Abstract
While the roles of 5-methyl-cytosine and 5-hydroxymethyl-cytosine in epigenetic regulation of gene expression are well-established, the functional effects of 5-formyl-cytosine and 5-carboxyl-cytosine in the genome on transcription are not clear. Here we report the first systematic study of the effects of five different forms of cytosine in DNA on mammalian and yeast RNA polymerase II transcription, providing new insights into potential functional interplay between cytosine methylation status and transcription.
Keywords: Epigenetics, DNA methylation, Pol II pausing, transcription elongation, transcriptional fidelity
At least five forms of cytosine have been identified in cells: cytosine, 5-methyl-cytosine (5mC), 5-hydroxymethyl-cytosine (5hmC), 5-formyl-cytosine (5fC), and 5-carboxyl-cytosine (5caC) (Fig. 1a). The 5mC and 5hmC modifications play critical roles in epigenetic regulation of gene expression and maintenance of cellular identity1,2. The two most recently identified cytosine derivatives, 5fC and 5caC3–5, are proposed to be part of the cytosine demethylation pathway catalyzed by TET (ten-eleven translocation) dioxygenases3,4,6. However, the functional effects of 5fC and 5caC in the genome on transcription are unclear and have not been reported (Fig. 1a).
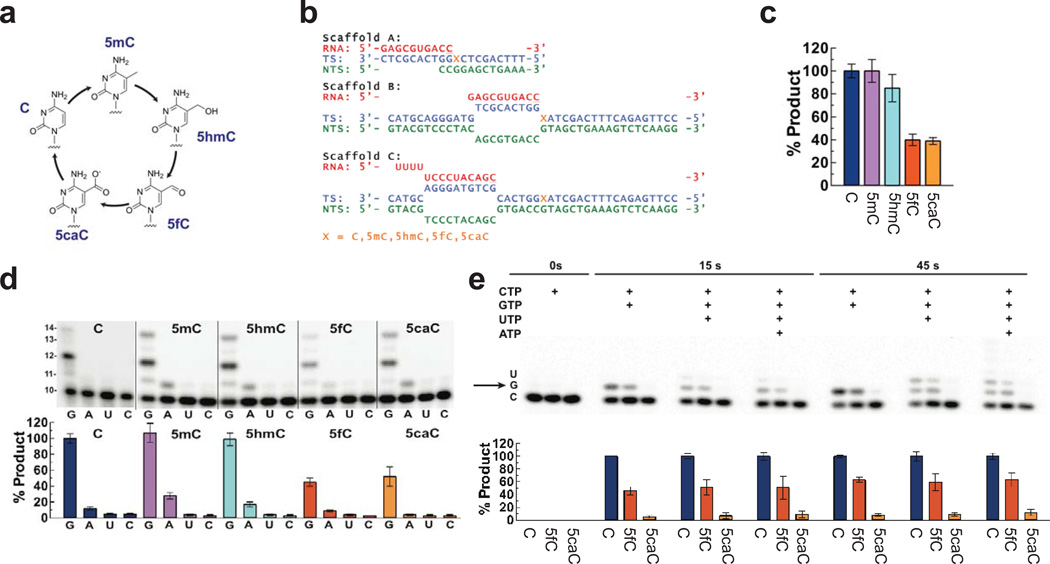
Fig. 1. Cytosine methylation status affects mammalian and yeast RNA polymerase II transcription.
(a) Five forms of cytosine residues. 5mC modification is catalyzed by DNA methyltransferase. Oxidation of 5mC by TET proteins produces 5hmC, 5fC, and 5caC, respectively. (b) Transcriptional elongation scaffolds for NTP incorporation on modified cytosine templates (denoted by X). RNA, template strand DNA (TS) and nontemplate strand DNA (NTS) are shown in red, cyan, and green, respectively. Modified cytosine residues are highlighted in orange. The 3’-ends of RNA residues are underlined. (c) Relative NTP incorporation efficiency on all five modified cytosine templates by mammalian Pol II (scaffold A). Data from C, 5mC, 5hmC, 5fC, and 5caC templates were shown in blue, magenta, cyan, red and orange, respectively. Data were normalized to GTP incorporation efficiency for C template at 15 sec. All error bars (standard deviation) are derived from three experiments. (d) Template-dependent NTP incorporation by yeast Pol II (scaffold A) and relative NTP incorporation efficiency on all five modified cytosine templates. Data were normalized to GTP incorporation efficiency for C template at 15 sec. All error bars (standard deviation) are derived from four experiments. Color codes are the same as (c). (e) Template-dependent NTP incorporation by yeast Pol II (scaffold B) and relative Pol II transcription efficiency on modified cytosine templates. Data were normalized to product formation for C template at the same experimental condition. All error bars (standard deviation) are derived from three experiments. Color codes are the same as (c). The band position corresponding to GTP incorporation opposite to modification site was depicted with an arrow in the gel.
DNA methylation is typically observed on CpG islands clustered around gene promoters as well as within gene bodies1,2,6,7. While much less abundant than 5mC and 5hmC, 5fC and 5caC are likely to occur at a frequency of between 104–5 per genome (Online Methods)3–5,8. This level is comparable to DNA damage caused by oxidative stress, which is commonly investigated in the context of transcription and DNA damage repair9. Their presence in the gene bodies raises an intriguing question of how these cytosine derivatives may affect RNA polymerase II (Pol II) transcription activity and transcription dynamics. To address this question, here we have systematically investigated the rate of Pol II elongation and substrate specificity (kcat/Km or kpol/Kd) for DNA templates containing a site-specific C, 5mC, 5hmC, 5fC, or 5caC, respectively.
We first measured in vitro Pol II elongation efficiency using purified mammalian Pol II with DNA templates containing a site-specific C, 5mC, 5hmC, 5fC, or 5caC, respectively (Fig. 1b). Our results show a striking difference in mammalian Pol II transcription efficiency among these five forms of cytosine. GTP incorporation efficiencies following a 15-sec incubation for 5fC- and 5caC-containing templates are significantly lower than that for the C template (40 ± 5 and 39 ± 3 percent, respectively), whereas no noticeable difference was observed among C, 5mC and 5hmC templates (Fig. 1c). We also observed similar pausing effects of 5fC and 5caC on transcription elongation efficiency using purified yeast Pol II (45 ± 5 and 52 ± 12 percent, respectively, Fig. 1d and Supplementary Fig. 1). Pol II has strictly conserved (almost identical) active site residues (Supplementary Fig. 2) and shares common enzymatic mechanisms for nucleotide incorporation from yeast to human10,11. Thus, it is not surprising that similar effects were observed for both mammalian and yeast Pol II, and yeast Pol II can be used as a model enzyme to study the effects of 5fC and 5caC templates on mammalian Pol II transcription elongation. It should be noted that we observed a similar trend of significant reduction of Pol II nucleotide incorporation and elongation efficiency on 5fC and 5caC templates from Pol II elongation complexes with different scaffolds (either with a full transcription bubble or downstream-edge of the transcription bubble) (Fig. 1b, 1c, 1d, 1e, and Supplementary Fig. 3), indicating that the observed reduction in transcription elongation efficiency is not due to specific sequence context or scaffold setting. Taken together, these results reveal that Pol II elongation efficiencies for 5fC and 5caC templates are significantly reduced relative to those for the C template.
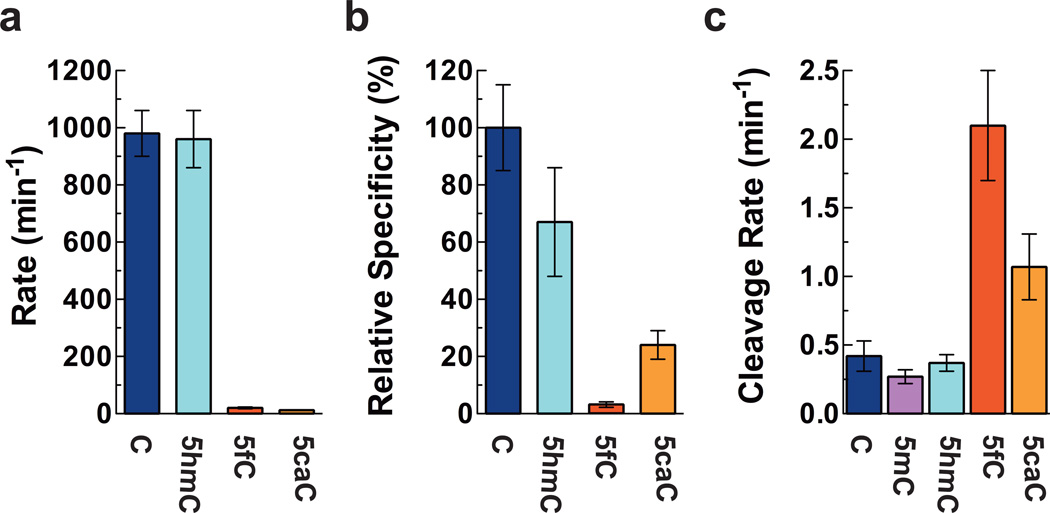
To further investigate the Pol II transcription elongation kinetics and quantitatively measure the effect of cytosine modifications on Pol II transcription elongation, we then determined the full pre-steady state single-turnover NTP incorporation kinetics for templates C, 5hmC, 5fC and 5caC. These single turnover experiments allow us to directly determine the key kinetic parameters kpol (catalytic rate constant) and Kd,app (the apparent substrate dissociation constant) of nucleotide incorporation at a single site. Furthermore, we can determine the ratio of kpol/Kd,app (substrate specificity), an important kinetic parameter for measuring Pol II enzymatic efficiency. The comparison of substrate specificity among these different forms of cytosine residues gives a quantitative measurement of the effect of these cytosine derivatives on Pol II transcription elongation. Yeast Pol II was used to overcome the technical difficulty in purification of sufficient amount of purified mammalian Pol II for extensive enzymology study. Intriguingly, Pol II polymerization rates (kpol) for GTP incorporation against 5fC and 5caC are significantly reduced to 2.0% and 1.3% of kpol of GTP incorporation against C template, respectively, whereas no noticeable changes are observed in the case of 5hmC compared with unmodified C template (Fig. 2a, Supplementary Fig. 4, and Supplementary Table 1). The Pol II specificity constants for GTP incorporation against 5fC and 5caC templates are decreased significantly by ~30.0- and 4.2-fold, respectively (Fig. 2b), whereas the specificity constant is only slightly reduced in the case of 5hmC compared with unmodified C template (Fig. 2b). Differences in both kpol and apparent Kd,app produce the altered GTP specificity (Supplementary Table 1). In addition, product formation was significantly more biphasic for 5fC and 5caC templates than C and 5hmC templates (Supplementary Table 2). The observation of distinct phases suggests multiple populations of Pol II elongation complexes (Supplementary Note): one population is poised for rapid GTP incorporation (observed as the fast phase of product formation, such as Pol II in post-translocation state); the second, slower phase represents a Pol II population that requires longer time for GTP incorporation (paused population, such as Pol II in the pre-translocation, frayed, or backtracked states). This slower phase may reflect a rate-limiting isomerization that must occur before GTP incorporation.
Fig. 2. Pol II polymerization rate and specificity of GTP incorporation for C, 5hmC, 5fC, and 5caC templates.
(a) Pol II polymerization rates for GTP incorporation on C, 5hmC, 5fC, and 5caC templates. (b) Relative Pol II substrate specificity of GTP incorporation for C, 5hmC, 5fC, and 5caC templates. The Pol II specificity (kpol/Kd,app) was normalized to that for C template. All error bars (standard deviation) are derived from three experiments. (c) Rates of RNA cleavage stimulated by Transcription Factor IIS for C, 5mC, 5hmC, 5fC, and 5caC templates. Error bars represent deviations in non-linear regression analysis. Color codes are the same as Fig. 1c.
To investigate whether the presence of 5fC and 5caC promotes the backtracking population of Pol II complex, we measured the rate of TFIIS-mediated backtracked RNA cleavage for all five scaffolds. The TFIIS-mediated cleavage rates for scaffolds containing 5fC and 5caC increase by 5.0- and 2.6-fold, respectively, whereas no noticeable changes are observed in the case of 5hmC and 5mC compared with unmodified C template (Fig. 2c). Taken together, these results indicate that the presence of 5fC and 5caC greatly shifts Pol II from an active population (poised for elongation) to a paused population and promotes Pol II stalling and RNA backtracking.
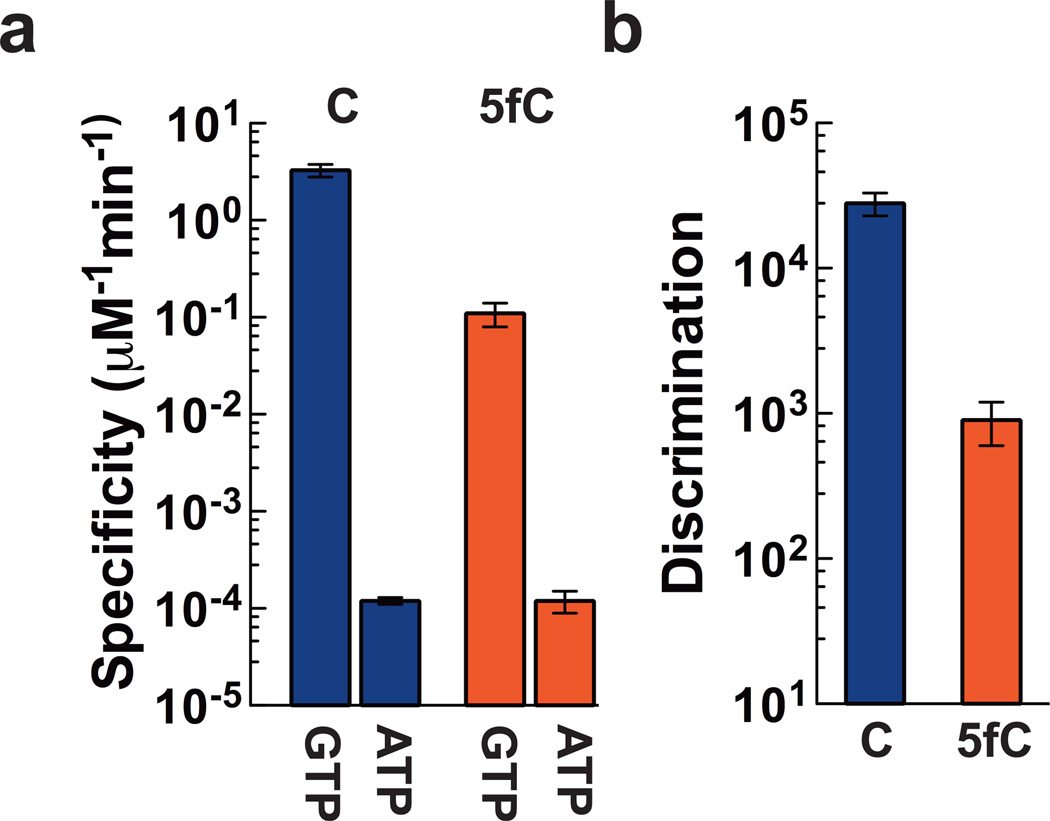
Finally, we investigated whether these cytosine derivatives cause significant changes in transcriptional fidelity. We first tested incorporation efficiency of individual NTP over templates with five different cytosine derivatives. We found that the misincorporation efficiency of ATP is significantly higher than that of UTP and CTP across all five templates (Fig. 1d and Supplementary Fig. 5). We then examined full pre-steady state single-turnover ATP misincorporation kinetics for C and 5fC templates. Notably, specificities for ATP misincorporation are significantly reduced by 103–4-fold than those for correct GTP incorporation, suggesting that ATP misincorporation is relatively inefficient. In contrast to GTP incorporation, there are no noticeable changes in specificities for ATP misincorporation between C and 5fC templates (Fig. 3a and Supplementary Table 3). To quantitatively evaluate the effect of 5fC on fidelity of nucleotide incorporation, we calculated the discrimination constants for GTP over ATP incorporation (defined as (kpol/Kd,app)GTP/(kpol/Kd,app)ATP) for C and 5fC templates (Fig. 3b and Supplementary Table 3). The discrimination constants of NTP incorporation for C and 5fC templates are 2.8 × 104 and 900, respectively. Therefore, substitution of C with 5fC in the template reduces the fidelity of nucleotide incorporation by ~30-fold (Fig. 3a and 3b, and Supplementary Table 3). This decreased discrimination is due to a reduced GTP incorporation specificity for the 5fC template.
Fig. 3. 5fC reduces Pol II substrate discrimination of GTP over ATP.
(a) Pol II substrate specificity of GTP and ATP incorporation for C and 5fC templates respectively. (b) Pol II substrate discrimination of GTP over ATP incorporation for C and 5fC templates. Data from C and 5fC templates are shown in blue and red, respectively. All error bars (standard deviation) are derived from three experiments.
The results that 5fC and 5caC affect the rates, substrate specificities and transcriptional fidelity of Pol II transcription are particularly intriguing because the positions of the formyl and carboxyl group of 5fC and 5caC, unlike most DNA modifications or lesions, are not expected to disrupt Watson-Crick base pairing or double helical structure. One attractive possibility is that specific interactions between the formyl or carboxyl groups of 5fC or 5caC bases on the DNA template and nearby Pol II residues (likely from switch 2 (Rpb1 328–340), fork loop 2 (Rpb2 499–510), bridge helix (Rpb1 812–841), or trigger loop (Rpb1 1059–1100)10,11) cause the modified cytosine base to deviate from the conventional template position (+1 position) for base pairing with incoming GTP. Further detailed structural investigation is required to reveal the molecular basis of the effects of 5fC or 5caC modifications on Pol II transcriptional efficiency and fidelity, and to reveal interactions between Pol II active site residues and 5fC (or 5caC) for a better understanding of these effects and their implications.
Cytosine modifications of 5fC and 5caC may play important roles in regulating Pol II transcription. Decreased rates of Pol II transcription that result from these templates induce Pol II transient pausing at these positions, which allows Pol II to interact differently with transcription elongation factors, Pol II CTD-interaction proteins, chromatin remodeling complexes, histone-modifying complexes, and the mRNA processing machinery. Changes in transcription rate have been shown to have a significant effect on transcriptional fidelity, mRNA processing such as splicing, polyadenylation, termination efficiency, chromatin remodeling, and many other transcription-related processes12–16. Thus, this Pol II transient pausing caused by 5fC and 5caC may add another layer of transcriptional regulation and also act to fine-tune transcriptional dynamics. In addition, Pol II pausing at 5fC and 5caC sites may serve as a signal for the recruitment of thymine DNA glycosylase (TDG) and base excision repair (BER) machinery to the sites. TDG, an enzyme that excises 5fC and 5caC sites through BER4,6,17,18, has been found to interact with several transcription factors, histone acetyltransferases (such as CBP/P300), de novo DNA methyltransferases, deaminase AID and damage response protein GADD45a6,18–20. It is possible that TDG is recruited to 5fC and 5caC sites during transcription elongation in a transcription-coupled manner through Pol II pausing. Future investigations into the effects of 5fC and 5caC on Pol II transcription may provide new insights into the functional interplay between cytosine methylation status and transcriptional regulation.
In summary, we have systematically investigated the effects of all known cytosine modifications on the rate of Pol II nucleotide incorporation and substrate specificity for DNA templates containing a site-specific C, 5mC, 5hmC, 5fC, or 5caC, respectively. We show that Pol II polymerization rates (kpol) and specificity constants (kpol/Kd) for GTP incorporation against 5fC and 5caC are reduced significantly in comparison with those for C template, whereas essentially no changes are observed for 5mC and 5hmC templates. We further reveal that the discrimination of GTP over ATP is reduced by ~30-fold for 5fC template in comparison with C template. Our results indicate that Pol II can read and distinguish subtle changes in cytosine modifications at the 5-position and process them differently. These findings provide an important new perspective on the potential functional interplay between modification status of cytosine and transcription.
Methods
Methods and any associated references are available in the online version of the paper.
Online Methods
These studies utilized both mammalian Pol II and yeast Pol II (as a model enzyme)10,11,21–24. Mammalian Pol II was purified from rat liver as described25. Saccharomyces cerevisiae Pol II was purified using established methods11. The RNA primer and DNA non-template oligonucleotides were purchased from IDT. Template DNA oligonucleotides containing site-specific DNA modification were synthesized and purified as described26. These modifications were confirmed by mass spectrometry. RNA primers were radiolabeled using 32P-γ-ATP and T4 Polynucleotide Kinase (NEB). The Pol II elongation complexes for transcription assays were assembled as described11.
Transcription elongation reactions with mammalian Pol II
An aliquot of mammalian Pol II purified from rat liver were pre-incubated with 1.25 µM DNA/RNA scaffold (RNA: 5’-GAGCGUGAC-3’, nontemplate strand: 5’-CCGGAGCTGAAA-3’, template strand: 5’-TTTCAGCTCXGGTCACGCTC-3’, X = C, 5mC, 5hmC, 5fC, 5caC) and 0.83 µM α-32P-CTP (3000 Ci/mmol) in reaction buffer (50 mM KCl, 25 mM Tris (pH = 7.5), 5 mM MgCl2 and 12.5 mM DTT) at room temperature for 20 min to form elongation complex and labeled RNA at 3’-end. The Pol II complex solution was then mixed with equal volumes of nucleotide solution containing 1 mM GTP in reaction buffer for 15 sec. The reactions were then quenched with one volume of 0.5 M EDTA solution (pH = 8.0). The products were analyzed by denaturing PAGE and then visualized using a storage phosphor screen and Pharos FX imager (Bio-Rad). Substrate and product bands were quantified using Image Lab 3.0.
Transcription elongation reactions with Saccharomyces cerevisiae Pol II
Transcription elongation reactions were performed by pre-incubating 100 nM scaffold (RNA: 5’-GAGCGUGACC-3’, nontemplate strand: 5’-CCGGAGCTGAAA-3’, template strand: 5’-TTTCAGCTCXGGTCACGCTC-3’, X = C, 5mC, 5hmC, 5fC, 5caC) with 400 nM Pol II in elongation buffer (40 mM KCl, 20 mM Tris pH = 7.5, and 10 mM DTT) at room temperature for 20 min before mixing with equal volumes of nucleotide solution containing various concentrations of NTP and 10 mM MgCl2 in elongation buffer. The final reaction conditions after mixing were 50 nM transcription scaffold, 200 nM enzymes, 5 mM MgCl2, and various final NTP concentrations in elongation buffer. The reactions were allowed to proceed for various times before quenching with one volume of 0.5 M EDTA solution (pH = 8.0). The products were analyzed by denaturing PAGE and then visualized using a storage phosphor screen and Pharos FX imager (Bio-Rad). Substrate and product bands were quantified using Image Lab 3.0.
Transcription elongation reactions with scaffold containing a full transcription bubble
The transcription elongation complex containing a full transcription bubble (scaffolds B or C in Fig. 1b) was assembled according to the protocol developed by Kashlev and co-workers with minor modifications described below27,28. Briefly, 1 pmol of Pol II was incubated with 0.3 pmol RNA:template DNA hybrid (see scaffolds B or C in Fig. 1b) for 10 min at room temperature. An aliquot of 1.6 pmol of non-template DNA and 0.8 µCi α-32P-CTP (3000 mCi/mmol, Perkin Elmer) was then added and the incubation was allowed to proceed for another 10 min at room temperature. The active Pol II elongation complex will have a 32P-labeled RNA at 3’-end. The pre-assembled transcription elongation complexes were mixed with equal volumes of nucleotide solution containing various concentrations of individual or mixed NTP (1 µM (Fig. 1e) or 10 µM (Supplementary Fig. 3) and 10 mM MgCl2 in elongation buffer. The final reaction conditions after mixing were 75 nM transcription scaffold, 250 nM enzyme, 5 mM MgCl2, and various final NTP concentrations in elongation buffer. The reactions were allowed to proceed for various times before quenching with one volume of 0.5 M EDTA solution (pH = 8.0). The products were analyzed by denaturing PAGE and then visualized using a storage phosphor screen and Pharos FX imager (Bio-Rad). Substrate and product bands were quantified using Image Lab 3.0.
Single-turnover nucleotide incorporation assays
The specificity constant (kcat/Km) governing GTP incorporation against C, 5hmC, 5fC, or 5caC templates was determined by rapid quench techniques. Yeast Pol II (200 nM) was pre-incubated with 50 nM scaffold in elongation buffer at room temperature for 20 min. The enzyme/scaffold solution was then mixed with increasing concentrations of GTP (C and 5hmC template: 10, 25, 50, 100, 250, 500, and 1000 µM; 5fC and 5caC template: 5, 10, 25, 50, 100, 250, 500, and 2500 µM) and 5mM MgCl2 in elongation buffer (all concentrations are final after mixing). Reactions were quenched at various time points by addition of one volume of 0.5 M EDTA (pH = 8.0). Reactions requiring quench time faster than 5 sec were performed using a RQF-3 Rapid Quench Flow (KinTek Corp.).
TFIIS cleavage assays
TFIIS cleavage assays were performed using methods similar to those described for single-turnover nucleotide incorporation assays. The enzyme-scaffold complex was rapidly mixed with a solution of elongation buffer, 10 mM DTT, 5 mM MgCl2, and 1.5 µM TFIIS (all concentrations final after mixing). Reactions were quenched from 0.25 to 120 minutes by addition of 0.5 M EDTA (pH = 8.0). Cleavage products were separated by denaturing PAGE.
Data fitting
Non-linear regression data fitting was performed using GraFit 5.0. The time dependence of product formation for various concentrations of GTP was fit to a double exponential equation (Equation 1). This fitting yielded two phases corresponding to fast and slow observed rates of product formation. The GTP concentration dependence of the fast phase was fit to a hyperbolic equation (Equation 2) to obtain values for kpol and Kd,app (Supplementary Fig. 4). Alternatively, the data were fit globally using Kinetic Global Explorer (KinTek Corp.)29,30. The specificity constant was determined by kcat/Km = kpol/Kd,app.
| (1) |
| (2) |
Occurrences of 5-formyl- and 5-carboxyl-cytosine per genome
Calculation of the number of occurrences of 5fC and 5caC per genome was performed by determining the total number of cytosine residues in the human genome. The diploid genome contains 1.2 × 1010 bases (2 (for sister chromosomes) × 2 (for two DNA strands) × 2.9 × 109 bases)31 and a GC content of 46 %32, resulting in 2.8 ×109 cytosine residues per diploid genome. 5hmC comprises 0.032% of all bases in genome (~4 × 106 bases per diploid genome)8. The recent discovery of 5fC and 5caC reported their abundance to be around 1.5 % of genomic 5hmC3,4. Therefore, we calculated the total number of 5fC occurrences to be 6 × 104 per genome. 5caC is estimated 1 × 103 – 9 × 103 per genome4. Similar levels of 5fC and 5caC were calculated based on data in recent reports from several laboratories. Ito et al reported there are about 20 5fC and 3 5caC in every 106 cytosine3. Thus, total 5fc and 5caC are estimated ~5.6 ×104 (20/106 × 2.8 × 109) and 8.4 × 103 (3/106 × 2.8 × 109) per genome in mouse ES cells, respectively. Pfaffeneder et al. reported that the highest 5fC level is up to 0.02% of total cytosine in mouse ES cells, which leads to an even higher level of 5fC (5.6 × 105 (0.02% × 2.8 ×109 cytosine) per genome)5. Conversely, oxidative DNA damage has been reported to occur transiently at a rate about 104 per day33.
Supplementary Material
Acknowledgments
D.W. acknowledges the NIH (GM085136), start-up funds from the Skaggs School of Pharmacy and Pharmaceutical Sciences, UCSD, and Kimmel Scholar Award from the Sidney Kimmel Foundation for Cancer Research. C.H. acknowledges the NIH (GM071440). We thank Drs. Joan Conaway and Ronald Conaway (Stowers Institute for Medical Research) for the generous gift of purified mammalian Pol II. We also thank Drs. E. Peter Geiduschek, James R. Halpert, Xiang-Dong Fu, Bing Ren, Yi Zhang and Kun Zhang for their insightful comments and critical reading of the manuscript.
Footnotes
Author Contributions
D.W. conceived the original idea. M.W.K., C.H. and D.W. designed the experiments. C.S. and X.L. carried out synthesis of DNA templates. J.C. and D.W. purified Pol II. M.W.K. and D.W. performed transcription assays. M.W.K. and D.W. carried out data analysis. M.W.K., J.C., C.H. and D.W. wrote the paper.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Jaenisch R, Bird A. Nat Genet. 2003;(33 Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 2.Law JA, Jacobsen SE. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ito S, et al. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He YF, et al. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pfaffeneder T, et al. Angew Chem Int Ed Engl. 2011;50:7008–7012. doi: 10.1002/anie.201103899. [DOI] [PubMed] [Google Scholar]
- 6.Wu H, Zhang Y. Genes Dev. 2011;25:2436–2452. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pastor WA, et al. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tahiliani M, et al. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindahl T, Barnes DE. Cold Spring Harb Symp Quant Biol. 2000;65:127–133. doi: 10.1101/sqb.2000.65.127. [DOI] [PubMed] [Google Scholar]
- 10.Cramer P, Bushnell DA, Kornberg RD. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 11.Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shukla S, et al. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pinto PA, et al. EMBO J. 2011;30:2431–2444. doi: 10.1038/emboj.2011.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Core LJ, Lis JT. Science. 2008;319:1791–1792. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de la Mata M, et al. Mol Cell. 2003;12:525–532. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 16.McDowell JC, Roberts JW, Jin DJ, Gross C. Science. 1994;266:822–825. doi: 10.1126/science.7526463. [DOI] [PubMed] [Google Scholar]
- 17.Maiti A, Drohat AC. J Biol Chem. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cortellino S, et al. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cortazar D, et al. Nature. 2011;470:419–423. doi: 10.1038/nature09672. [DOI] [PubMed] [Google Scholar]
- 20.Tini M, et al. Mol Cell. 2002;9:265–277. doi: 10.1016/s1097-2765(02)00453-7. [DOI] [PubMed] [Google Scholar]
References
- 21.Werner F, Grohmann D. Nat Rev Microbiol. 2011;9:85–98. doi: 10.1038/nrmicro2507. [DOI] [PubMed] [Google Scholar]
- 22.Nudler E. Annu Rev Biochem. 2009;78:335–361. doi: 10.1146/annurev.biochem.76.052705.164655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conaway JW, Shilatifard A, Dvir A, Conaway RC. Trends Biochem Sci. 2000;25:375–380. doi: 10.1016/s0968-0004(00)01615-7. [DOI] [PubMed] [Google Scholar]
- 24.Shilatifard A, Conaway JW, Conaway RC. Curr Opin Genet Dev. 1997;7:199–204. doi: 10.1016/s0959-437x(97)80129-3. [DOI] [PubMed] [Google Scholar]
- 25.Serizawa H, Conaway RC, Conaway JW. J Biol Chem. 1993;268:17300–17308. [PubMed] [Google Scholar]
- 26.Dai Q, He C. Org Lett. 2011;13:3446–3449. doi: 10.1021/ol201189n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sidorenkov I, Komissarova N, Kashlev M. Mol Cell. 1998;2:55–64. doi: 10.1016/s1097-2765(00)80113-6. [DOI] [PubMed] [Google Scholar]
- 28.Kireeva ML, Komissarova N, Waugh DS, Kashlev M. J Biol Chem. 2000;275:6530–6536. doi: 10.1074/jbc.275.9.6530. [DOI] [PubMed] [Google Scholar]
- 29.Johnson KA, Simpson ZB, Blom T. Anal Biochem. 2009;387:20–29. doi: 10.1016/j.ab.2008.12.024. [DOI] [PubMed] [Google Scholar]
- 30.Johnson KA, Simpson ZB, Blom T. Anal Biochem. 2009;387:30–41. doi: 10.1016/j.ab.2008.12.025. [DOI] [PubMed] [Google Scholar]
- 31.Venter JC, et al. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 32.Romiguier J, Ranwez V, Douzery EJ, Galtier N. Genome Res. 2010;20:1001–1009. doi: 10.1101/gr.104372.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Senturker S, et al. FEBS Lett. 1997;416:286–290. doi: 10.1016/s0014-5793(97)01226-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.