus tokodaii contains intrachain and interchain disulfide bonds. J Mol Biol. 2007;372:1293–304. doi: 10.1016/j.jmb.2007.07.051. [DOI] [PubMed] [Google Scholar]
- 26.Jawhari A, et al. Structure and oligomeric state of human transcription factor TFIIE. EMBO Rep. 2006;7:500–5. doi: 10.1038/sj.embor.7400663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo J, Fishburn J, Hahn S, Ranish J. An Integrated Chemical Cross-linking and Mass Spectrometry Approach to Study Protein Complex Architecture and Function. Mol Cell Proteomics. 2012;11:M111 008318. doi: 10.1074/mcp.M111.008318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim TK, Ebright RH, Reinberg D. Mechanism of ATP-dependent promoter melting by transcription factor IIH. Science. 2000;288:1418–22. doi: 10.1126/science.288.5470.1418. [DOI] [PubMed] [Google Scholar]
- 29.Miller G, Hahn S. A DNA-tethered cleavage probe reveals the path for promoter DNA in the yeast preinitiation complex. Nat Struct Mol Biol. 2006;13:603–10. doi: 10.1038/nsmb1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grohmann D, et al. The initiation factor TFE and the elongation factor Spt4/5 compete for the RNAP clamp during transcription initiation and elongation. Mol Cell. 2011;43:263–74. doi: 10.1016/j.molcel.2011.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grunberg S, Bartlett MS, Naji S, Thomm M. Transcription factor E is a part of transcription elongation complexes. J Biol Chem. 2007;282:35482–90. doi: 10.1074/jbc.M707371200. [DOI] [PubMed] [Google Scholar]
- 32.Coin F, Bergmann E, Tremeau-Bravard A, Egly JM. Mutations in XPB and XPD helicases found in xeroderma pigmentosum patients impair the transcription function of TFIIH. EMBO J. 1999;18:1357–66. doi: 10.1093/emboj/18.5.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin YC, Choi WS, Gralla JD. TFIIH XPB mutants suggest a unified bacterial-like mechanism for promoter opening but not escape. Nat Struct Mol Biol. 2005;12:603–7. doi: 10.1038/nsmb949. [DOI] [PubMed] [Google Scholar]
- 34.Luo J, Hahn S, Ranish J. An integrated chemical crosslinking and mass spectrometry approach to study protein complex architecture and function. Mol Cell Proteomics. 2011 doi: 10.1074/mcp.M111.008318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Datwyler SA, Meares CF. Protein-protein interactions mapped by artificial proteases: where sigma factors bind to RNA polymerase. Trends Biochem Sci. 2000;25:408–14. doi: 10.1016/s0968-0004(00)01652-2. [DOI] [PubMed] [Google Scholar]
- 36.Chen HT, Hahn S. Binding of TFIIB to RNA polymerase II: Mapping the binding site for the TFIIB zinc ribbon domain within the preinitiation complex. Mol Cell. 2003;12:437–47. doi: 10.1016/s1097-2765(03)00306-x. [DOI] [PubMed] [Google Scholar]
- 37.Ranish JA, Yudkovsky N, Hahn S. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev. 1999;13:49–63. doi: 10.1101/gad.13.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu X, Bushnell DA, Silva DA, Huang X, Kornberg RD. Initiation complex structure and promoter proofreading. Science. 2011;333:633–7. doi: 10.1126/science.1206629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Douziech M, et al. Mechanism of promoter melting by the xeroderma pigmentosum complementation group B helicase of transcription factor IIH revealed by protein-DNA photo-cross-linking. Mol Cell Biol. 2000;20:8168–77. doi: 10.1128/mcb.20.21.8168-8177.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caruthers JM, McKay DB. Helicase structure and mechanism. Curr Opin Struct Biol. 2002;12:123–33. doi: 10.1016/s0959-440x(02)00298-1. [DOI] [PubMed] [Google Scholar]
- 41.Goel S, Krishnamurthy S, Hampsey M. Mechanism of start site selection by RNA polymerase II: interplay between TFIIB and Ssl2/XPB helicase subunit of TFIIH. J Biol Chem. 2012;287:557–67. doi: 10.1074/jbc.M111.281576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chin JW, et al. An expanded eukaryotic genetic code. Science. 2003;301:964–7. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]
- 43.Kettenberger H, Armache KJ, Cramer P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol Cell. 2004;16:955–65. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 44.Naji S, Grunberg S, Thomm M. The RPB7 orthologue E′ is required for transcriptional activity of a reconstituted archaeal core enzyme at low temperatures and stimulates open complex formation. J Biol Chem. 2007;282:11047–57. doi: 10.1074/jbc.M611674200. [DOI] [PubMed] [Google Scholar]
- 45.Werner F, Grohmann D. Evolution of multisubunit RNA polymerases in the three domains of life. Nat Rev Microbiol. 2011;9:85–98. doi: 10.1038/nrmicro2507. [DOI] [PubMed] [Google Scholar]
- 46.Vannini A, Cramer P. Conservation between the RNA Polymerase I, II, and III Transcription Initiation Machineries. Mol Cell. 2012;45:439–46. doi: 10.1016/j.molcel.2012.01.023. [DOI] [PubMed] [Google Scholar]
- 47.Bischler N, et al. Localization of the yeast RNA polymerase I-specific subunits. Embo J. 2002;21:4136–44. doi: 10.1093/emboj/cdf392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu CC, Lin YC, Chen HT. The TFIIF-like Rpc37/53 dimer lies at the center of a protein network to connect TFIIIC, Bdp1, and the RNA polymerase III active center. Mol Cell Biol. 2011;31:2715–28. doi: 10.1128/MCB.05151-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jennebach S, Herzog F, Aebersold R, Cramer P. Crosslinking-MS analysis reveals RNA polymerase I domain architecture and basis of rRNA cleavage. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks220. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okamoto T, et al. Analysis of the role of TFIIE in transcriptional regulation through structure-function studies of the TFIIEbeta subunit. J Biol Chem. 1998;273:19866–76. doi: 10.1074/jbc.273.31.19866. [DOI] [PubMed] [Google Scholar]
- 51.Brun I, Sentenac A, Werner M. Dual role of the C34 subunit of RNA polymerase III in transcription initiation. Embo J. 1997;16:5730–41. doi: 10.1093/emboj/16.18.5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vannini A, et al. Molecular basis of RNA polymerase III transcription repression by Maf1. Cell. 2010;143:59–70. doi: 10.1016/j.cell.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 53.Coin F, et al. Mutations in the XPD helicase gene result in XP and TTD phenotypes, preventing interaction between XPD and the p44 subunit of TFIIH. Nat Genet. 1998;20:184–8. doi: 10.1038/2491. [DOI] [PubMed] [Google Scholar]
- 54.Lin YC, Gralla JD. Stimulation of the XPB ATP-dependent helicase by the beta subunit of TFIIE. Nucleic Acids Res. 2005;33:3072–81. doi: 10.1093/nar/gki623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Treutlein B, et al. Dynamic Architecture of a Minimal RNA Polymerase II Open Promoter Complex. Mol Cell. 2012;46:136–46. doi: 10.1016/j.molcel.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 56.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–8. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eswar N, Eramian D, Webb B, Shen MY, Sali A. Protein structure modeling with MODELLER. Methods Mol Biol. 2008;426:145–59. doi: 10.1007/978-1-60327-058-8_8. [DOI] [PubMed] [Google Scholar]
- 58.Colovos C, Yeates TO. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 1993;2:1511–9. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luthy R, Bowie JU, Eisenberg D. Assessment of protein models with three-dimensional profiles. Nature. 1992;356:83–5. doi: 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
- 60.Davis IW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–83. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.