Abstract
We have developed a multiplex sequencing-by-synthesis method combining terminal-phosphate labeled fluorogenic nucleotides (TPLFNs) and resealable microreactors. In the presence of phosphatase, the incorporation of a non-fluorescent TPLFN into a DNA primer by DNA polymerase results in a fluorophore. We immobilize DNA templates within polydimethylsiloxane (PDMS) microreactors, sequentially introduce one of the four identically labeled TPLFNs, seal the microreactors, allow template-directed TPLFN incorporation, and measure the signal from the fluorophores trapped in the microreactors. This workflow allows sequencing in a manner akin to pyrosequencing but without constant monitoring of each microreactor. With cycle times of <10 minutes, we demonstrate 30 base reads with ∼99% raw accuracy. “Fluorogenic pyrosequencing” combines benefits of pyrosequencing, such as rapid turn-around, native DNA generation, and single-color detection, with benefits of fluorescence-based approaches, such as highly sensitive detection and simple parallelization.
High throughput sequencing methods are revolutionizing biology and promise to significantly impact the future of medicine. However, further reductions in cost and improvements in turn-around time and sample preparation will be necessary for sequencing to realize its scientific and diagnostic promise. Commercially available clonal sequencing-by-synthesis technologies can be divided into two major classes.
First, in pyrosequencing schemes, typified by the 454 Genome Sequencer (Roche)1, native nucleotide species are serially introduced to sequencing reactions, and a transient bioluminescence signal is detected upon nucleotide incorporation1,2. The semiconductor sequencing scheme implemented in the Ion Torrent Personal Genome Machine (Life Technologies) also employs serial introduction of native nucleotides but avoids the use of an enzymatic cascade by detecting released hydrogen ions electrochemically3. Second, in the fluorescence detection sequencing scheme, employed by Illumina's HiSeq 2000, DNA polymerase incorporates a fluorescently labeled, reversible terminator nucleotide, resulting in the conjugation of a different fluorophore to DNA for each nucleotide species4. The identity of the base is indicated by the color of the resulting fluorescence, and subsequent base incorporations require chemical removal of the fluorophore and terminating moiety. Similar chemistry is employed in the HeliScope (Helicos) for single molecule sequencing5. In competing fluorescence-based sequencing-by-ligation technologies, such as SOLiD (Life Technologies)6, Polonator (Danaher)7, and cPAL (Complete Genomics)8, short, fluorescently labeled oligonucleotide probes are hybridized, ligated to anchor strands, and imaged to identify bases in the DNA template. The workflow for sequencing-by-ligation also involves a distinct step to remove labeled components, and the introduction of many different labeled and unlabeled oligonucleotides.
Each class of sequencing methodology has distinct advantages and disadvantages. The native nucleotides used in pyrosequencing and semiconductor sequencing allow the synthesis of native DNA and require no chemical removal step subsequent to incorporation to continue the sequencing process. Therefore, sequencing can progress as fast as the nucleotides can be introduced, incorporated, detected, and replaced, leading to relatively rapid turn-around times on the order of hours1. In addition, longer read lengths have been achieved with pyrosequencing1. However, the detection of a transient luminescence or electrochemical signal requires constant monitoring of every clonal population, which has thus far severely limited throughput. In addition, the luminescence and electrochemical detection schemes employed in pyrosequencing and semiconductor sequencing, respectively, are less sensitive when compared to fluorescence detection.
Fluorescence-based schemes achieve massive throughput per run because the fluorescence signal of an incorporated base-labeled nucleotide can be probed at any time, allowing for a scanning-based readout of an area not limited by detector size4-8. The scalability of these methods allows the generation of >1011 bases/run8, while pyro-detection sequencers are currently limited to ∼109 bases/run1. Finally, the inherent sensitivity of fluorescence allows fluorescence-based sequencing methods to use >103 times fewer DNA templates than pyro-detection methods1,3, reducing reagent volumes and costs4-8. However, the multiple chemical steps required in each sequencing cycle results in a more complex workflow than pyrosequencing, limiting sequencing speed and read lengths.
The single molecule real-time (SMRT) approach developed by Pacific Biosciences uses four terminal phosphate-labeled nucleotides (TPLNs), each of which is conjugated to a different colored fluorophore9. This method allows for a fluorescence-based readout along with generation of native DNA and extremely rapid turn-around times9. However, the transient fluorescence signal requires constant monitoring with high sampling rates and single molecule sensitivity and the single-molecule nature of the readout, as well as the fact that binding rather than incorporation is observed, leads to higher single read error rates compared to clonal methods9.
Here we describe fluorogenic pyrosequencing, an approach that uses fluorogenic TPLNs (TPLFNs)10-12 to allow for a pyrosequencing workflow with a non-transient, fluorescence-based signal, combining benefits of both classes of clonal sequencers. Fluorogenic pyrosequencing combines scalability with fast cycle times and, unlike commercially available fluorescence-based sequencers, uses a simple one-color, wide field imaging system. The microreactor flow cell platform is ideal for low reagent consumption and can be readily integrated into conventional microfluidic devices for sample preparation.
Results
Fluorogenic Pyrosequencing Workflow
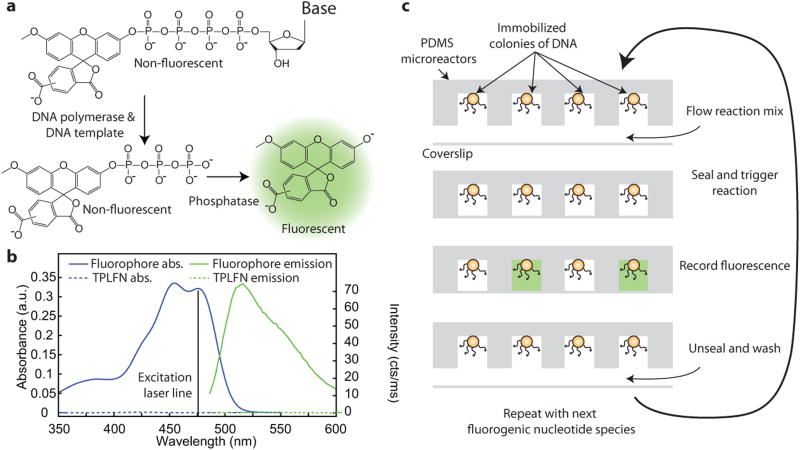
In fluorogenic pyrosequencing, each nucleotide species is labeled at the terminal phosphate position with an identical dye moiety (Fig. 1a), forming a non-fluorescent substrate (Fig. 1b). Upon incorporation of a TPLFN into an immobilized DNA template by DNA polymerase, native DNA is generated and a labeled polyphosphate molecule is released. This polyphosphate chain is rapidly digested by phosphatase, yielding a fluorophore (Fig. 1a,b) that is trapped in resealable PDMS microreactors and detected by a CCD camera. This process is repeated for all four TPLFNs, each of which has the same label.
Figure 1.
Fluorogenic pyrosequencing chemistry and workflow. (a) A non-fluorescent terminal phosphate-labeled fluorogenic nucleotide (TPLFN) is incorporated by DNA polymerase into a DNA primer/template, releasing a labeled, non-fluorescent polyphosphate which is digested by phosphatase to generate a fluorescent product. (b) Absorption (blue) and fluorescence emission (green) spectra of the TPLFN (dotted line) and fluorescent product (solid line). Excitation wavelength of 476 nm is shown. (c) DNA is immobilized on beads in PDMS microreactors which are loaded with a reaction mixture containing DNA polymerase, phosphatase, and one of four TPLFNs at low temperature, sealed, and heated to trigger primer extension. Fluorophores are generated in microreactors that contain DNA templates in which the base adjacent to the primer is complementary to the introduced TPLFN. The array is imaged with a fluorescence microscope, unsealed, and washed before the cycle is repeated.
Upon incorporation of a TPLFN, the detectable species released by DNA polymerase diffuses away, destroying the spatial correspondence between base incorporation and the presence of fluorophores.. To solve this problem, DNA templates are spatially arranged in individual, sealable microreactors fabricated using soft lithography in PDMS (Fig. 1c). These reactors can be sealed reversibly against a coverglass, isolating individual sequencing reactions and trapping the generated fluorophores13. The microreactors, each of which contains ∼5,000 copies of a primed DNA template, are cooled to a temperature where DNA polymerase is relatively inactive and loaded with DNA polymerase, phosphatase, and one of the four species of TPLFNs. TPLFNs are excellent substrates for DNA polymerase14, and because TPLFNs are non-fluorescent (Fig. 1b), they can be present in the sample at relatively high concentrations without concern for fluorescence background. The microreactors are then sealed, and the temperature of the sample is raised to allow nucleotide incorporation. If the introduced TPLFN is complementary to the DNA template base adjacent to the primer, fluorophores will be generated and trapped in the microreactor (Fig 1c). In homopolymeric regions with multiple, identical bases in the DNA template adjacent to the primer, primer extension will continue until the identity of the template base changes. Hence, the number of fluorophores generated ought to be proportional to the length of the homopolymer. After the nucleotide incorporation reaction has gone to completion, the microreactor array is imaged with a one-color fluorescence microscope, unsealed, and washed. This cycle can be automated and repeated within a large array to sequence multiple DNA templates in parallel. Each sequencing cycle results in the generation of physically trapped fluorophores in the microreactors which can be detected with high sensitivity at any time, obviating the need for real-time monitoring.
Fluorogenic Pyrosequencing Chemistry
TPLN synthesis15-17 is simple compared to that of many other nucleotide analogs, and the same conjugation strategy can be applied to all four nucleotides10,11,14. Although TPLFNs, in which the label is attached to the γ-phosphate, are rather poor substrates for DNA polymerase10,11, extension of the phosphate chain to increase the distance between the labeling moiety and the polymerase active site, and recover the native charge of a nucleotide triphosphate has led to significant improvements10,11. To demonstrate fluorogenic pyrosequencing, we synthesized a set of four δ-labeled nucleotide tetraphosphates (Fig. 1a) which are incorporated by Bst DNA polymerase. We conjugated all four nucleotides to 3′-O-methyl-5(6)-carboxyfluorescein, which is a fluorogenic dye excitable at 476 nm with extremely high fluorogenic contrast (Figure 1b) that does not diffuse appreciably into PDMS under the conditions of our experiment. It is easily synthesized from the commercially available fluorophore 5(6)-carboxyfluorescein, and can be phosphorylated with standard phosphoramidite chemistry18 allowing conjugation to all four dNTPs10,11,14 (Supplementary Note 1, Supplementary Figs. 1 and 2) to generate TPLFNs (Fig. 1a).
We used Bst DNA polymerase (large fragment) because it incorporates TPLFNs efficiently, has good strand displacement activity, a very temperature-sensitive incorporation rate (high above 50 °C but low below 5 °C), and no exonuclease activity. After the microreactors are sealed, we trigger primer extension by raising the temperature to 55°C, driving TPLFN incorporation and release of 3′-O-methyl-5(6)-carboxyfluorescein triphosphate to completion within two minutes. We introduce calf intestinal alkaline phosphatase (CIAP) along with the polymerase to rapidly digest the released 3′-O-methyl-5(6)-carboxyfluorescein triphosphate, generating fluorescent product.
PDMS Microreactor Flow Cell
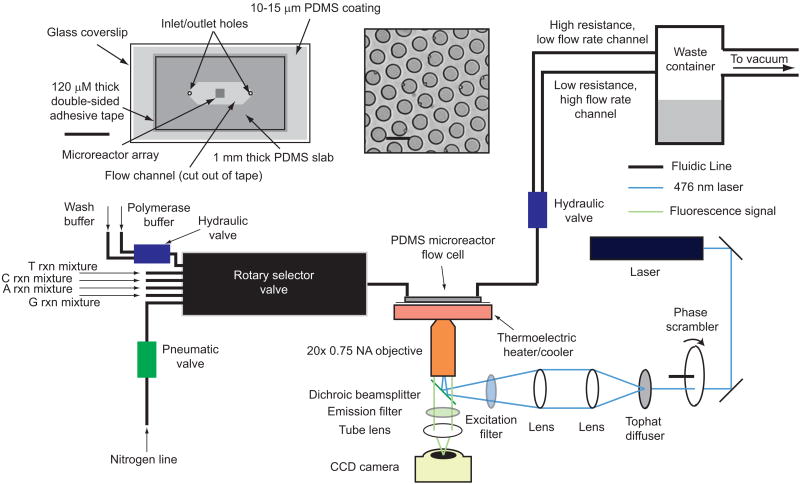
Fluorogenic pyrosequencing occurs within a flow cell that facilitates rapid fluid exchange while minimizing sample volume1,3-8. The instrumentation required for fluorogenic pyrosequencing consists of three modules: imaging, fluidics, and temperature control (Fig. 2, Online Methods).Flow cells are constructed from PDMS19(see Online Methods). We generated arrays of micron-sized pillars on a silicon wafer using conventional photolithography (see Online Methods), and used this silicon master to generate PDMS microreactor arrays with soft lithography13,19. The regular array format has the potential for greatly simplified data acquisition and analysis while providing a straightforward means of increasing sample density and throughput8.
Figure 2.
Schematic of the fluorogenic pyrosequencing system. The sequencer comprises of three modules: imaging, fluidics, and temperature control. The imaging module is a one-color epifluorescence microscope composed of a laser, halogen lamp, CCD camera, microscope frame, filter set, and a 20× air objective with some additional optics (phase scrambler and tophat diffuser) to flatten the field-of-view. The lamp provides bright field transmission images of the microreactor array that are used for maintaining the proper focal plane. A rotary selector valve, hydraulic and pneumatic valves, a nitrogen line, and a vacuum line comprise the fluidics module which controls sealing, reagent flow, and washing. The vacuum line is used to both deliver reagents to the sample and also seal the PDMS microreactor array against the lower surface of the flow cell. The thermoelectric temperature controller cools the microreactor array while reagents are loaded into the reactors to avoid significant nucleotide incorporation prior to sealing. This module then heats the array after sealing to allow rapid primer extension. (inset) A schematic of a PDMS microreactor flow cell and a bright field image of the PDMS microreactors. The microreactors are ∼5 μm in diameter, 3.8 μm in height with center-to-center distance of 7.5 μm. Some of the microreactors contain 1 μm polystyrene beads. Scale bars are 1 cm (left) and 7.5 μm (right).
PDMS is largely transparent to visible light and has low autofluorescence relative to other polymers used in microfluidics20, making it compatible with fluorescence microscopy. Although PDMS is largely inert, it can be treated with oxygen plasma to form reactive groups for functionalization19. This feature allows biotinylation21 of PDMS microreactors, facilitating capture of streptavidin-coated beads to which DNA templates are bound (see Online Methods). PDMS is also a flexible elastomer that can seal reversibly to flat surfaces13. Similar platforms involving silica microreactors sealed to a silicone gasket have also been applied to single molecule enzymology22. We constructed PDMS microreactor flow cells consisting of a microreactor array-containing slab, a spacer, and a glass coverslip coated with a thin layer of PDMS (Fig. 2). Sealing is accomplished by applying vacuum to the outlet of the flow cell, causing the elastomeric PDMS slab that forms the top of the flow cell to deform and seal to the PDMS coating on the glass surface (see Online Methods, Supplementary Fig. 3). This process is repeated in every sequencing cycle to facilitate microreactor loading, fluorescent product trapping, and washing.
Fluorogenic Pyrosequencing Performance
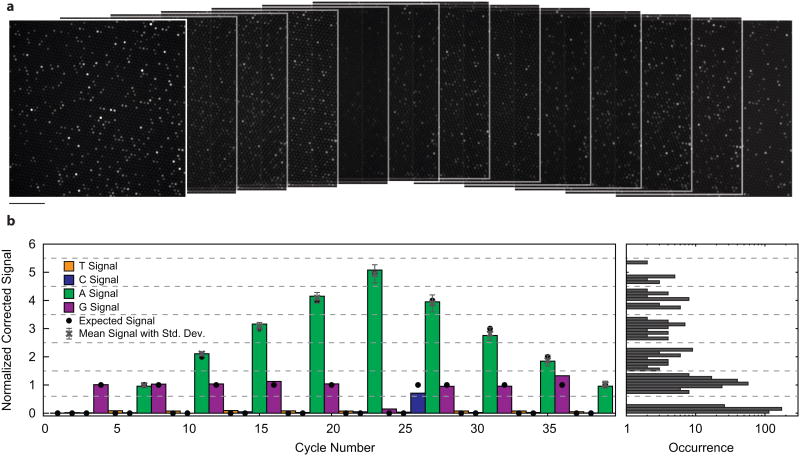
A subset of the fluorescence images obtained during a 60-cycle fluorogenic pyrosequencing run are shown in Fig. 3a, with each image corresponding to the measured fluorescence generated after one TPLFN reaction mixture was introduced to the sample. A fraction of the microreactors were loaded with 1 μm polystyrene beads (not visible in the fluorescence images) each with ∼5,000 copies of immobilized DNA template. Each of the beads was decorated with one of four different DNA oligo test sequences (Supplementary Table 1). The first test sequence is a “homopolymer ladder” (HL) oligo (Fig. 3b), and the other three are randomers with very different sequences (R1, R2, and R3) (Fig. 4, Supplementary Table 1). Data presented in Figs. 3 and 4 are from the same run.
Figure 3.
Fluorogenic pyrosequencing images and demonstration of signal linearity. (a) Sequential fluorescence images of a PDMS microreactor array obtained during the first 13 cycles of a 60 cycle fluorogenic pyrosequencing run. Signal heterogeneity between reactors is due either to the presence of multiple beads, or a homopolymeric region of the DNA template, which leads to the generation of more than one fluorophore per DNA copy. Scale bar 74 μm. (b) Representative sequence of homopolymeric DNA. The colored bars represent a corrected (see Online Methods) sequencing trace from a single microreactor containing the “homopolymer ladder” (HL) sequence. Thresholds (dotted lines) indicate the intensity ranges used to determine homopolymer lengths. Signal intensities expected from this template sequence (black dots), show that this sequencing trace is error-free. [The black dots are the correct sequence]The average and standard deviation of the intensities for all analyzed traces (N=15, grey points) all fall within the expected intensity range. Intensity levels for all sequencing cycles from the analyzed HL traces were histogramed (right).
Figure 4.
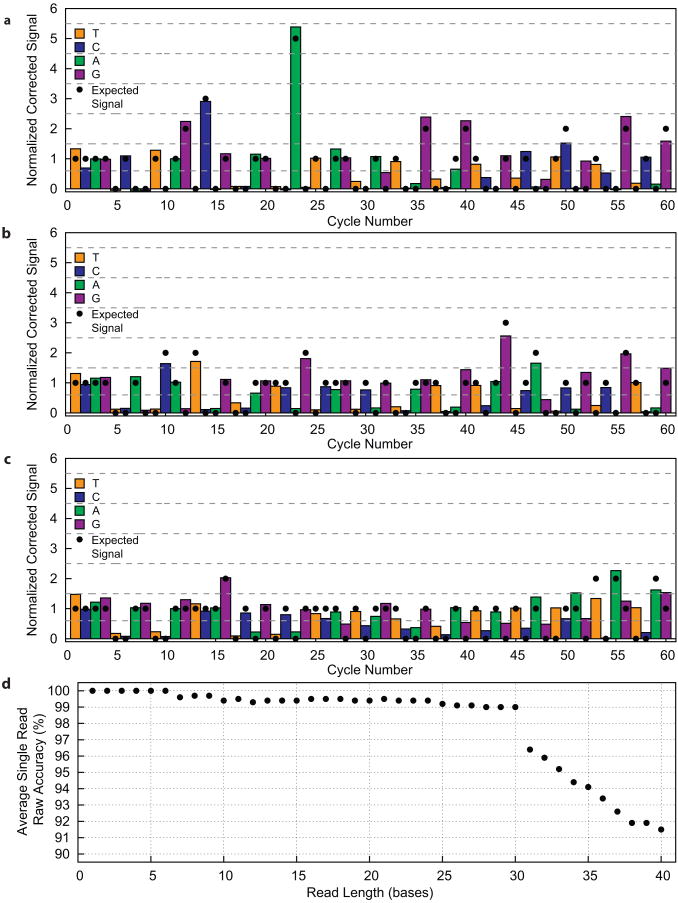
Sequencing traces of quasi-random sequences, and analysis of global errors (a)-(c) Corrected sequencing traces from individual microreactors containing the R1, R2, and R3 DNA templates, respectively. Signal intensities expected from this template sequence (black dots) and threshold lines (dotted) that denote the intensity ranges for different homopolymer lengths are shown. (a) Sequencing trace from R1sequence that is error-free to 36 bases with one error in the 42-base read. (b) Sequencing trace from R2 sequence that is error free for the entire 41-base read. (c) Sequencing trace from R3 sequence that is error-free to 30 bases with 6 errors in the entire 40-base read. (d) Global analysis of accuracy as a function of read length for all analyzed R1, R2, and R3 traces (N = 36). 30-base reads have an average raw accuracy of 99%.
The HL oligo sequence consists of a series of poly-T homopolymers (up to five bases long) interrupted by single G or C bases.Unlike reversible terminator chemistry, in which primer extension cannot proceed beyond the first base in a homopolymer within a single cycle, primer extension continues through the whole length of a homopolymer in pyrosequencing. The fidelity of fluorogenic pyrosequencing depends critically on the ability to quantitatively differentiate the levels of signal generated at homopolymers of different lengths1-3. Ideally, the signal is directly proportional to homopolymer length. Homopolymer sequencing accuracy is one of the major challenges of methods with a pyrosequencing-type workflow such as fluorogenic pyrosequencing. The system achieved a single read raw accuracy of >99%, and 80% of these HL sequencing traces are error-free. The origins of this accuracy can be visualized from the average and standard deviations (Fig. 3b, grey points) of the signal generated in each cycle for all HL oligo-containing microreactors analyzed from this run. In addition, a histogram of the signal distribution for all analyzed HL oligo-containing microreactors across all sequencing cycles shows that the signal levels corresponding to zero, one, two, three, four, and five bases are well-separated (Fig. 3b, right).
In order to test this sequencing methodology on arbitrary sequences, we designed three quasi-random DNA templates with very different sequence characteristics, and generated individual sequencing traces from the three oligos (R1 in Fig. 4a, R2 in Fig. 4b, and R3 in Fig 4c). A global analysis of all analyzed traces for all three random oligos sequenced in this experiment (36 traces, 12 for each sequence, Fig. 4d) demonstrates that our current system maintains a mean single read raw accuracy of at least 99% (that is an error rate of 1% or less) to a read length of 30 bases.
The reduction in accuracy beyond ∼30 bases (Fig. 4d) is due to signal decay and dephasing which occur in many clonal sequencing systems1,4,7. Data presented in Figs. 3 and 4 were background subtracted and corrected for signal decay (see Online Methods, Supplementary Fig. 4), but not corrected for dephasing. However, we can reasonably expect that substantial improvements in read length and accuracy could be achieved by more sophisticated means of data analysis.
Discussion
Fluorogenic pyrosequencing has the potential for dramatically improved performance over this proof-of-principle demonstration. The current cycle times of <10 minutes can be reduced by increasing the speed of nucleotide incorporation and the rate of fluidic exchange (for example by using positive pressure and optimized flow-cell geometries). TPLNs with longer phosphate chains have been shown to impart better catalytic efficiency compared to the δ-labeled nucleotides used here, making incorporation more rapid9-11,14. The time required for washing the system can be further reduced with more sophisticated fluidics and enzymes that thoroughly digest excess nucleotides between cycles1,2. Finally, more advanced data processing algorithms to correct for de-phasing will increase the tolerance of our system to faster cycle times1,23,24.
The observed signal decay can occur for a number of reasons including DNA loss resulting from misincorporation, primer-template melting, digestion by enzymatic impurities, or even DNA dissociation. De-phasing occurs as a result of either incomplete extension, in which primer extension for a sub-population of DNA templates falls behind, or carry-forward, in which incomplete washing causes a sub-population of DNA templates to advance ahead of the main population1,4,23,24. Commercial sequencing platforms employ computer algorithms to model and correct for dephasing in combination with sophisticated image processing, leading to significant increases in read length23,24. We expect these improvements to our sequencing chemistry, wash cycle efficiency, and data analysis to yield increases in read length and accuracy.
Because real-time monitoring is unnecessary, DNA samples in two flow cells may be sequenced simultaneously with one sample undergoing wash and TPLFN incorporation cycles while the other is imaged. Although imaging a very large array will inevitably lead to increased cycle times, further increases in throughput can be realized by fabrication of smaller microreactors and the implementation of high-speed fluorescence imaging schemes such as time delay integration (TDI) line-scanning. We have demonstrated fluorogenic pyrosequencing in arrays of ∼2.5 μm diameter microreactors (Supplementary Fig. 5), each of which contain only ∼2,000 copies of DNA template. These microreactors are ∼8× smaller in volume than those shown in Figs. 2 and 3, and the arrays are also >2.3-fold denser. Higher density arrays may be combined with more efficient sample loading methods that are not limited by Poisson statistics to increase the fraction of DNA-containing microreactors1,8 (Supplementary Fig. 6). Increases in array density and loading efficiency will lead to decreases in reagent consumption per base (see Supplementary Note 2 for discussion). The off-the-shelf polymerase and phosphatase enzymes are currently the dominant reagent cost. Fabrication and material costs of the PDMS microreactor flow cells are extremely low and highly scalable. Despite having similar chemistry, fluorogenic pyrosequencing has the potential for much lower reagent costs than conventional pyrosequencing due to the relatively small DNA copy number (thousands vs. ∼10 million for pyrosequencing)1 and feature size required in fluorescence-based approaches. At the same time, TPLFN chemistry is considerably simpler than that employed in commercial fluorescence-based platforms.
Rapid reduction in the reagent costs per sequenced base in commercial sequencers has made up-front instrumentation cost increasingly important. For sequencing systems that employ an optical readout, the illumination and detection equipment, including lasers, optics, and cameras, contribute substantially to instrument cost. However, fluorogenic pyrosequencing requires only one-color fluorescence detection. Furthermore, DNA colonies in fluorogenic pyrosequencing are arranged in a regular array8 allowing more efficient use of the detector than in most commercial platforms, where colonies are immobilized randomly4-7. The realization of high-speed, scanning-based fluorescence detection is substantially simplified if spectral separation is unnecessary and the sample can be spatially registered with the detector.
Sample preparation costs are also becoming more critical, particularly as diagnostic applications are contemplated. Fluorogenic pyrosequencing is, in principle, compatible with all commercially available methods for library preparation and clonal amplification. We have demonstrated the sequencing of DNA oligos immobilized on polymer beads, making clonal amplification by emulsion PCR a natural extension of our current technique25. However, because the number of template DNAs required for sequencing is small (<104), less efficient but simpler on-chip amplification methods such as bridge PCR4 or solid-state rolling circle amplification (RCA)26 are also applicable. Additionally, our PDMS microreactor platform can be readily integrated into PDMS microfluidic devices for processing complex samples, single cell manipulation, extraction of genetic material, whole genome amplification, and targeted amplification on-chip27-29 (see Supplementary Note 3). This potential for seamless microfluidic integration opens the door to sample-to-sequence analysis of raw biomaterials on a unitary microfluidic device.
Supplementary Material
Supplementary Figure 1: Synthesis of 3′-O-methyl-5(6)-carboxyfluorescein-monophosphate
Supplementary Figure 2: Synthesis of dG4P-δ-3′-O-methyl-5(6)-carboxyfluorescein
Supplementary Figure 3: Vacuum-based sealing of PDMS microreactor array
Supplementary Figure 4: Sample sequencing data processing
Supplementary Figure 5: Demonstration of fluorogenic pyrosequencing in high density microreactor arrays.
Supplementary Figure 6: Super-Poissonian loading of PDMS microreactors
Supplementary Table 1: DNA Sequences used in fluorogenic pyrosequencing experiments
Supplementary Note 1: Synthesis of terminal phosphate-labeled fluorogenic nucleotides
Supplementary Note 2: The scalability of fluorogenic pyrosequencing
Supplementary Note 3: Potential for microfluidic integration of fluorogenic pyrosequencing
Acknowledgments
This work was supported by the United States National Institutes of Health National Human Genome Research Institute Grant (HG005097-01) to X.S.X. and the United States National Institutes of Health Natinoal Human Genome Research Institute Recovery Act Grand Opportunities Grant (1RC2HG005613-01) to X.S.X. W.J.G. was supported by the Damon Runyon Cancer Research Foundation (DRG-2000-09 to W.J.G.). We would like to acknowledge J. Foley, S. Song, L. Song, G. Holtom, and K. Fiala for valuable discussions and technical assistance. This work was performed in part at the Center for Nanoscale Systems, a member of the National Nanotechnology Infrastructure Network, which is supported by the National Science Foundation award no. ECS-0335765. The Center for Nanoscale Systems is part of Harvard University.
Footnotes
Author Contributions: P.A.S., W.J.G., and X.S.X. conceived the fluorogenic pyrosequencing concept. P.A.S. and W.J.G. constructed the sequencing apparatus. H.D. synthesized the TPLFNs. P.A.S. collected and analyzed the data. W.J.G. carried out the microfabrication. P.A.S., W.J.G., H.D., and X.S.X. wrote the manuscript.
Competing Financial Interests: Harvard University has filed patent applications based on this work.
References
- 1.Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ronaghi M, Karamohamed S, Pettersson B, Uhlen M, Nyren P. Real-time DNA sequencing using detection of pyrophosphate release. Anal Biochem. 1996;242:84–89. doi: 10.1006/abio.1996.0432. [DOI] [PubMed] [Google Scholar]
- 3.Rothberg JM, et al. Methods and apparatus for measuring analytes using large scale FET arrays. 2010 WO 2010/008480 A2. [Google Scholar]
- 4.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harris TD, et al. Single molecule DNA sequencing of a viral genome. Science. 2008;320:106–109. doi: 10.1126/science.1150427. [DOI] [PubMed] [Google Scholar]
- 6.McKernan KJ, et al. Sequence and structure variation in a human genome uncovered by short-read, massively parallel ligation sequencing using two-base encoding. Genome Res. 2009;19:1527–1541. doi: 10.1101/gr.091868.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shendure J, et al. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 8.Drmanac R, et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 9.Eid J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 10.Kumar S, et al. Terminal phosphate-labeled nucleotides: synthesis, applications, and linker effect on incorporation by DNA polymerases. Nucleosides, Nucleotides, Nucleic Acids. 2005;24:401–408. doi: 10.1081/ncn-200059823. [DOI] [PubMed] [Google Scholar]
- 11.Sood A, et al. Terminal phosphate-labeled nucleotides with improved substrate properties for homogeneous nucleic acid assays. J Am Chem Soc. 2005;127:2394–2395. doi: 10.1021/ja043595x. [DOI] [PubMed] [Google Scholar]
- 12.Kozlov M, Bergendahl V, Burgess R, Goldfarb A, Mustaev A. Homogeneous fluorescent assay for RNA polymerase. Anal Biochem. 2005;342:206–213. doi: 10.1016/j.ab.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 13.Rondelez Y, et al. Microfabricated arrays of femtoliter chambers allow single molecule enzymology. Nat Biotechnol. 2005;23:361–365. doi: 10.1038/nbt1072. [DOI] [PubMed] [Google Scholar]
- 14.Korlach J, et al. Long, processive enzymatic DNA synthesis using 100% dye-labeled terminal phosphate-linked nucleotides. Nucleosides, Nucleotides, Nucleic Acids. 2008;27:1072–1083. doi: 10.1080/15257770802260741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ankilova VN, Knorre DG, Kravchenko VV, Lavrik OI, Nevinsky GA. Investigation of the phenylalanyl-tRNA synthetase modification with gamma-(p-azidoanilide-)-ATP. FEBS Lett. 1975;60:172–175. doi: 10.1016/0014-5793(75)80445-5. [DOI] [PubMed] [Google Scholar]
- 16.Yarbrough LR. Synthesis and properties of a new fluorescent analog of ATP: adenosine-5′-triphosphoro-γ-1-(5-sulfonic acid) napthylamidate. Biochem Biophys Res Commun. 1978;81:35–41. doi: 10.1016/0006-291x(78)91627-3. [DOI] [PubMed] [Google Scholar]
- 17.Grachev MA, Zaychikov EF. ATP gamma-anilidate: a substrate of DNA-dependent RNA-polymerase of Escherichia coli. FEBS Lett. 1974;49:163–166. doi: 10.1016/0014-5793(74)80502-8. [DOI] [PubMed] [Google Scholar]
- 18.Takakusa H, Kikuchi K, Urano Y, Kojima H, Negano T. A novel design method of ratiometric fluorescent probes based on fluorescence resonance energy transfer switching by spectral overlap. Chem Eur J. 2003;9:1479–1485. doi: 10.1002/chem.200390167. [DOI] [PubMed] [Google Scholar]
- 19.McDonald JC, Whitesides GM. Poly(dimethylsiloxane) as a material for fabricating microfluidic devices. Acc Chem Res. 2002;35:491–499. doi: 10.1021/ar010110q. [DOI] [PubMed] [Google Scholar]
- 20.Piruska A, et al. The autofluorescence of plastic materials and chips measured under laser irradiation. Lab Chip. 2005;5:1348–1354. doi: 10.1039/b508288a. [DOI] [PubMed] [Google Scholar]
- 21.Liu D, Perdue RK, Sun L, Crooks RM. Immobilization of DNA onto poly(dimethylsiloxane) surfaces and application to a microelectrochemical enzyme-amplified DNA hybridization assay. Langmuir. 2004;20:5905–5910. doi: 10.1021/la049605p. [DOI] [PubMed] [Google Scholar]
- 22.Gorris HH, Rissin DM, Walt DR. Stochastic inhibitor release and binding from single enzyme molecules. Proc Natl Acad Sci USA. 2007;104:17680–17685. doi: 10.1073/pnas.0705411104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leamon JH, Rothberg JM. Cramming more sequencing reactions onto microreactor chips. Chem Rev. 2007;107:3367–3376. doi: 10.1021/cr068297s. [DOI] [PubMed] [Google Scholar]
- 24.Erlich Y, et al. Alta-cyclic: a self-optimizing base caller for next-generation sequencing. Nat Methods. 2008;5:679–682. doi: 10.1038/nmeth.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dressman D, Yan H, Traverso G, Kinzler KW, Vogelstein B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc Natl Acad Sci USA. 2003;100:8817–8822. doi: 10.1073/pnas.1133470100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hatch A, Sano T, Misasi J, Smith CL. Rolling circle amplification of DNA immobilized on solid surfaces and its application to multiplex mutation detection. Genetic Analysis: Biomolecular Engineering. 1999;15:35–40. doi: 10.1016/s1050-3862(98)00014-x. [DOI] [PubMed] [Google Scholar]
- 27.Marcus JS, Anderson WF, Quake SR. Microfluidic single-cell mRNA isolation and analysis. Anal Chem. 2006;78:3084–3089. doi: 10.1021/ac0519460. [DOI] [PubMed] [Google Scholar]
- 28.Fan HC, Wang J, Potanina A, Quake SR. Whole genome molecular haplotyping of single cells. Nature Biotechnol. 2011;29:51–57. doi: 10.1038/nbt.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tewhey R, et al. Microdroplet-based PCR enrichment for large-scale targeted sequencing. Nature Biotechnol. 2009;27:1025–1031. doi: 10.1038/nbt.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frimat, et al. Plasma stenciling methods for cell patterning. Anal Bioanal Chem. 2009;395:601–609. doi: 10.1007/s00216-009-2824-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Synthesis of 3′-O-methyl-5(6)-carboxyfluorescein-monophosphate
Supplementary Figure 2: Synthesis of dG4P-δ-3′-O-methyl-5(6)-carboxyfluorescein
Supplementary Figure 3: Vacuum-based sealing of PDMS microreactor array
Supplementary Figure 4: Sample sequencing data processing
Supplementary Figure 5: Demonstration of fluorogenic pyrosequencing in high density microreactor arrays.
Supplementary Figure 6: Super-Poissonian loading of PDMS microreactors
Supplementary Table 1: DNA Sequences used in fluorogenic pyrosequencing experiments
Supplementary Note 1: Synthesis of terminal phosphate-labeled fluorogenic nucleotides
Supplementary Note 2: The scalability of fluorogenic pyrosequencing
Supplementary Note 3: Potential for microfluidic integration of fluorogenic pyrosequencing