Abstract
Studies carried out in recent years have established that growth under conditions of reduced gravity enhances Salmonella enterica serovar Typhimurium virulence. To analyze the possibility that this microgravity-induced increase in pathogenicity could involve alterations in the ability of Salmonella to withstand oxidative stress, we have compared the resistance to hydrogen peroxide of various Salmonella enterica strains grown under conditions of low shear modeled microgravity (LSMMG) or normal gravity (NG). We have found that growth in LSMMG significantly enhances hydrogen peroxide resistance of all the strains analyzed. This effect is abolished by deletion of the genes encoding for the catalases KatG and KatN, whose activity is markedly modulated by growth in LSMMG. In addition, we have observed that Salmonella enterica serovar Typhimurium strains lacking Hfq, RpoE, RpoS or OxyR are still more resistant to oxidative stress when grown in LSMMG than in NG conditions, indicating that these global gene regulators are not responsible for the microgravity-induced changes in KatG and KatN activity. As Salmonella likely encounters low shear conditions in the intestinal tract, our observations suggest that alterations in the relative activity of KatG and KatN could enhance Salmonella resistance to the reactive oxygen species produced also during natural infections.
Keywords: Hydrogen peroxide resistance, Catalase, Microgravity, Bacterial virulence, Salmonella; HARV Bioreactor.
1. INTRODUCTION
In spite of all the efforts carried out to generate and maintain microbiologically-controlled environments, several microorganisms, likely derived by contaminants and crew-member microflora, have been commonly isolated in the aircrafts employed in spaceflights [1]. Moreover, a few reports have shown that microgravity conditions induce various physiological responses in bacteria, which might threaten human health during space missions. These include the enhancement of virulence [2], an increase in resistance to antibiotics [3] and the induction of microbial biofilms [4]. These observations are a cause of concern as, during spaceflights, the human body experiences different kinds of stress, including changes in gravitational forces, space radiations, altered nutritional status and anxiety, which may adversely affect functionality of the immune system [5]. Such condition exposes astronauts to the risk of infections, including those sustained by opportunistic bacterial pathogens, whose proliferation is normally controlled in healthy humans in a gravitational environment.
In recent years, several studies have focused on the effects of modeled microgravity on well characterized microorganisms such as Salmonella enterica serovar Typhimurium and Escherichia coli. It has been reported that Salmonella grown under low-shear modeled microgravity (LSMMG) displays lower LD50, shortened host time-to-death, and increased colonization of liver and spleens in infected mice [2]. Enhanced Salmonella pathogenicity is associated to higher resistance to acid, thermal and osmotic stress, increased ability to grow in cultivated macrophages and significant alterations in global gene expression [6-8]. A recent spaceflight experiment has confirmed these phenotypes and revealed a significant role of the global regulator Hfq in the genomic and proteomic Salmonella response to reduced gravity [9]. Moreover, it has been observed that microgravity stimulate formation of Salmonella biofilms [9]. LSMMG induces alterations in gene transcription [10] and biofilm formation [11] also in E. coli. However, no similarities have been found in LSMMG-dependent gene regulation between E. coli and S. Typhimurium, thus suggesting that different microorganisms may dramatically differ in their responses to low-shear and space environments [10].
Although limited studies have been carried out on the effects of LSMMG on Salmonella and E. coli ability to withstand the toxic effects of reactive oxygen species, a few investigations have suggested that microgravity may affect the oxidative stress response of different bacteria. Enhanced superoxide dismutase activity and decreased glutathione content have been observed in Anabaena sp. PCC 7120 cells under simulated microgravity [12]. In addition, H2O2-resistant Bacillus pumilus vegetative cells and spores were repeatedly isolated from various surface locations of a spacecraft assembly facility at the Jet Propulsion Laboratory [13], whereas H2O2 resistance was exhibited also by a number of other spore-forming bacteria isolated from the Mars Odyssey spacecraft and the clean room facilities at Kennedy Space Center [14, 15]. Moreover, it has been recently reported that LSMMG significantly increases P. aeruginosa resistance to hydrogen peroxide [16].
To analyze the possibility that LSMMG could influence Salmonella ability to withstand oxidative stress, we have compared hydrogen peroxide resistance of various Salmonella strains grown under LSMMG or normal gravity (NG) conditions. Our results indicate that most Salmonella strains exhibit higher resistance to hydrogen peroxide when cultivated under LSMMG and that the catalases KatG and KatN concur to this phenotype.
2. MATERIALS AND METHODS
2.1. Bacterial Strains and Growth Conditions
Bacterial strains are listed in Table 1. Bacterial preinocula were grown in Luria Bertani (LB) broth as static cultures at 37°C for 7-8 hours, reaching an optical density of 1.5-2 at 600 nm. Cultures were then diluted 1:100 into 40 ml of LB broth and subsequently introduced into two 10 ml High-Aspect-Ratio Vessels (HARV, Synthecon, Inc. Houston, Tex), a special kind of rotating bioreactors designed at the Johnson Space Center (Houston, Tex.) [19]. Care was taken to ensure that the bioreactors were completely filled with culture medium, avoiding any bubbles, to create a low-shear environment. The bioreactors were oriented to grow cells under conditions of modeled microgravity (its axis of rotation is perpendicular to gravity) or normal gravity (its axis of rotation is parallel to gravity vector). All incubations in the HARV were done at 37°C with a rotation rate of 25 rpm for 20 hours. Gas exchange occurs by perfusion through a permeable membrane and is sufficient to maintain an aerobic environment. Cell growth was monitored spectrophotometrically at 600 nm.
Table 1.
Bacterial Strains
| Name | Relevant Genotype | Source |
|---|---|---|
| S. Typhimurium ATCC14028 | ATCC± | |
| S.Typhimurium DT104 | [29] | |
| S.Typhimurium LT2 | lab collection | |
| S.Enteritidis LK5‡ | [17] | |
| S.Choleraesuis A50 | [32] | |
| MA7224§ | sodCI::3XFLAG-kan ilv3305::Tn10dTAC-cat-43::3XFLAG-kan | [23] |
| MA7225 | sodCII:: 3XFLAG-kan ilv3305::Tn10dTAC-cat-43::3XFLAG-kan | [23] |
| MA8029 | eptB115::MudK hfq67::cat | [18] |
| PF108 | dps::3XFLAG-kan ilv3305::Tn10dTAC-cat-43::3XFLAG-kan | [46] |
| PF114 | katG::cat | this work |
| PF121b† | katE:: 3xFLAG-kan ilv3305::Tn10dTAC-cat-43::3XFLAG-scar | this work |
| PF119 | katG::3xFLAG-kan ilv3305::Tn10dTAC-cat-43::3XFLAG-kan | this work |
| PF120 | katN::3XFLAG-kan ilv3305::Tn10dTAC-cat-43::3XFLAG-kan | this work |
| PF125 | hfq67::cat | this work |
| PF126 | katE::cat | this work |
| PF132 | katG::cat katN::kan | this work |
| PF133 | oxyR::cat | this work |
| PF134 | katN::kan | this work |
| PF137 | rpoE::kan | this work |
| PF139 | katE::cat katN::kan | this work |
| PF141† | katE::cat katG::scar | this work |
| PF148 | rpoS::cat | this work |
| SA211 | sodA::3XFLAG | this work |
ATCC, American Type Culture Collection.
The strain was obtained by S. Maloy via L. Bossi.
All mutant strains are derived from Salmonella enterica serovar Typhimurium strain ATCC14028, with the exception of strain MA8029 which is a derivative of strain LT2.
The term “scar” refers to the DNA sequence remaining after excision of antibiotic-resistance cassette (20)
2.2. Construction of Deletion Mutants
Mutations were constructed in S. Typhimurium ATCC14028 using the method of Datsenko and Wanner [20]. The coding sequences of katG, katE, oxyR and rpoS genes were replaced with the chloramphenicol resistance cassette amplified with the primers listed in Table 2, using the pKD3 plasmid [20] as a template for PCR amplification. The coding sequences of katN and rpoE genes were replaced with the kanamycin resistance cassette amplified with the primers listed in Table 2, using the pKD4 plasmid [20] as a template for PCR amplification. The constructs were verified by PCR analysis. Mutations were transferred into a clean S. Typhimurium ATCC14028 background by P22 transduction. The new strains are listed in Table 1. The double mutants katEkatN and katGkatN were generated by transduction of katN::kan allele from strain PF134 into PF126 (katE::cat) and PF114 (katG::cat), obtaining strains PF139 and PF132, respectively. To construct the double mutant katEkatG, the resistance cassette was eliminated from strain PF114 (katG::cat) by homologous recombination between the two flanking FRT sites using plasmid pCP20 [20, 21], thereby obtaining strain PF140 (katG::scar). Subsequently, the katE::cat was introduced in the PF140 by P22 transduction, to obtain strain PF141 (katE::cat, katG::scar).
Table 2.
Oligonucleotides Used in This Study
| Name | Sequence | Mutant |
|---|---|---|
| oPF10 | 5'-atgggtgaaagtgatgaacctggaccgtttcgatctgcaagactacaaagaccatgacgg-3' | katG::3xFLAG |
| oPF11 | 5'-tggtatccctgaacgtcacgcctgaaggggtgccgctgcatatgaatatcctccttag-3' | katG::3XFLAG |
| oPF14 | 5'-gcgcagaactaggctgcggcgaaccgaaagaagacaaggactacaaagaccatgacgg-3 | katN::3xFLAG |
| oPF15 | 5'-tcgcgtgatgttgtgacgaaaaaagaccccgccaggcgcatatgaatatcctccttag-3' | katN::3xFLAG |
| oPF18 | 5'-tgtctggtcgcgtgcggggaaaattaacgcgattcctgcagactacaaagaccatgacgg-3' | katE::3xFLAG |
| oPF19 | 5'-gccagagcgcctattgttacccccgcttaactgaactggacatatgaatatcctccttag-3 | katE::3xFLAG |
| oPF8 | 5'-actttaaaagggagctgagatatgagcacgaccgacgattgtaggctggagctgcttcg-3 | katG::cat |
| oPF9 | 5'-actttaaaagggagctgagatatgagcacgaccgacgattgtaggctggagctgcttcg-3 | katG::cat |
| oPF12 | 5'-tctttcggttcgccgcagcctagttctgcgccagtgagagtgtaggctggagctgcttcg-3' | katN::kan |
| oPF13 | 5’-cgtaaaacaacttcaatatactgtgcgagtgagcgaaccatatgaatatcctccttagtt-3' | katN::kan |
| oPF16 | 5'-gagttcaatgtcgcataatgagaaatccccccatcagtcctgtaggctggagctgcttcg-3' | katE::cat |
| oPF17 | 5'-aatcgcgttaattttccccgcacgcgaccagacacggtgtcatatgaatatcctccttagtt-3' | katE::cat |
| oPF20 | 5'-atgagcgagcagttaacggaccaggtcctggttgaacgtgtaggctggagctgcttcg-3 | rpoE::kan |
| oPF21 | 5'-gtcaacgcctgataagcggttgaactttattatcaatagctcatatgaatatcctccttagtt-3' | rpoE::kan |
| oPF22 | 5'-ttactcgcggaacagcgcttcgatattcagcccctgcgtgtaggctggagctgcttcg-3' | rpoS::cat |
| oPF23 | 5'-gtcagaatacgctgaaagttcatgatttaaatgaagacgccatatgaatatcctccttagtt-3' | rpoS::cat |
| oPF24 | 5'-atgaatattcgtgatcttgaatatctggtggcgttagccgtgtaggctggagctgcttcg-3' | oxyR::cat |
| oPF25 | 5'-tgttttaacgccttgtcgaaatggccatccattgcgccaccatatgaatatcctccttagtt-3' | oxyR::cat |
| oli105 | 5’-gaactgggacgaagcagcagcgcgtttcgccgctaaaaaagactacaaagaccatgacgg-3’ | sodA::3xFLAG |
| oli106 | 5’-gacccagctcggtcagcataagttcgccatcggcctgcaccatatgaatatcctccttag-3’ | sodA::3xFLAG |
2.3. Epitope Tagging and Immunodetection
Epitope tagging of KatE, KatG, KatN and SodA was obtained by adding a 3xFLAG at the 3’-terminus of the genes, following a described procedure [22]. DNA fragments were amplified on pSUB11 with primers oPF10/11 (for katG::3xFLAG), oPF14/15 (for katN::3xFLAG), oPF18/19 (for katE::3xFLAG) and oli105/106 (for sodA::3xFLAG) listed in Table 2. DNA was electroporated in S. Typhimurium 14028 containing plasmid pKD46 [20]. Transformants were selected and recombination was confirmed by PCR. Subsequently, the 3xFLAG mutations were transferred into a clean 14028 background by P22 transduction. To have an internal standard for western blot analysis, a 3xFLAG epitope-tagged chloramphenicol acetyl transferase gene (cat) constitutively expressed under control of the tac promoter, was transduced from strain MA7223 [23], obtaining the doubly tagged strains listed in Table 1.
Strains carrying the epitope-tagged genes were grown in the bioreactors as described above. After 20 hours of growth, 108 cells were removed immediately and harvested by centrifugation for the analysis of protein accumulation by Western blotting. The pellets were boiled for 10 min in Laemmli lysis buffer and the proteins were separated by 12% SDS-PAGE and blotted onto nitrocellulose membrane (Hybond ECL, Amersham). The epitope flagged proteins were revealed by the use of an anti-FLAG M2 monoclonal antibody (Sigma-Aldrich) as primary antibody, and an anti-mouse HRP-conjugated IgG (Sigma-Aldrich) as secondary antibody. Detection was performed by enhanced chemiluminescence (ECL Advance, Amersham). Densitometric analysis of the western blots was carried out with the Gel-Pro Analyzer software (Media Cybernetics) to evaluate differences in protein accumulation in NG and LSMMG.
2.4. Resistance to Hydrogen Peroxide Stress
Bacteria grown under LSMMG or NG were evaluated for their ability to survive to hydrogen peroxide stress. After 20 hours of growth under conditions described above, a period sufficient to reach the stationary phase, bacterial cultures were immediately (within 5 min) diluted to 107 cells/ml in phosphate buffered saline (PBS) and incubated statically at room temperature with 17.4 mM hydrogen peroxide, accordingly to a previous investigation [6]. Aliquots were removed after 30 minutes and the number of viable cells was determined by serial dilution and plating onto LB agar. The percent survival following hydrogen peroxide exposure was calculated for each strain by dividing the number of CFU (Colony Forming Units) obtained upon incubation in hydrogen peroxide by the number of CFU obtained upon incubation in PBS alone. Each assay was repeated at least three times, and standard deviations were calculated. A time course of resistance to 17.4 mM hydrogen peroxide of LSMMG and NG grown cells was carried out under the same conditions, by removing bacterial aliquots after 15, 30 and 45 minutes.
2.5. Resistance to Acid Stress
To determine susceptibility to acid stress, bacteria grown in LSMMG and NG conditions for 20 hours were diluted to 2x107 cells/ml in LB medium and subsequently to 107 cells/ml by adding a citrate buffer adjusted to pH 3.0. and incubated statically for 1 hour at room temperature. Aliquots were removed after 1 hour and the number of viable cells was determined by serial dilution and plating onto LB agar. The percent survival following acid exposure was calculated for each strain by dividing the number of CFU obtained upon incubation in acid citrate buffer (pH 3.0.) by the number of CFU obtained upon incubation in citrate buffer (pH 7.12). Each assay was repeated at least three times, and standard deviations were calculated.
2.6. Preparation of Crude Extracts and Measurement of Proteins
Bacteria grown in LSMMG and NG conditions were immediately pelleted at 4°C, resuspended in a phosphate buffer (5 mM potassium phosphate pH 7,5 mM disodium EDTA and 10% glycerol) and lysed by sonication. To avoid inactivation of heat-labile catalases during sonication, cells were given four rounds of 15-s pulses with a Branson B-12 sonicator and a microtip at a power setting of 6. Tubes were kept on ice between rounds. Debris was pelleted by centrifugation at 4°C for 20 minutes at 17000 x g. Total protein concentration was determined by the method of Lowry et al. [24].
2.7. Catalase and Peroxidase Activity Assays
Catalases present in crude extracts were separated by electrophoresis through a 9% nondenaturing polyacrylamide gel and visualized according to the method of Woodbury et al. [25]. Briefly, the gels are washed in distilled water and then incubated with 0.003% hydrogen peroxide for 10 minutes. The gels are next briefly rinsed with distilled water and subsequently placed in a solution containing 1% ferric chloride and 1% potassium ferricyanide (III) for 5 minutes. Achromatic zones against the dark background are due to the activity of catalase, which prevent the H2O2-dependent reduction of ferricyanide to ferrocyanide. Densitometric analyses of gels were carried out to obtain a semiquantitative evaluation of the activity of the different catalase in NG and LSMMG. Measurements of total catalase activity in cell extracts were carried according to described procedures [26]. The peroxidase activity of KatG was visualized on native gel by a previously described procedure [27].
3. RESULTS
3.1. Low-shear Modeled Microgravity Induces Hydrogen Peroxide Resistance in Different Salmonella Enterica Serovars
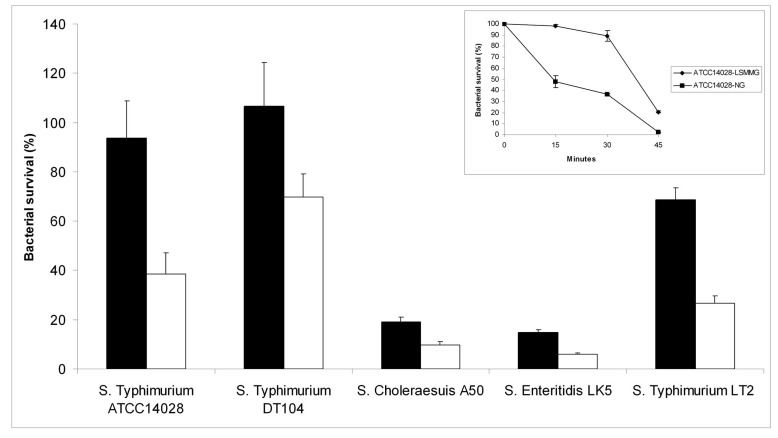
To understand the effects of microgravity on Salmonella enterica, we have tested the susceptibility to hydrogen peroxide of Salmonella enterica serovar Typhimurium ATCC-14028 cultures grown in low shear modeled microgravity (LSMMG) and in normal gravity (NG) conditions. Bacteria were grown for 20 hours in HARV bioreactors rotating on a horizontal axis to obtain a LSMMG environment or on a vertical axis to grow bacteria under NG, accordingly to established procedures [28]. The harvested cultures were immediately diluted in PBS and challenged with the addition of 17.4 mM hydrogen peroxide for 30 min at room temperature. Fig. (1) shows that bacteria grown under LSMMG are significantly more resistant to hydrogen peroxide than bacteria grown under NG. Similar results were obtained also with bacteria cultivated for 10 hours in bioreactors. Even though under these conditions the difference in hydrogen peroxide-resistance of bacteria cultivated in LSMMG and NG was less striking, bacteria grown under reduced gravitational force still exhibited higher resistance to H2O2 (data not shown). The inset of Fig. (1) shows that up to 30 min incubation with hydrogen peroxide (which corresponds to less than 35 min from the moment of extraction of bacteria from the bioreactors) there is a sharp difference in bacterial survival between bacteria grown in LSMMG and NG. At 45 min of incubation with hydrogen peroxide bacteria grown in LSMMG show a significant increase in mortality, which may be explained by the transient nature of the phenotypic changes induced by microgravity [6].
Fig. (1).
Resistance of different S. enterica strains to hydrogen peroxide. Results are expressed as percent survival after 30 minutes incu-bation with 17.4 mM hydrogen peroxide at room temperature. The S. Typhimurium LT2 strain was incubated with 8.7 mM hydrogen perox-ide, due to its higher susceptibility to this agent. The different serovars were grown in both LSMMG (solid bars) and NG (open bars) condi-tions. The percent survival was calculated for each strain by dividing the number of CFU/ml obtained after incubation in the presence of hydrogen peroxide by the number of CFU/ml obtained after incubation in PBS alone. The results are the average of three trials and represent mean ± standard errors. The inset shows a time course of S. Typhimurium ATCC14028 survival to 17.4 mM hydrogen peroxide, grown in LSMMG (diamond) and NG (square) conditions. The results are the average of three samples and represent mean ± standard errors.
To evaluate the general significance of these results, we analyzed the susceptibility of some other Salmonella strains to hydrogen peroxide and acid stress after growth in LSMMG and NG. We tested a virulent DT104 S. Typhimurium strain from our laboratory collection [29], S. Typhimurium LT2, an attenuated strain characterized by low levels of RpoS production [30, 31], S. enterica serovar Choleraeasuis A50 [32] and S. enterica serovar Enteritidis LK5 (lab collection).
Fig. (1) shows that these strains exhibited differential susceptibility to hydrogen peroxide, S. Typhimurium DT104 showing the highest resistance. S. Typhimurium LT2 proved to be particularly sensitive to this agent and, therefore, the data concerning this strain reported in Fig. (1) were obtained by incubating bacteria with a lower concentration of hydrogen peroxide. Interestingly, cultivation under LSMMG enhanced resistance to hydrogen peroxide of all these strains, suggesting that this is a common feature of most Salmonella strains.
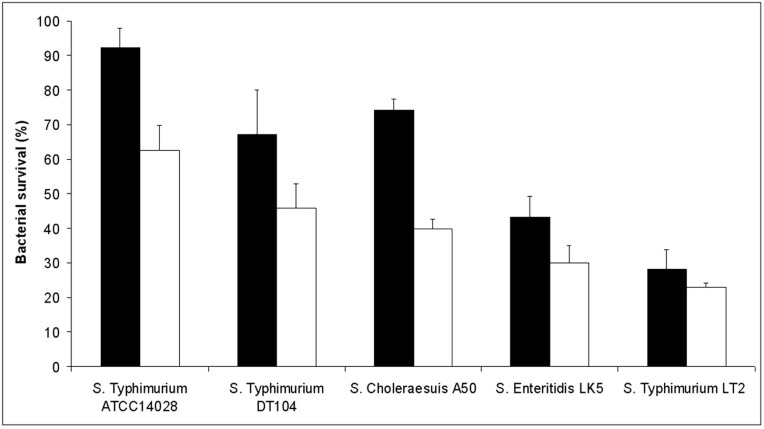
As previous studies carried out with S. Typhimurium SL1344 have indicated that LSMMG enhances Salmonella resistance to acid stress [2], we have also tested the acid stress resistance of all the Salmonella strains employed in this work. To this aim, we grew bacteria in HARV bioreactors in both LSMMG and NG for 20 hours and then exposed the cells to pH 3.0 for 1 hour at room temperature. In agreement with previous studies, all the strains examined displayed a marked increase in survival at pH 3.0 when grown in LSMMG condition (Fig. 2). The only exception is represented by the S. Typhimurium LT2 strain, which did not show a significant difference (according to a Student's t-test analysis) in survival between cells cultivated in LSMMG and NG.
Fig. (2).
Resistance of different S. enterica strains to acid stress. Results are expressed as percent survival after 1 hour incubation with citrate buffer (pH 3.0.) at room temperature. The different serovars are grown under conditions of LSMMG (solid bars) and NG (open bars). The percent survival was calculated for each strain by dividing the number of CFU/ml obtained after incubation in citrate buffer (pH 3.0.) by the number of CFU/ml obtained after incubation in citrate buffer at pH 7.12. The results are the average of three independent experiments and represent mean ± standard errors.
3.2. Analysis of Catalase Activity in Bacteria Grown in LSMMG and NG Conditions
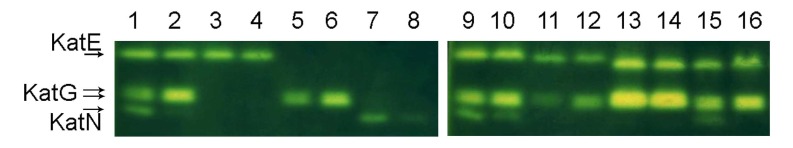
In consideration of the higher resistance to hydrogen peroxide of Salmonella strains grown in LSMMG, we looked for alterations in the activity of bacterial enzymes involved in H2O2 detoxification. Three different catalases, KatG and the RpoS-regulated catalases KatE and KatN, have been identified in Salmonella [33]. KatE and KatG are heme-containing catalases, whereas KatN is likely a manganese-containing enzyme. The activity of these proteins in bacterial extracts was assayed in non-denaturing 9% polyacrylamide gels stained for catalase activity (Fig. 3). Bacterial extracts were prepared following a previously described method that prevents KatG and KatN inactivation [26]. In fact, high temperatures and prolonged sonication readily inactivate KatG [26] and we have found that similar conditions disrupt KatN activity (data not shown). Moreover, we have also observed that this catalase is inactivated by extraction procedures involving the use of a detergent such as deoxycholic acid. Fig. (3) shows that KatN activity is increased in the extracts from S. Typhimurium ATCC14028 cells cultivated in LSMMG conditions (see lanes 1 and 2). Such an increase in KatN activity is clearly appreciable also in the isogenic mutant strain PF141, which is deleted of the katE and katG genes (Fig. 3, lanes 7 and 8). A densitometric analysis of the gel indicates that KatN activity in S. Typhimurium is 5-10 folds higher in bacteria cultivated in LSMMG. Moreover, Fig. (3) also shows a decrease (close to the 50% according to the densitometric analysis) in the intensity of the two KatG bands in extracts from bacteria cultivated in LSMMG, compared to extracts from bacteria grown in NG. A comparable modification in KatG activity can be observed also in the mutant strain PF139, which lacks the katE and katN genes (lanes 5 and 6). As KatG is a bifunctional catalase-peroxidase [34] we also stained gels for peroxidase activity, confirming the decrease in KatG activity in LSMMG (data not shown). The microgravity-induced changes in KatG and KatN activity were confirmed by quantitative spectrophotometric assays carried out on soluble protein extracts on S. Typhimurium ATCC 14028 strains (Table 3). Although the absolute values of catalase activity showed significant variations from experiment to experiment, we were never able to appreciate significant changes in total catalase activity in cell extracts from bacteria grown in LSMMG and NG deriving from the same preinoculum. Similarly, we did not observe differences in KatE activity between the mutant strain PF132 grown in LSMMG and NG conditions. In contrast, in good agreement with the densitometric analysis, the LSMMG environment induced a 2-fold decrease in the KatG activity of the mutant strain PF139. We were unable to measure the activity of KatN in NG condition, possibly because it is below the lower limit of the assay (0.05 units) [35]. These observations suggest that the increase in KatN activity largely compensates for the reduction of KatG activity observed in extracts from LSMMG-cultivated bacteria.
Fig. (3).
Visualization of catalase activity on polyacrylamide gels. The activity bands due to the three different Salmonella catalases were detected in native 9% polyacrylamide gels using the method of Woodbury et al. (1971). 50 µg of proteins from total cellular extracts were loaded in each lane: 1 and 2, ATCC 14028; 3 and 4, PF132 (katG::cat katN::kan); 5 and 6, PF139 (katE::cat katN::kan); 7 and 8, PF141 (katE::cat katG::scar); 9 and 10, DT104; 11 and 12, LT2; 13 and 14, S. Choleraesuis A50; 15 and 16, S. Enteritidis LK5. The odd lanes contain extracts from LSMMG grown bacteria. In the even lanes extracts from NG grown bacteria were loaded.
Table 3.
Catalase activity (U/mg) in S. Typhimurium ATCC 14028
| Strain | LSMMG | NG |
|---|---|---|
| S. Typhimurium ATCC 14028 | 7.58 ± 0.06 | 7.28 ± 0.26 |
| PF132 (katGkatN) | 3.5 ± 0.2 | 3.5 ± 0.2 |
| PF139 (katEkatN) | 0.46 ± 0.09 | 0.95 ± 0.15 |
| PF141(katEkatG) | 0.73 ± 0.14 | N.D. |
This Table reports measurements carried out in triplicate on the samples loaded on the gel reported in Figure 2. N.D.= activity below the detection limit of the assay.
A comparable variation in the catalase activity pattern between bacteria grown in LSMMG and NG was observed in extracts from S. Typhimurium DT104 (lanes 9 and 10) and S. Enteritidis LK5 (lanes 15 and 16). In contrast, the KatN activity band was not observed in extracts from S. Typhimurium LT2 (lanes 11 and 12). This observation, together with an apparent reduction of KatE activity, is likely related to the mutation in the rpoS allele that characterizes this strain [36]. Nevertheless, also in S. Typhimurium LT2 KatG activity is higher in bacteria grown in NG than in LSMMG. In S. Choleraesuis, where KatG is expressed at much higher levels than in the other strains and the activity band corresponding to KatN is hardly visible (lanes 13 and 14), we were not able to identify alterations in the activity of catalases between bacteria grown in the two conditions. It should be noted that this strain is less resistant to hydrogen peroxide stress than S. Typhimurium DT104 and S. Typhimurium ATCC 14028, despite its catalase activity is quite high (25,7 U/mg in LSMMG and 26,2 U/mg in NG). This observation indicates that additional factors are responsible for the intrinsic susceptibility of the different strains to hydrogen peroxide.
To analyze the possibility that changes in catalase activity bands reflect modifications in the intracellular concentration of proteins, we inserted a sequence encoding the 3xFLAG epitope at the 3’-terminus of katN, katG and katE genes and of a few other genes involved in oxidative stress response (dps, sodCI, sodCII, sodA) producing, respectively the ferritin-like protein Dps (DNA-binding proteins from starved cells), Cu,Zn superoxide dismutases I and II (SodCI and SodCII) and Mn superoxide dismutase (SodA). We observed a repeatable 15-20% reduction in the accumulation of KatG in LSMMG (Fig. 4), which partially accounts for the above described reduction in enzyme activity. In contrast, we did not observe appreciable variations in the intracellular concentration of Dps, SodCI, SodCII, SodA, KatE and KatN in bacteria cultivated in LSMMG and NG conditions (data not shown).
Fig. (4).
KatG accumulation in S. Typhimurium ATCC14028. A) Strain PF114 was grown in both LSMMG and NG conditions for 20 hours and then 108 cells were immediately removed and treated as described in Materials and Methods for protein immunodetection. Cat (Chloramphenicol acetyl transferase) accumulation is shown as an internal standard. B) densitometric analysis of the Western blot presented in panel A; relative OD were calculated as Cat/KatG ratios.
3.3. Involvement of Catalases in Hydrogen Peroxide Resistance in LSMMG
To evaluate the effective contribution of genes coding for catalases to the enhanced hydrogen peroxide resistance of Salmonella strains grown in LSMMG, we constructed mutant strains of S. Typhimurium ATCC14028 lacking the single katG, katE or katN genes and the double mutants katEkatG, katGkatN and katEkatN. All these mutants were grown either in LSMMG or in NG conditions and the harvested bacteria were incubated with hydrogen peroxide. The single katE, katG and katN mutants were more resistant to killing by hydrogen peroxide when grown in LSMMG, although their survival was slightly lower than that of the wild type strain (data not shown). A similar behaviour was observed for the double katEkatN (PF139) and katEkatG mutants (PF141) (Fig. 5). In contrast, the resistance to hydrogen peroxide of the katGkatN (PF132) mutant strain was identical in bacteria cultivated in LSMMG or NG conditions. This result demonstrates that both KatG and KatN concur to the higher hydrogen peroxide resistance of the Salmonella strains grown in LSMMG. Interestingly, the katGkatN (PF132) mutant strain still exhibited higher resistance to acid stress when cultivated in LSMMG than in NG conditions (data not shown). This observation indicates that KatG and KatN are not involved in the enhanced resistance to low pH induced by microgravity.
Fig. (5).
Hydrogen peroxide susceptibility of wild type and mutant S. Typhimurium ATCC14028 strains lacking the kat genes.. Results are expressed as percent survival after 30 minutes incubation with 17.4 mM hydrogen peroxide at room temperature. The different strains [wt; PF139 (katE::cat katN::kan); PF141 (katE::cat katG::scar); PF132 (katG::cat katN::kan)] were grown in both LSMMG (solid bars) and NG (open bars) conditions. The percent survival was calculated for each strain by dividing the number of CFU/ml obtained after incubation with hydrogen peroxide by the number of CFU/ml obtained after incubation in PBS alone. The results are the average of three experiments and represent mean ± standard errors.
3.4. Hydrogen Peroxide and Acid Resistance of Mutant S. Typhimurium Strains Defective in Global gene Regulators
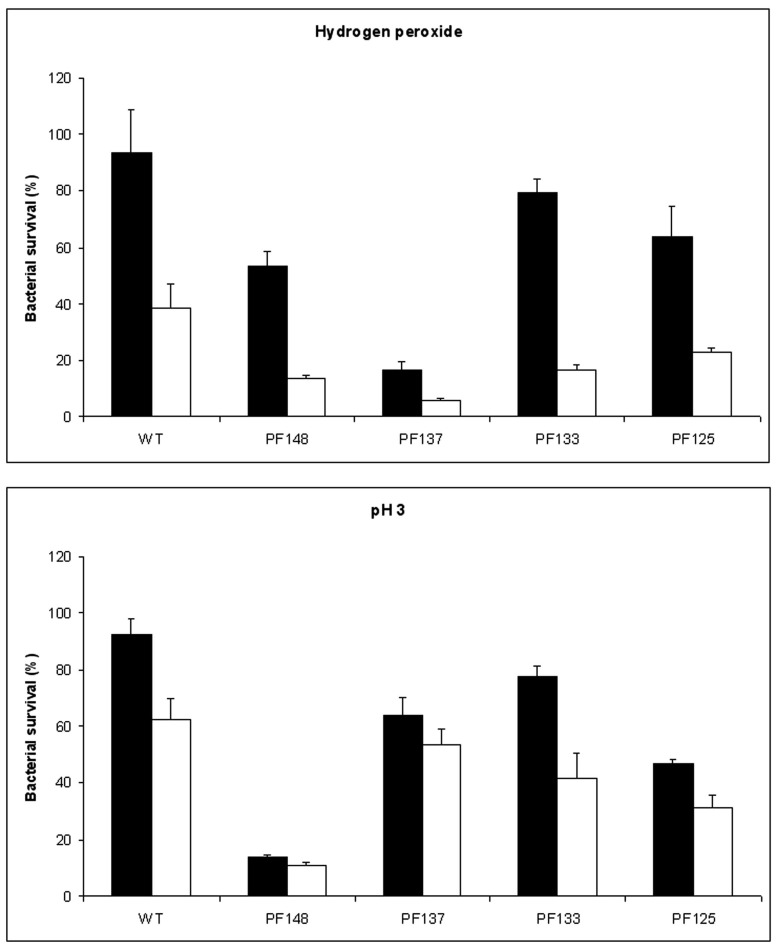
Recent studies have suggested that the global transcriptional regulators Hfq could play a major role in the alterations in gene expression induced by LSMMG or by spaceflights in S. Typhimurium [9] as well as in P. aeruginosa [37]. Therefore, we have analyzed the effect on catalases expression of mutations in hfq and other global regulators: rpoE, which modulates Salmonella virulence and resistance to oxidative stress through the regulation of several genes involved in the envelope stress response [38], oxyR, a global regulator of genes involved in bacterial response to hydrogen peroxide [39] and rpoS, the master regulator of a variety of stationary-phase genes, required for Salmonella virulence in mice [40-42].
As expected, Salmonella strains unable to produce Hfq, OxyR, RpoE or RpoS were much more susceptible to hydrogen peroxide than the wild type strain (Fig. 6). In particular, the hfq, oxyR and rpoS mutants were almost completely killed upon treatments with 17.4 mM hydrogen peroxide (data not shown). Therefore, the survival data reported in Fig. (6) for these three mutant strains were obtained upon incubation with a lower amount of hydrogen peroxide (5 mM). Independently of the higher susceptibility to oxidative stress, all these mutant strains proved to be more resistant to hydrogen peroxide when cultivated in LSMMG than in NG. This observation indicates that such global regulators are not responsible of the increased resistance to peroxide stress observed in Salmonella strains grown in microgravity conditions. Finally, we assayed the susceptibility of these mutants to pH acid, after a growth in NG and in LSMMG condition (Fig. 6). Interestingly, deletion of rpoE reduces the LSMMG-induced resistance to acid pH.
Fig. (6).
Susceptibility of wild type and mutant S. Typhimurium ATCC14028 strains deleted of the global gene regulatores rpoS, rpoE, oxyR and hfq to hydrogen peroxide and acid stress. Strains PF148(rpoS), PF137 (rpoE), PF133 (oxyR) and PF125 (hfq) were grown in both LSMMG (solid bars) and NG (open bars) conditions and then exposed to hydrogen peroxide (upper panel) or to acid stress (lower panel). Wild type and PF137 strains were incubated with 17.4 mM hydrogen peroxide whereas the PF148, PF133 and PF125 strains were incubated with 5 mM hydrogen peroxide. Results are expressed as percent survival after 30 minutes incubation with hydrogen peroxide at room tem-perature (upper panel) and after 1 hour incubation with citrate buffer at pH 3.0. (lower panel). The percent survival was calculated as de-scribed in the legends to figure 1 and 2. The results are the average of three independent experiments and represent mean ± standard errors.
DISCUSSION
Previous studies have established that either spaceflights or growth in ground-based spaceflight analog bioreactors induce phenotypic modifications and relevant alterations in gene expression in S. Typhimurium. In particular, these investigations have revealed that growth under microgravity enhances Salmonella virulence, suggesting that this well characterized microorganism may represent a suitable model to analyze bacterial pathogens to this environmental condition. Studies on the Salmonella response to LSMMG may provide useful information on the pathogenic mechanisms of this organism as it encounters low fluid shear areas during the infectious process, such as between brush border microvilli in the intestinal tract [28].
During infections, pathogenic bacteria are exposed to high concentrations of reactive oxygen species generated by the respiratory burst of phagocytic cells. To investigate the possibility that microgravity could enhance Salmonella ability to withstand oxidative stress, we have analyzed the effect of LSMMG on Salmonella enterica resistance to hydrogen peroxide. Our findings indicate that Salmonella is significantly more resistant to hydrogen peroxide stress when cultivated in LSMMG than in NG (Fig. 1). Although this observation is in disagreement with a previous experiment carried out with the S. Typhimurium strain SL1344 [6], our results indicate that this response is common to several S. Typhimurium strains as well as to other Salmonella strains belonging to different serotypes. Interestingly enough, a comparable increase in resistance to oxidative stress has been recently observed also in P. aruginosa [15].
To evaluate the possibility that LSMMG might influence the antioxidant apparatus involved in H2O2 detoxification, we monitored the activity of Salmonella catalases in extracts from bacteria grown in LSMMG and NG. Staining of gels for catalase activity revealed changes in the activity of KatN and KatG. In fact, KatN activity was 5-10 folds increased in extracts from S. Typhimurium ATCC14028, DT104 and S. Enteritidis LK5 grown in LSMMG, whereas, under the same conditions, it was possible to appreciate a decrease in the activity of KatG in all the strains except for S. Choleraesuis.
The role of KatN in Salmonella has been poorly investigated, but a possible involvement of this enzyme in virulence has been suggested by studies showing that overexpression of this catalase increases Salmonella lethality in flies mutated in the NF-kB pathway and is sufficient to render a non-pathogenic E. coli strain highly virulent to NF-kB pathway mutant flies [43]. Expression of katN is under control of the alternative sigma factor RpoS [33], although it can be induced independently of rpoS in the presence of bile [44]. The absence of a KatN band in the extracts from S. Typhimurium LT2 can be supposedly attributed the rpoS mutation characterizing this strain. In support of this view, we have observed that KatN activity can not be detected in extracts from the rpoS mutant strain PF148 (data not shown). We have not investigated if this gene is expressed in S. Choleraesuis. Interestingly, the increase in KatN activity in S. Typhimurium grown in LSMMG is not due to an enhanced intracellular concentration of the enzyme, as we failed to observe a variation in the intracellular accumulation of the 3xFLAG epitope-tagged protein. Therefore, we suggest that the explanation for the higher KatN activity in bacteria grown in microgravity should be looked for in post-translational events. For example, an increase in KatN activity could be related to the efficiency of manganese insertion in the enzyme active site or to microgravity-dependent differences in the susceptibility of this protein to oxidative inactivation.
In contrast, the decrease in KatG activity in bacteria grown under LSMMG, is paralleled by a slight change in protein accumulation (Fig. 3), thus suggesting that it is at least partially due to gravity-dependent alterations in gene regulation. No differences were observed in the intracellular accumulation of KatE, nor of other proteins involved in bacterial resistance to oxidative stress, including Dps, SodCI, SodCII, and SodA. The involvement of KatG and KatN in the hydrogen peroxide resistance induced by the LSMMG environment was confirmed by the analysis of single and double mutant strains of S. Typhimurium ATCC14028 devoid of katE, katG and katN genes. Although the mutant strains were more sensitive than the wild type strains to hydrogen peroxide, all the single mutants and the double katE/katG and katE/katN mutant strains displayed higher resistance to hydrogen peroxide when grown in LSMMG. In contrast, the double katG/katN mutant exhibited an identical susceptibility to killing by H2O2 either in LSMMG or in NG (Fig. 5). Solution assays revealed that the catalase activity of extracts from bacteria grown in LSMMG and NG is very similar, indicating that the higher resistance to hydrogen peroxide of cells grown in LSMMG is not simply due to enhanced catalase activity. In contrast, our results suggest that the higher resistance to hydrogen peroxide killing is associated to variations in the relative pattern of activity of KatG and KatN. This finding suggests that the three Salmonella catalases, while possessing a similar ability to scavenge hydrogen peroxide, play distinct physiological roles in vivo. This possibility is further supported by the observation that Salmonella possesses also other enzymes able to scavenge hydrogen peroxide, including two alkyl hydroperoxide reductases [45] and the Dps protein [46]. The presence of such a high number of enzymes with apparently redundant functions is likely justified by the significant toxicity of hydrogen peroxide [45], but also suggests that each of these proteins plays additional roles which are not completely overlapping with those of the other antioxidant enzymes. It is known that the different enzymes belonging to the catalase family differ significantly in their overall and active-site architecture and in the mechanism of reaction [47] and recent studies have shown that KatG is also able to catalyze the breakdown of peroxynitrite, suggesting that each catalase could play different roles in the detoxification of oxidative stresses [48]. In addition, the three catalases could be differently implicated in the mechanisms of adaptation to a variety of stimuli and conditions, as previously proposed to explain other known examples of apparent genetic redundancy of antioxidant enzymes. A well known example is represented by SodCI and SodCII, the two periplasmic Cu,Zn superoxide dismutases of S. Typhimurium. These enzymes catalyze the same reaction and share a very similar three-dimensional structure, but show well distinct functional roles: SodCI facilitate survival within the infected host by protecting bacteria from the phagocytic oxidative burst [46], whereas SodCII plays a role in protection from the reactive oxygen species produced endogenously during aerobic growth [39]. Recent studies have established that structural and regulatory differences accounts for the different role of the two enzymes [49]. Likewise, the sodA and sodB genes, encoding for a manganese- and an iron-containing cytoplasmic superoxide dismutases respectively, are differently regulated: the former is finely regulated to favor its expression under aerobic and oxidative stress conditions, while the latter is constitutively expressed, even under anaerobic conditions [50, 51]. In addition, it has been shown that MnSOD can associate with DNA and preferentially localizes with the nucleoid, while FeSOD is concentrated in the periphery of the cells, near the inner membrane [52]. Whereas KatG possess a peroxidative activity that is not present in KatE, the studies of bacterial superoxide dismutases suggest that additional subtle differences in regulation, catalytic properties or intracellular distribution could provide a rationale for the presence of different catalases in Salmonella. In this connection, the finding that microgravity induces changes in the relative activity of KatN and KatG, which significantly influence Salmonella ability to withstand hydrogen peroxide stress, support the hypothesis that the various catalases play roles that are not interchangeable.
We have also evaluated the involvement of the global transcriptional regulators OxyR, RpoE, RpoS and Hfq in the increased resistance to hydrogen peroxide stress of LSMMG-grown cells. OxyR coordinates the induction of genes required for survival to hydrogen peroxide stress, including KatG, the alkyl hydroperoxidase reductase, the Dps protein and glutathione reductase [53, 54]. RpoE is an alternative sigma factor that regulates genes required for the maintenance of membrane and periplasmic homeostasis in response to extracytoplasmic stress, which controls antioxidant defences and is critically important for S. Typhimurium virulence [38, 55]. RpoS, an alternative sigma factor, plays a key role in the survival of bacteria during starvation or exposure to various stress conditions and is required for the expression of many genes in the stationary phase of growth [41, 56]. It is also known that it is involved in Salmonella virulence in mice [40, 42]. Hfq is a RNA chaperone that binds small regulatory RNAs and facilitate bacterial responses to different kinds of stress [57]. Recent studies have suggested that Hfq plays a central role in the Salmonella response to reduced gravity [7]. Also, a regulatory cascade involving RpoE, Hfq and other transcriptional regulators controls the expression of the RpoS stress regulon [58].
As expected, we have found that S. Typhimurium strains deleted of such regulators show increased susceptibility to H2O2, both in LSMMG and in NG conditions. In addition, in agreement with a recent study showing that rpoE is induced by acidic conditions and plays an important role in resistance to low pH in vitro and in the intracellular vacuole [59], we have observed that the inactivation of rpoE reduced the acid stress resistance of cells cultivated in LSMMG. However, our results clearly indicate that OxyR, RpoE, RpoS and Hfq are not directly responsible for the increase of S. Typhimurium resistance to hydrogen peroxide induced by LSMMG. In fact, the S. Typhimurium strains lacking these genes were all more resistant to H2O2 when cultivated in LSMMG than in NG (Fig. 6). These observations rule out the possibility that Hfq is the general regulator of all the phenotypic changes of Salmonella in response to microgravity [7] and suggest that multiple regulatory pathways are involved in the bacterial adaptation to such condition.
Taken together, our results indicate that microgravity conditions induce alterations in the activity of KatG and KatN that enhance Salmonella ability to withstand H2O2-mediated oxidative stress. Besides contributing to understand the mechanisms that modulate bacterial virulence during spaceflights, our observations suggest that low shear conditions such as those encountered in the intestine between the brush border microvilli may induce changes in the relative activity of catalases, which may favor the natural course of Salmonella infections.
ACKNOWLEDGMENTS
The authors are grateful to Lionello Bossi (CNRS, Gif Sur Ivette, France) for providing the hfq mutant allele used in this work. A fellowship to F.P. was supported by a grant from the Italian Space Agency (ASI contract I/06/006/0). We are indebted to Prof. Francesco Lacquaniti, director of the Center for Space Biomedicine at the University of Rome Tor Vergata, for his constant encouragements and for providing us the HARV bioreactor. Thanks are due to Serena Ammendola for collaborating in the construction of some of the mutant strains used in this work.
REFERENCES
- 1.Song B, Leff LG. Identification and characterization of bacterial isolates from the Mir space station. Microbiol Res. 2005;160:111–7. doi: 10.1016/j.micres.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Nickerson CA, Ott MC, Mister SJ, Morrow BJ, Burns-Keliher L, Dierson DL. Microgravity as a novel environmental signal affecting Salmonella enterica serovar Typhimurium virulence. Infect Immun. 2000;68:3147–52. doi: 10.1128/iai.68.6.3147-3152.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klaus DM, Howard HM. Antibiotic efficacy and microbial virulence during space flight. Trends Biotechnol. 2006;24:131–6. doi: 10.1016/j.tibtech.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 4.Leys NM, Hendrickx L, De Boever P, Baatout S, Mergeay M. Spaceflights effects on bacterial physiology. J Biol Regul Homeost Agents. 2004;18:193–9. [PubMed] [Google Scholar]
- 5.Sonnenfeld G, Shearer WT. Immune function during space flight. Nutrition. 2002;18:899–903. doi: 10.1016/s0899-9007(02)00903-6. [DOI] [PubMed] [Google Scholar]
- 6.Wilson JW, Ott CM, Ramamurthy R, et al. Low-shear modeled microgravity alters the Salmonella enterica serovar Typhimurium stress response in an RpoS-independent manner. App1 Environ Microbiol. 2002;68:5408–16. doi: 10.1128/AEM.68.11.5408-5416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson JW, Ramamurthy R, Porwollik S, et al. Microarray analysis identifies Salmonella genes belonging to the low-shear modeled microgravity regulon. Proc Natl Acad Sci USA. 2002;99:13807–12. doi: 10.1073/pnas.212387899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chopra V, Fadl AA, Sha J, Chopra S, Galindo CL, Chopra AK. Alteration in the virulence potential of enteric pathogens and bacterial-host cell interactions under simulated microgravity conditions. J Toxicol Environ Health A. 2006;69:1345–70. doi: 10.1080/15287390500361792. [DOI] [PubMed] [Google Scholar]
- 9.Wilson JW, Ott CM, Höner zu Bentrup K, et al. Space flight alters bacterial gene expression and virulence and reveals a role for global regulator Hfq. Proc Natl Acad Sci USA. 2007;104:16299–304. doi: 10.1073/pnas.0707155104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tucker DL, Ott MC, Huff S, et al. Characterization of Escherichia coli MG1655 grown in a low-shear modeled microgravity environment. BMC Microbiol. 2007;7:7–15. doi: 10.1186/1471-2180-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lynch SV, Mukundakrishnan K, Benoit MR, Ayyaswamy PS, Matin A. Escherichia coli biofilms formed under low-shear modeled microgravity in a ground-based system. Appl Environ Microbiol. 2006;72:7701–10. doi: 10.1128/AEM.01294-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li GB, Liu YD, Wang GH, Song LR. Reactive oxygen species and antioxidant enzymes activity of Anabaena sp. PCC 7120 (Cyano-bacterium) under simulated microgravity. Acta Astronautica. 2004;55:953–7. doi: 10.1016/j.actaastro.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 13.Kemp MJ, Chen F, Kern R, Venkateswaran K. Recurrent isolation of hydrogen peroxide-resistant spores of Bacillus pumilus from a spacecraft assembly facility. Astrobiology. 2005;5:391–405. doi: 10.1089/ast.2005.5.391. [DOI] [PubMed] [Google Scholar]
- 14.La Duc MT, Nicholson W, Kern R, Venkateswaran K. Microbial characterization of the Mars Odyssey spacecraft and its encapsulation facility. Environ Microbiol. 2003;5:977–85. doi: 10.1046/j.1462-2920.2003.00496.x. [DOI] [PubMed] [Google Scholar]
- 15.La Duc MT, Dekas A, Osman S, Moissl C, Newcombe D, Venkateswaran K. Isolation and characterization of bacteria capable of tolerating the extreme conditions of clean room environments. Appl Environ Microbiol. 2007;73:2600–11. doi: 10.1128/AEM.03007-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crabbé A, Pycke B, Van Houdt R, et al. Response of Pseudomonas aeruginosa PAO1 to low shear modelled microgravity involves AlgU regulation. Environ Microbiol. 2010;12:1545–64. doi: 10.1111/j.1462-2920.2010.02184.x. [DOI] [PubMed] [Google Scholar]
- 17.Edwards RA, Matlock BC, Heffernan BJ, Maloy SR. Genomic analysis and growth-phase-dependent regulation of the SEF14 fimbriae of Salmonella enterica serovar Enteritidis. Microbiology. 2001;147:2705–15. doi: 10.1099/00221287-147-10-2705. [DOI] [PubMed] [Google Scholar]
- 18.Figueroa-Bossi N, Lemire S, Maloriol D, Balbontín R, Casadesús J, ossi L. Loss of Hfq activates the sigmaE-dependent envelope stress response in Salmonella enteric. Mol Microbiol. 2006;62:838–2. doi: 10.1111/j.1365-2958.2006.05413.x. [DOI] [PubMed] [Google Scholar]
- 19.Schwarz RP, Wolf DA. Rotating bioreactor cell culture apparatus. US patent 4988623. 1991.
- 20.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–5. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cherapanov PP, Wackernagel W. Gene disruption in Escherichia coli, TeR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 22.Uzzau S, Figueroa-Bossi N, Rubino S, Bossi L. Epitope tagging of chromosomal genes in Salmonella. Proc Natl Acad Sci USA. 2001;98:15264–9. doi: 10.1073/pnas.261348198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uzzau S, Bossi L, Figueroa-Bossi N. Differential accumulation of Salmonella Cu,Zn superoxide dismutases SodCI and SodCII in intracellular bacteria, correlation with their relative contribution to pathogenicity. Mol Microbiol. 2002;46:147–156. doi: 10.1046/j.1365-2958.2002.03145.x. [DOI] [PubMed] [Google Scholar]
- 24.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–75. [PubMed] [Google Scholar]
- 25.Woodbury W, Spencer AK, Stahmann MA. An improved procedure using ferrycianide for detecting catalase isozymes. Anal Biochem. 1971;44:301–5. doi: 10.1016/0003-2697(71)90375-7. [DOI] [PubMed] [Google Scholar]
- 26.Visick JE, Clarke S. RpoS- and OxyR-independent induction of HPI catalase at stationary phase in Escherichia coli and identifica-tion of rpoS mutations in common laboratory strains. J Bacteriol. 1997;179:4158–63. doi: 10.1128/jb.179.13.4158-4163.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pacello F, Langford PR, Kroll JS, et al. A novel heme protein, the Cu,Zn-superoxide dismutase from Haemophilus ducreyi. J Biol Chem. 2001;276:30326–34. doi: 10.1074/jbc.M010488200. [DOI] [PubMed] [Google Scholar]
- 28.Nickerson CA, Ott CM, Wilson JW, Ramamurthy R, Pierson DL. Microbial responses to microgravity and other low-shear environments. Microbiol Mol Biol Rev. 2004;68:345–61. doi: 10.1128/MMBR.68.2.345-361.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pasquali P, Ammendola S, Pistoia C, et al. Attenuated Salmonella enterica serovar Typhimurium lacking the ZnuABC transporter confers immune-based protection against challenge infections in mice. Vaccine. 2008;26:3421–6. doi: 10.1016/j.vaccine.2008.04.036. [DOI] [PubMed] [Google Scholar]
- 30.Lee IS, Lin J, Hall HK, Bearson B, Foster JW. The stationary-phase sigma factor sigma S (RpoS) is required for a sustained acid tolerance response in virulent Salmonella typhimurium. Mol Microbiol. 1995;17:155–67. doi: 10.1111/j.1365-2958.1995.mmi_17010155.x. [DOI] [PubMed] [Google Scholar]
- 31.Wilmes-Riesenberg MR, Foster JW, Curtiss R. An altered rpoS allele contributes to the avirulence of Salmonella typhimurium LT2 Infect Immun. 1997;65:203–10. doi: 10.1128/iai.65.1.203-210.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watson PR, Paulin SM, Jones PW, Wallis TS. Interaction of Salmonella serovars with porcine macrophages in vitro does not correlate with virulence. Microbiology. 2000;146:1639–49. doi: 10.1099/00221287-146-7-1639. [DOI] [PubMed] [Google Scholar]
- 33.Robbe-Saule V, Coynault C, Ibanez-Ruiz M, Hermant D, Norel F. Identification of a non-haem catalase in Salmonella and its regulation by RpoS (σs) Mol Microbiol. 2001;39:1533–45. doi: 10.1046/j.1365-2958.2001.02340.x. [DOI] [PubMed] [Google Scholar]
- 34.Loewen PC, Stauffer GV. Nucleotide sequence of katG of Salmonella typhimurium LT2 and characterization of its product, hydroperoxidase I. Mol Gen Genet. 1990;224:147–51. doi: 10.1007/BF00259461. [DOI] [PubMed] [Google Scholar]
- 35.Beers RF, Sizer IW. A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J Biol Chem. 1952;195:133–40. [PubMed] [Google Scholar]
- 36.Robbe-Saule V, Lopes MD, Kolb A, Norel F. Physiological effects of Crl in Salmonella are modulated by σs level and promoter specificity. J Bacteriol. 2007;189:2976–87. doi: 10.1128/JB.01919-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crabbé A, Schurr MJ, Monsieurs P, et al. Transcriptional and proteomic responses of Pseudomonas aeruginosa PAO1 t,o spaceflight conditions involve Hfq regulation and reveal a role for oxygen. Appl Environ Microbiol. 2011;77:1221–30. doi: 10.1128/AEM.01582-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Testerman TL, Vazquez-Torres A, Xu Y, Jones-Carson J, Libby SJ, Fang FC. The alternative sigma factor sigmaE controls antioxidant defences required for Salmonella virulence and stationary-phase survival. Mol Microbiol. 2002;43:771–82. doi: 10.1046/j.1365-2958.2002.02787.x. [DOI] [PubMed] [Google Scholar]
- 39.Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem. 2008;77:755–76. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fang FC, Libby SJ, Buchmeier NA, et al. The alternative sigma factor KatF (RpoS) regulates Salmonella virulence. Proc Natl Acad Sci USA. 1992;89:11978–82. doi: 10.1073/pnas.89.24.11978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ishihama A. Adaptation of gene expression in stationary phase bacteria. Curr Opin Genet Dev. 1997;7:582–8. doi: 10.1016/s0959-437x(97)80003-2. [DOI] [PubMed] [Google Scholar]
- 42.Nickerson CA, Curtiss R. Role of sigma factor RpoS in initial stages of Salmonella typhimurium infection. Infect Immun. 1997;65:1814–23. doi: 10.1128/iai.65.5.1814-1823.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ryu JH, Ha EM, Oh CT, et al. An essential complementary role of NF-kappaB pathway to microbicidal oxidants in Drosophila gut immunity. EMBO J. 2006;25:3693–701. doi: 10.1038/sj.emboj.7601233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prouty AM, Brodsky IE, Manos J, Belas R, Falkow S, Gunn JS. Transcriptional regulation of Salmonella enterica serovar Typhimurium genes by bile. FEMS Immunol Med Microbiol. 2004;41:177–85. doi: 10.1016/j.femsim.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 45.Hébrard M, Viala JP, Méresse S, Barras F, Aussel L. Redundant hydrogen peroxide scavengers contribute to Salmonella virulence and oxidative stress resistance. J Bacteriol. 2009;191: 4605–14. doi: 10.1128/JB.00144-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pacello F, Ceci P, Ammendola S, Pasquali P, Chiancone E, Battistoni A. Periplasmic Cu, Zn superoxide dismutase and cytoplasmic Dps concur in protecting Salmonella enterica serovar Typhimurium from extracellular reactive oxygen species. Biochem Byophys Acta. 2008;1780:226–32. doi: 10.1016/j.bbagen.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 47.Zamocky M, Furtmuller PG, Obinger C. Evolution of catalases from bacteria to humans. Antioxid Redox Signal. 2008;10:527–47. doi: 10.1089/ars.2008.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McLean S, Bowman LA, Poole RK. KatG from Salmonella Typhimurium is a peroxynitritase. FEBS Lett. 2010;584:1628–32. doi: 10.1016/j.febslet.2010.03.029. [DOI] [PubMed] [Google Scholar]
- 49.Ammendola S, Pasquali P, Pacello F, et al. Regulatory and structural differences in the Cu,Zn-superoxide dismutases of Salmonella enterica and their significance for virulence. J Biol Chem. 2008;283:13688–99. doi: 10.1074/jbc.M710499200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Compan I, Touati D. Anaerobic activation of arcA transcription in Escherichia coli, roles of Fnr and ArcA. Mol Microbiol. 1994;11:955–64. doi: 10.1111/j.1365-2958.1994.tb00374.x. [DOI] [PubMed] [Google Scholar]
- 51.Fee JA. Regulation of sod genes in Escherichia coli, relevance to superoxide dismutase function. Mol Microbiol. 1991;5:2599–610. doi: 10.1111/j.1365-2958.1991.tb01968.x. [DOI] [PubMed] [Google Scholar]
- 52.Steinman HM, Weinstein L, Brenowitz M. The manganese superoxide dismutase of Escherichia coli K-12 associates with DNA. J Biol Chem. 1994;269:28629–34. [PubMed] [Google Scholar]
- 53.Christman MF, Morgan RW, Jacobson FS, Ames BN. Positive control of a regulon for defenses against oxidative stress and some heat-shock proteins in Salmonella typhimurium. Cell. 1985;41:753–62. doi: 10.1016/s0092-8674(85)80056-8. [DOI] [PubMed] [Google Scholar]
- 54.Pomposiello PJ, Demple B. Redox-operated genetic switches, the SoxR and OxyR transcription factors. Trends Biotechnol. 2001;19:109–14. doi: 10.1016/s0167-7799(00)01542-0. [DOI] [PubMed] [Google Scholar]
- 55.Humphreys S, Stevenson A, Bacon A, Weinhardt AB, Roberts M. The alternative sigma factor, σE, is critically important for the virulence of Salmonella typhimurium. Infect Immun. 1999;67:1560–156. doi: 10.1128/iai.67.4.1560-1568.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Loewen PC, Hu B, Strutinsky J, Sparling R. Regulation in the rpoS regulon of Escherichia coli. Can J Microbiol. 1998;44:707–17. doi: 10.1139/cjm-44-8-707. [DOI] [PubMed] [Google Scholar]
- 57.Valentin-Hansen P, Eriksen M, Udesen C. The bacterial Sm-like protein Hfq, a key player in RNA transactions. Mol Microbiol. 2004;51:1525–33. doi: 10.1111/j.1365-2958.2003.03935.x. [DOI] [PubMed] [Google Scholar]
- 58.Bang I-S, Frye JE, McClelland M, Velayudhan J, Fang FC. Alternative sigma factor interactions in Salmonella, σE and σH promote antioxidant defences by enhancing σS levels. Mol Microbiol. 2005;56:811–23. doi: 10.1111/j.1365-2958.2005.04580.x. [DOI] [PubMed] [Google Scholar]
- 59.Muller C, Bang I-S, Velayudhan J, et al. Acid stress activation of the σE stress response in Salmonella enterica serovar Typhimurium. Mol Microbiol. 2009;71:228–38. doi: 10.1111/j.1365-2958.2009.06597.x. [DOI] [PMC free article] [PubMed] [Google Scholar]