Abstract
The three-dimensional organization of the enormously long DNA molecules packaged within metaphase chromosomes has been one of the most elusive problems in structural biology. Chromosomal DNA is associated with histones and different structural models consider that the resulting long chromatin fibers are folded forming loops or more irregular three-dimensional networks. Here, we report that fragments of chromatin fibers obtained from human metaphase chromosomes digested with micrococcal nuclease associate spontaneously forming multilaminar platelike structures. These self-assembled structures are identical to the thin plates found previously in partially denatured chromosomes. Under metaphase ionic conditions, the fragments that are initially folded forming the typical 30-nm chromatin fibers are untwisted and incorporated into growing plates. Large plates can be self-assembled from very short chromatin fragments, indicating that metaphase chromatin has a high tendency to generate plates even when there are many discontinuities in the DNA chain. Self-assembly at 37°C favors the formation of thick plates having many layers. All these results demonstrate conclusively that metaphase chromatin has the intrinsic capacity to self-organize as a multilayered planar structure. A chromosome structure consistent of many stacked layers of planar chromatin avoids random entanglement of DNA, and gives compactness and a high physical consistency to chromatids.
Introduction
Each chromosome contains a single DNA molecule (1) that is complexed with histones, forming a chromatin filament composed of many nucleosomes (2). It is known that during mitosis DNA is densely packed (3), but the organization of chromatin in metaphase chromosomes has been a long-standing problem (4,5). From early electron micrographs of histone-depleted chromosomes (6), it was suggested that chromatin is coiled into loops attached to a protein scaffold. The models based on loops bound to a helical scaffold (7–9) were widely accepted, but more recently, stretching experiments demonstrated that chromosomes do not have a mechanically contiguous protein scaffold (10), suggesting that chromatin associated with nonhistone proteins forms an irregular network. Immunofluorescence and cryo-electron microscopy studies (11,12) also suggested that chromatin is irregularly folded in mitotic chromosomes.
Unexpectedly, it was observed that incubation of metaphase chromosomes at 37°C produced the emanation of many multilayered plates clearly visible in the electron microscope (13). Atomic force microscopy in aqueous media, polarizing microscopy, electron tomography, and cryo-electron microscopy results indicated that nucleosomes in the plates are irregularly oriented, allowing the interdigitation of layers that have an apparent thickness of ∼6 nm (14,15). Friction force measurements at the nanoscale (nanotribology) showed that DNA is the main component responsible for the mechanical strength of the flexible two-dimensional chromatin network that forms the layers (16). All these findings led to the proposal of the thin-plate model, in which it is considered that chromosomes are built by many stacked chromatin layers orthogonal to the chromatid axis (14,15,17).
Loop-scaffold (6–9) and irregular folding models (10–12) have a straightforward relationship with the linear geometry of DNA. Essentially, in these models it is considered that a linear chromatin fiber is folded forming a three-dimensional network that fills the chromatid. In contrast, in the thin-plate model (14,15,17), the linear chromatin filament is transformed into a two-dimensional network; the stacking of many layers of this planar structure occupies the entire volume of the chromatid. This model is based on the structural and mechanical properties of the multilayered plates emanated from partially denatured chromosomes under metaphase ionic conditions (14–16). Furthermore, it is in agreement with the experimental observation that chromatin fibers are only visible in metaphase chromosomes treated with buffers without cations (13). Nevertheless, whereas different in vitro experiments have demonstrated that in the presence of intermediate cation concentrations the chromatin filament is folded forming fibers of ∼30 nm in diameter having different compaction degrees (17–25), there is no direct evidence of the transformation of the chromatin filament into planar structures. Here we show that, under metaphase ionic conditions, chromatin fragments obtained from human metaphase chromosomes associate spontaneously, forming large multilayered plates. The results obtained are discussed in the light of present knowledge of chromatin structure and supramolecular self-assembly of micrometer-scale materials.
Materials and Methods
HeLa cells cultured in Nunc (Roskilde, Denmark) TripleFlasks (3–6 for each experiment) were blocked in metaphase with colcemid and chromosomes were isolated from mitotic cells (at a concentration of 8 × 106 cells/mL) and purified on sucrose step gradients containing 5 mM PIPES (pH 7.2), 5 mM NaCl, and 5 mM MgCl2 as previously described in Caravaca et al. (13). To obtain chromatin fragments of different lengths, metaphase chromosomes were digested in the same buffer (complemented with 1 mM CaCl2) with micrococcal nuclease (Sigma, St Louis, MO; 3 × 10−3 units per mL of chromosome suspension) for 5–15 min at 37°C. Digested chromosomes were dialyzed overnight at 4°C against 10 mM PIPES (pH 7.2) and 10 mM EDTA, and centrifuged at 2500 × g for 5 min. In the typical experiments (Figs. 1 and 2, and see Fig. S1 and Fig. S2 in the Supporting Material), the supernatant containing the chromatin fragments was directly dialyzed for 5–6 h at 4°C against the self-assembly solution (10 mM PIPES, pH 7.2, 120 mM KCl, 20 mM NaCl, and 17 mM MgCl2), and finally spread on carbon-coated electron microscope grids.
Figure 1.
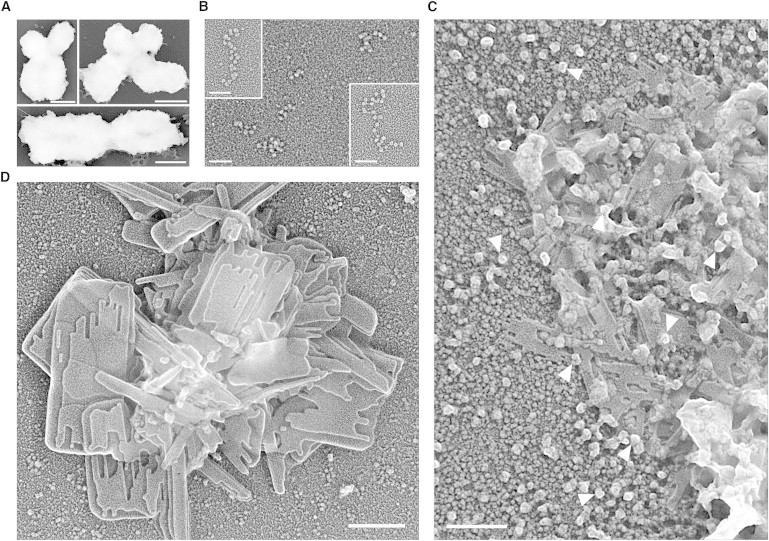
Self-assembly of metaphase chromatin fragments into multilayered plates. (A) Transmission electron microscopy (TEM) images of native metaphase chromosomes. (B) TEM images of chromatin fragments from nuclease-digested chromosomes; the sample was diluted 10- to 20-fold with a low ionic strength buffer (1 mM PIPES, pH 7.2, and 1 mM EDTA) before spreading. (C and D) Examples of TEM images corresponding to planar structures formed by association of metaphase chromatin fragments dialyzed at 4°C against the self-assembly solution (10 mM PIPES, pH 7.2, 120 mM KCl, 20 mM NaCl, and 17 mM MgCl2) before spreading. (Head arrows in C) 30-nm circular structures corresponding to chromatin fragments folded as short interdigitated solenoids (26,28,29) oriented with their axes perpendicular to carbon and plate surfaces. Scale bars: 500 nm (A); 100 nm (B); 200 nm (C and D).
Figure 2.
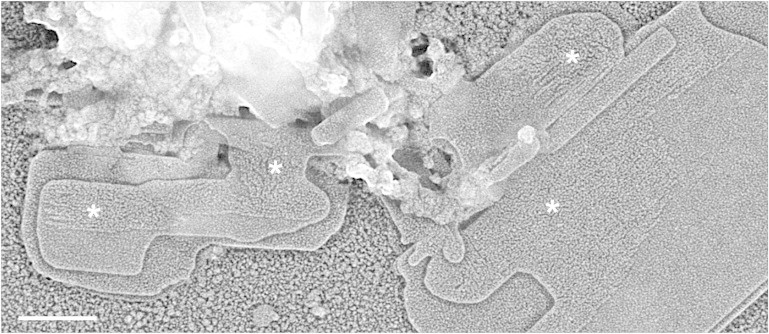
Self-assembled chromatin plates having partially unfolded layers. In some experiments metaphase chromatin fragments extensively dialyzed at 4°C against the self-assembly solution produced plates having regions with a granular texture in the upper layers (indicated by asterisks); the dimensions of the granules suggest that they correspond to unorganized nucleosomes in chromatin filaments not yet incorporated into well-defined layers. Scale bar: 200 nm.
In some experiments (see Fig. 4 and Fig. S4), the chromatin fragments were first spread on a carbon-coated grid by centrifugation, then the grid and 95 μL of the supernatant were placed on a dialysis cup (Slide-A-Lyzer; Thermo Scientific, Waltham, MA) and dialyzed against the self-assembly solution for different times (1–24 h) and temperatures (4, 25, and 37°C). In other experiments, chromatin fragments were purified by nondenaturing electrophoresis (26) before the dialysis against the self-assembly solution (Fig. 3 and see Fig. S3). In this case, chromatin fragments were electrophoresed on 0.5% agarose gels in 90 mM Tris-borate buffer; excised bands in 1.5 mL of Tris-borate were dialyzed overnight against this buffer, and finally placed on an Ultrafree-CL filter (Millipore, Billerica, MA) and centrifuged at 2500 × g for 30 min; the eluted chromatin fragments were concentrated (using Amicon Ultra-4 filters; Millipore) and dialyzed first for 1 h at 4°C against 10 mM PIPES (pH 7.2) and 10 mM EDTA, and then for 6 h at 4°C against 10 mM PIPES (pH 7.2), 120 mM KCl, 20 mM NaCl, and 17 mM MgCl2; part of the sample was used to analyze DNA lengths on agarose gels.
Figure 4.
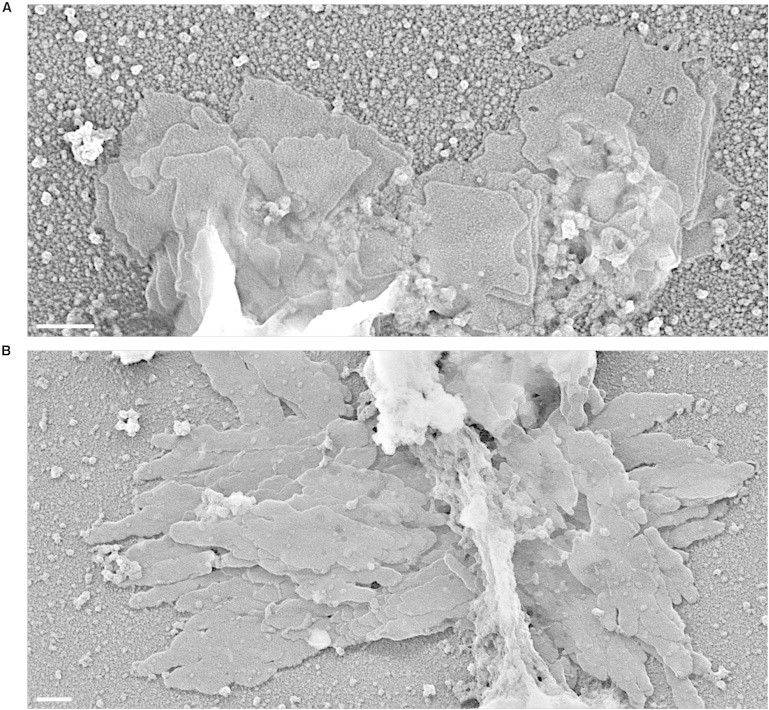
Self-assembly of chromatin plates in experiments performed at different temperatures. Metaphase chromatin fragments were spread on carbon-coated grids by centrifugation, then the grids immersed in a solution of chromatin fragments were dialyzed for different times against the self-assembly solution at different temperatures. TEM images corresponding to samples incubated at 25°C for 24 h (A) and at 37°C for 7 h (B). Scale bars: 200 nm.
Figure 3.
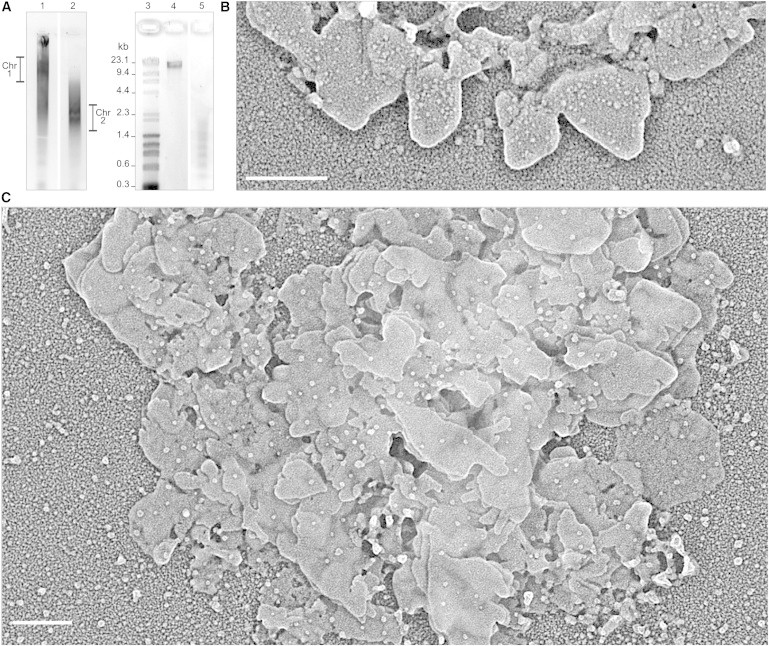
Self-assembly of plates from metaphase chromatin fragments having different length. (A) Chromatin fragments were purified by nondenaturing electrophoresis on agarose gels; fragments extracted from excised bands Chr 1 (lane 1) and Chr 2 (lane 2) were concentrated, dialyzed for 6 h at 4°C against the self-assembly solution, and finally analyzed in the electron microscope (see B and C); part of the sample was used to measure DNA lengths on agarose gels (lanes 4 and 5 correspond to the DNA of the chromatin bands Chr 1 and Chr 2, respectively; the size of some DNA length markers is indicated in lane 3). (B) Plates formed by self-assembly from chromatin fragments (extracted from band Chr 1) having a DNA length (∼18 kb) corresponding to ∼95 nucleosomes. (C) Plates obtained with chromatin fragments (extracted from band Chr 2) having a DNA length (∼0.6–1.3 kb) corresponding to 3–7 nucleosomes. Scale bars: 200 nm.
For spreading, the sample (90 μL) above the carbon-coated grid (placed in the cap of an inverted tube) was centrifuged for 10 min at 1500 × g on a swinging-bucket rotor; extensions were fixed with glutaraldehyde and shadowed with platinum as described in Gállego et al. (14) and Castro-Hartmann et al. (15). Images were obtained using a Jeol (Tokyo, Japan) JEM-1400 microscope. Micrographs are presented in reverse contrast to facilitate the visualization of plate edges.
Results
In vitro self-assembly of chromatin plates
We have obtained chromatin fragments (Fig. 1 B) from HeLa metaphase chromosomes (Fig. 1 A) digested with micrococcal nuclease, and we have analyzed in the electron microscope the structures formed by these fragments in ionic conditions approaching to those observed in metaphase. Considering the analytical results obtained by scanning ion microscopy (27) indicating that metaphase chromosomes contain physiological concentrations of K+ and Na+ and 12–22 mM Mg2+ distributed homogeneously in the chromatids, our experiments were performed in a buffer containing 120 mM K+, 20 mM Na+, and 17 mM Mg2+. Dialysis of chromatin fragments against this buffer resulted in the formation of amorphous aggregates and planar structures of different sizes. The electron micrographs presented in Figs. 1 D and 2 and in Fig. S1, C–F, and Fig. S2 show examples of the large multilayered plates produced by the association of chromatin fragments under these ionic conditions.
The structural properties (i.e., flat and smooth surface; well-defined edges; stacking of several layers) of the self-assembled plates are identical to those observed in the plates emanated directly from metaphase chromosomes (13–15). Furthermore, the height of the monolayer plates obtained by self-assembly (6.8 ± 1.0 nm, n = 15) determined in unidirectional shadowing experiments (see Fig. S4 H) is equal to the height of the plates emanated from chromosomes (∼6 nm) determined using different techniques (17).
Chromatin fibers are unfolded and incorporated into growing plates
Moreover, in the micrographs of self-assembled plates, we observed circular structures of ∼30 nm in diameter (some of them are indicated by head arrows in Fig. 1 C and Fig. S1, A and B). According to previous studies performed with chicken erythrocyte chromatin (26,28), these structures correspond to short fragments of the 30-nm fiber folded as compact interdigitated solenoids (29). The fragments have only few solenoidal turns and are short enough to be oriented perpendicular to the carbon film surface (i.e., the observed circular images correspond to the top view of short fibers). More recently, the same compact 30-nm chromatin fibers were found in studies performed with reconstituted nucleosome arrays (30), and atomic-scale modeling allowed the study of the structural flexibility and dynamic properties of these interdigitated solenoid structures (31,32).
We observed these compact chromatin fiber fragments surrounding the plates but also closely associated with them. The short fibers in contact with the plates were often structurally altered; they are seen partially unfolded in some micrographs (see for instance Fig. 1 C), suggesting that they were in the process of being associated with the growing plates when the sample was spread on the carbon-coated grid. In agreement with this association mechanism, involving fiber unfolding during their incorporation into plates, it was found that for the oligomerization of nucleosome arrays (33) and for the lateral association of chicken erythrocyte chromatin fragments (14) it was necessary the unfolding of the fibers formed by the reconstituted or native chromatin filaments used in these previous studies. Finally, also in agreement with the requirement of fiber unfolding for plate formation, in the upper layer of some plates, we observed regions having a grainy texture (see examples indicated by asterisks in Fig. 2 and Fig. S2); the size of the granules suggests that these regions correspond to nucleosomes of fully unfolded filaments not completely organized as well-defined plates at the time of spreading.
All these results demonstrate that fragments of chromatin filaments from metaphase chromosomes can associate forming plates, but they can also suffer an intramolecular folding that gives rise to short compact 30-nm fibers. These fibers were observed in previous studies performed using a relatively low chromatin concentration in the presence of intermediate concentrations of Mg2+ (1–2 mM; see (17,26,28–30)). However, the high chromatin concentration and metaphase ionic conditions (27) used in the experiments presented here favor the association reaction. Even the fragments that are initially folded as compact fibers can be untwisted and incorporated into growing planar structures that produce the huge multilayered plates observed in the self-assembly experiments presented in this work. Our results also suggest that the apparently amorphous chromatin aggregates that are often seen associated with planar aggregates could eventually be transformed into multilayered plates.
Large plates can be self-assembled from very short chromatin fragments
In the self-assembly experiments, generally we have used chromatin fragments of different length extracted from chromosomes after the digestion with micrococcal nuclease, but in some experiments we have used chromatin fragments of more defined length obtained by preparative nondenaturing electrophoresis on agarose gels. As can be seen in Fig. 3 and Fig. S3, we obtained large plates independently on the length of the fragments used in different self-assembly experiments. The observed plates are formed by the typical thin layers and there is no correlation between fragment length and the total plate thickness. Note, in particular, that in the experiment with very short fragments containing 3–7 nucleosomes, we also observed well-defined plates (Fig. 3 C). These results indicate that chromatin at metaphase ionic conditions has a high tendency to adopt a planar organization even when there are many discontinuities in the DNA chain.
Self-assembly of thick plates with many layers
In the experiments described above, solutions containing the chromatin fragments were directly dialyzed at 4°C against the self-assembly solution, and then the resulting structures were spread on carbon-coated grids. In other experiments, chromatin fragments were first spread on a carbon-coated grid, and then (in the presence of a solution of chromatin fragments) were dialyzed against the self-assembly solution for different times and temperatures (see Fig. 4 and Fig. S4). The results obtained showed that the initial adsorption of chromatin to the carbon film does not increase the yield of plate formation, indicating that the generation of plates does not require a planar substrate. Plates observed in experiments performed at 25°C (Fig. 4 A and Fig. S4, A–C) are indistinguishable from those obtained at 4°C, but plates formed at 37°C are often very thick (see examples in Fig. 4 B and Fig. S4 F). We measured the height of these plates using unidirectional shadowing (Fig. S4 G); the values found for 14 plates range from 15 to 85 nm. Taking into account that the images obtained show that these thick plates are also formed by thin layers, these results indicate that the self-assembled plates can contain many stacked layers.
Discussion
The results obtained in the self-assembly experiments demonstrate conclusively that chromatin filaments obtained from metaphase chromosomes have intrinsic structural and cohesive properties for the formation of multilayered planar structures, which are identical to those emanated directly from condensed chromosomes (see schematic summary in Fig. 5). The spontaneous formation of planar metaphase chromatin opens completely new possibilities for the experimental and theoretical study of the structure and function of chromosomes. Our results favor the thin-plate model for chromatin folding in metaphase chromosomes. According to genomic results showing that there is a direct correlation between the DNA sequence of each human chromosome and its cytogenetic map (1,34), in this model it is considered that plates are oriented perpendicularly to the chromatid axis and that the successive chromatin layers fill the three-dimensional space of the chromatid progressively from one telomere to the other (14).
Figure 5.
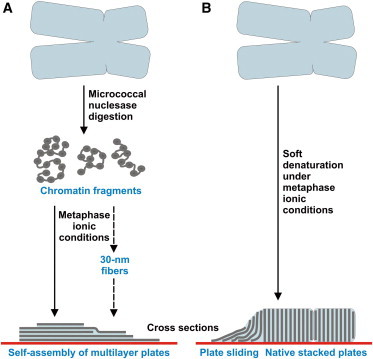
Summary and relationship to previous studies. (A) Chromatin fragments obtained by nuclease digestion of chromosomes associate spontaneously forming multilayer plates (represented as parallel shaded lines in the cross section; the red line corresponds to the carbon film substrate used for TEM imaging); part of the chromatin fragments form short 30-nm fibers that can be unfolded and incorporated into the growing plates (this association reaction is indicated with a discontinuous line). (B) The multilayered planar structures that were found in previous studies (13–17) sliding from partially denatured chromosomes were clearly visible in the electron micrographs (schematized in the left part of the chromatid cross section) and have the same structural characteristics as the laminar aggregates produced by self-assembly. The results obtained in this study demonstrating the spontaneous formation of planar metaphase chromatin strengthens the hypothesis that condensed chromosomes are formed by many stacked chromatin layers (represented in the right part of the chromatid cross section). The different structural elements schematized in this figure are not drawn at the same scale.
Considering that in the self-assembly experiments we have frequently observed huge plates (see Figs. 1 D, 2, and 4, and Fig. S1, C and F, Fig. S2, and Fig. S4, C and F) and that the plates emanating from partially denatured chromosomes are split but they are often very large (15), it can be suggested that planar chromatin may form an extensive structure in native chromosomes. As discussed elsewhere (15,17), taking into account early studies indicating that chromatids are helically coiled (7–9,35), it is possible that each chromatid is formed by a single helicoidal plate. The assembly in the cell of such a large structure made of a thin sheet of chromatin seems, in principle, unlikely. Nevertheless, nanotechnology research has demonstrated, using different macromolecules and larger components, that self-assembly processes are able to build, spontaneously, extremely complex meso- and macroscale materials (36–38), including impressive helical structures (39–42).
Also in favor of a continuous helicoidal chromatin structure within chromatids, it has to be taken into account that the flat plates seen in our micrographs and a hypothetical helicoidal plate are topologically equivalent and can be converted into each other without changing their mean curvature; the plane and the helicoid are both minimal surfaces (i.e., their mean curvature is zero; see Hyde et al. (43)). In addition, it is known that the formation of helical structures in the self-assembly reactions is favored by the chiral geometry of the building blocks (39,42) and, in the case of chromatin, both DNA and nucleosomes are chiral (44). Finally, note that the compactness of a helicoidal chromatin sheet with closely stacked turns is equivalent to that of the self-assembled plates formed by layers in close contact to each other. The resulting compaction degree is higher than that obtained with the typical chromatin fibers considered in different chromosome models (3), and is compatible with the constraint imposed by the high DNA density found experimentally in metaphase chromosomes of different species (45).
Typical chromatids are cylinders having a diameter of ∼500 nm and a length of several micrometers. Self-assembly studies have shown that hierarchical association starting from building blocks having nanometer-scale dimensions can produce micrometric structures (41,42). Nucleosomes are nanometer-scale particles (∼11 nm diameter), but in contrast to other self-assembly systems, these chromatin building blocks are joined by a molecular bridge (the linker DNA connecting successive nucleosome core particles in the chromatin filament). This network organization may be useful for nanotechnology research because it could inspire the design of self-assembling materials containing long polymers of technological interest. In particular, note that in native chromosomes all the building blocks are connected by a single DNA molecule that has a nanometric diameter (∼2 nm) but a macroscopic length (several centimeters in the case of human chromosomes). A continuous helicoidal plate may allow the packaging of this giant molecule according to a homogeneous geometry along most of the chromosome length. The anomalous conformation of nucleosomes in the centromere (46,47) could produce an alteration in the structure of planar chromatin that could be related with the constriction observed in this region.
From a nanotechnological point of view, the self-assembly of planar chromatin is equivalent to the transformation of threads having a nanometric diameter into nonwoven fabriclike sheets with a nanometric thickness and a large surface area; such a transformation precludes the random entanglement of threads and improves the mechanical properties of the whole structure. In planar chromatin, these basic properties of fabriclike materials have been observed experimentally. First, the absence of entanglement is consistent with the observation that stacked chromatin layers in plates can easily slide relative to each other (14,15). Second, whereas the stretching force required for DNA breakage is ∼1 nN (48), nanotribology studies (16) showed that larger forces (∼5 nN) can be applied during the friction measurements without producing any damage to the plates. These remarkable structural and nanomechanical properties of planar chromatin facilitate enormously the complex topological problem of DNA packaging and give a much higher physical consistency to the chromosome than the different chromatin organizations suggested in fibrillar models. Furthermore, after decondensation of mitotic chromatin, the remains of this planar structure could facilitate the unentanglement between the DNA chains of different chromosomes that is observed during interphase (49).
Nucleosomes in the plates do not form regular columns; they are irregularly oriented allowing the interdigitation of adjacent layers (14,15). This justifies that the apparent height of each layer is lower than the nucleosome diameter and allows face-to-face and lateral side-by-side interactions between nucleosomes of successive layers. The same interactions were observed in dense phases (arcs and helices; columnar hexagonal and bilayers) formed by isolated nucleosome core particles (50–52). However, whereas in these aggregates of core particles the strongest interaction is produced between nucleosome faces (53), in plates this interaction is weaker than the forces holding the nucleosomes within each layer. The presence of linker DNA in planar chromatin may be responsible for these differences. The relatively high concentration of Mg2+ in metaphase may also strengthen this interaction. Furthermore, as it is known that mitotic histones H1 and H3 are phosphorylated (54) and that the structure of nucleosomes is dynamic (55), the bulk of mitotic nucleosomes could be structurally altered favoring the observed affinity differences.
Since plates self-assembled from chromatin fragments are indistinguishable from native plates emanated directly from metaphase chromosomes containing undigested DNA, it is very likely that the architecture of planar chromatin is capable of placing the discontinuous DNA in the plates following the same pattern as the long DNA molecules of native chromosomes. Moreover, the self-assembly of well-defined plates containing very small chromatin fragments observed in this study (Fig. 3 C) suggests that, in the cell, planar metaphase chromatin can maintain the structural integrity of chromosomes even if many double-strand breaks are produced during mitosis.
Acknowledgments
The authors thank Pere Puigdomènech, Benjamí Piña, and David B. Amabilino for useful comments, and the staffs of the Serveis de Microscòpia and Cultius Cel·lulars (UAB) for their assistance.
This work was supported in part by research grants BFU2008-04514-E and BFU2010-18939 from the Ministerio de Ciencia e Innovación, and a predoctoral fellowship to M.M. by the Ministerio de Ciencia e Innovación.
Footnotes
This is an Open Access article distributed under the terms of the Creative Commons-Attribution Noncommercial License (http://creativecommons.org/licenses/by-nc/2.0/), which permits unrestricted noncommercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Supporting Material
References
- 1.Lander E.S., Linton L.M., Chen Y.J., International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Luger K., Mäder A.W., Richmond T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 3.Daban J.R. High concentration of DNA in condensed chromatin. Biochem. Cell Biol. 2003;81:91–99. doi: 10.1139/o03-037. [DOI] [PubMed] [Google Scholar]
- 4.Belmont A.S. Mitotic chromosome structure and condensation. Curr. Opin. Cell Biol. 2006;18:632–638. doi: 10.1016/j.ceb.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 5.Maeshima K., Hihara S., Eltsov M. Chromatin structure: does the 30-nm fiber exist in vivo? Curr. Opin. Cell Biol. 2010;22:291–297. doi: 10.1016/j.ceb.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 6.Paulson J.R., Laemmli U.K. The structure of histone-depleted metaphase chromosomes. Cell. 1977;12:817–828. doi: 10.1016/0092-8674(77)90280-x. [DOI] [PubMed] [Google Scholar]
- 7.Rattner J.B., Lin C.C. Radial loops and helical coils coexist in metaphase chromosomes. Cell. 1985;42:291–296. doi: 10.1016/s0092-8674(85)80124-0. [DOI] [PubMed] [Google Scholar]
- 8.Boy de la Tour E., Laemmli U.K. The metaphase scaffold is helically folded: sister chromatids have predominantly opposite helical handedness. Cell. 1988;55:937–944. doi: 10.1016/0092-8674(88)90239-5. [DOI] [PubMed] [Google Scholar]
- 9.Saitoh Y., Laemmli U.K. Metaphase chromosome structure: bands arise from a differential folding path of the highly AT-rich scaffold. Cell. 1994;76:609–622. doi: 10.1016/0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- 10.Poirier M.G., Marko J.F. Mitotic chromosomes are chromatin networks without a mechanically contiguous protein scaffold. Proc. Natl. Acad. Sci. USA. 2002;99:15393–15397. doi: 10.1073/pnas.232442599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kireeva N., Lakonishok M., Belmont A.S. Visualization of early chromosome condensation: a hierarchical folding, axial glue model of chromosome structure. J. Cell Biol. 2004;166:775–785. doi: 10.1083/jcb.200406049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eltsov M., Maclellan K.M., Dubochet J. Analysis of cryo-electron microscopy images does not support the existence of 30-nm chromatin fibers in mitotic chromosomes in situ. Proc. Natl. Acad. Sci. USA. 2008;105:19732–19737. doi: 10.1073/pnas.0810057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caravaca J.M., Caño S., Daban J.R. Structural elements of bulk chromatin within metaphase chromosomes. Chromosome Res. 2005;13:725–743. doi: 10.1007/s10577-005-1008-3. [DOI] [PubMed] [Google Scholar]
- 14.Gállego I., Castro-Hartmann P., Daban J.R. Dense chromatin plates in metaphase chromosomes. Eur. Biophys. J. 2009;38:503–522. doi: 10.1007/s00249-008-0401-1. [DOI] [PubMed] [Google Scholar]
- 15.Castro-Hartmann P., Milla M., Daban J.R. Irregular orientation of nucleosomes in the well-defined chromatin plates of metaphase chromosomes. Biochemistry. 2010;49:4043–4050. doi: 10.1021/bi100125f. [DOI] [PubMed] [Google Scholar]
- 16.Gállego I., Oncins G., Daban J.R. Nanotribology results show that DNA forms a mechanically resistant 2D network in metaphase chromatin plates. Biophys. J. 2010;99:3951–3958. doi: 10.1016/j.bpj.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daban J.R. Electron microscopy and atomic force microscopy studies of chromatin and metaphase chromosome structure. Micron. 2011;42:733–750. doi: 10.1016/j.micron.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 18.Hansen J.C. Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annu. Rev. Biophys. Biomol. Struct. 2002;31:361–392. doi: 10.1146/annurev.biophys.31.101101.140858. [DOI] [PubMed] [Google Scholar]
- 19.Robinson P.J.J., Rhodes D. Structure of the ‘30 nm’ chromatin fiber: a key role for the linker histone. Curr. Opin. Struct. Biol. 2006;16:336–343. doi: 10.1016/j.sbi.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 20.Tremethick D.J. Higher-order structures of chromatin: the elusive 30 nm fiber. Cell. 2007;128:651–654. doi: 10.1016/j.cell.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 21.van Holde K.E., Zlatanova J. Chromatin fiber structure: where is the problem now? Semin. Cell Dev. Biol. 2007;18:651–658. doi: 10.1016/j.semcdb.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 22.Mozziconacci J., Lavelle C. Chromatin fiber: 30 years of models. In: Russe A.S., editor. Computational Biology: New Research. Nova Science Publishers; Hauppauge, NY: 2009. pp. 147–163. [Google Scholar]
- 23.Schlick T., Hayes J., Grigoryev S. Toward convergence of experimental studies and theoretical modeling of the chromatin fiber. J. Biol. Chem. 2012;287:5183–5191. doi: 10.1074/jbc.R111.305763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grigoyev S.A., Woodcock C.L. Chromatin organization: The 30 nm fiber. Exp. Cell Res. 2012 doi: 10.1016/j.yexcr.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 25.Rippe K. The folding of the nucleosome chain. In: Rippe K., editor. Genome Organization and Function in the Cell Nucleus. Wiley-VCH; Weinheim, Germany: 2012. pp. 139–167. [Google Scholar]
- 26.Bartolomé S., Bermúdez A., Daban J.R. Electrophoresis of chromatin on nondenaturing agarose gels containing Mg2+. Self-assembly of small chromatin fragments and folding of the 30-nm fiber. J. Biol. Chem. 1995;270:22514–22521. doi: 10.1074/jbc.270.38.22514. [DOI] [PubMed] [Google Scholar]
- 27.Strick R., Strissel P.L., Levi-Setti R. Cation-chromatin binding as shown by ion microscopy is essential for the structural integrity of chromosomes. J. Cell Biol. 2001;155:899–910. doi: 10.1083/jcb.200105026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bartolomé S., Bermúdez A., Daban J.R. Internal structure of the 30 nm chromatin fiber. J. Cell Sci. 1994;107:2983–2992. doi: 10.1242/jcs.107.11.2983. [DOI] [PubMed] [Google Scholar]
- 29.Daban J.R., Bermúdez A. Interdigitated solenoid model for compact chromatin fibers. Biochemistry. 1998;37:4299–4304. doi: 10.1021/bi973117h. [DOI] [PubMed] [Google Scholar]
- 30.Robinson P.J.J., Fairall L., Rhodes D. EM measurements define the dimensions of the “30-nm” chromatin fiber: evidence for a compact, interdigitated structure. Proc. Natl. Acad. Sci. USA. 2006;103:6506–6511. doi: 10.1073/pnas.0601212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong H., Victor J.M., Mozziconacci J. An all-atom model of the chromatin fiber containing linker histones reveals a versatile structure tuned by the nucleosomal repeat length. PLoS ONE. 2007;2:e877. doi: 10.1371/journal.pone.0000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kepper N., Foethke D., Rippe K. Nucleosome geometry and internucleosomal interactions control the chromatin fiber conformation. Biophys. J. 2008;95:3692–3705. doi: 10.1529/biophysj.107.121079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou J., Fan J.Y., Tremethick D.J. The nucleosome surface regulates chromatin compaction and couples it with transcriptional repression. Nat. Struct. Mol. Biol. 2007;14:1070–1076. doi: 10.1038/nsmb1323. [DOI] [PubMed] [Google Scholar]
- 34.Cheung V.G., Nowak N., Trask B.J., BAC Resource Consortium Integration of cytogenetic landmarks into the draft sequence of the human genome. Nature. 2001;409:953–958. doi: 10.1038/35057192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohnuki Y. Demonstration of the spiral structure of human chromosomes. Nature. 1965;208:916–917. doi: 10.1038/208916a0. [DOI] [PubMed] [Google Scholar]
- 36.Whitesides G.M., Boncheva M. Beyond molecules: self-assembly of mesoscopic and macroscopic components. Proc. Natl. Acad. Sci. USA. 2002;99:4769–4774. doi: 10.1073/pnas.082065899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whitesides G.M., Grzybowski B. Self-assembly at all scales. Science. 2002;295:2418–2421. doi: 10.1126/science.1070821. [DOI] [PubMed] [Google Scholar]
- 38.Antonietti M., Ozin G.A. Promises and problems of mesoscale materials chemistry or why meso? Chemistry. 2004;10:28–41. doi: 10.1002/chem.200305009. [DOI] [PubMed] [Google Scholar]
- 39.Sierra T. Expression of chilarity in polymers. In: Amabilino D.B., editor. Chilarity at the Nanoscale. Wiley-VCH; Weinheim, Germany: 2009. pp. 115–189. [Google Scholar]
- 40.Nguyen T.D., Glotzer S.C. Switchable helical structures formed by the hierarchical self-assembly of laterally tethered nanorods. Small. 2009;5:2092–2098. doi: 10.1002/smll.200900168. [DOI] [PubMed] [Google Scholar]
- 41.Chung W.J., Oh J.W., Lee S.W. Biomimetic self-templating supramolecular structures. Nature. 2011;478:364–368. doi: 10.1038/nature10513. [DOI] [PubMed] [Google Scholar]
- 42.Gibaud T., Barry E., Dogic Z. Reconfigurable self-assembly through chiral control of interfacial tension. Nature. 2012;481:348–351. doi: 10.1038/nature10769. [DOI] [PubMed] [Google Scholar]
- 43.Hyde S., Andersson S., Ninham B.W. The Role of Curvature in Condensed Matter: Physics, Chemistry and Biology. Elsevier; Amsterdam, The Netherlands: 1997. The language of shape; pp. 1–42. [Google Scholar]
- 44.Leforestier A., Bertin A., Livolant F. Expression of chilarity in columnar hexagonal phases of DNA and nucleosomes. C. R. Chim. 2008;11:229–244. [Google Scholar]
- 45.Daban J.R. Physical constraints in the condensation of eukaryotic chromosomes. Local concentration of DNA versus linear packing ratio in higher order chromatin structures. Biochemistry. 2000;39:3861–3866. doi: 10.1021/bi992628w. [DOI] [PubMed] [Google Scholar]
- 46.Furuyama T., Henikoff S. Centromeric nucleosomes induce positive DNA supercoils. Cell. 2009;138:104–113. doi: 10.1016/j.cell.2009.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sekulic N., Bassett E.A., Black B.E. The structure of (CENP-A-H4)2 reveals physical features that mark centromeres. Nature. 2010;467:347–351. doi: 10.1038/nature09323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bustamante C., Smith S.B., Smith D. Single-molecule studies of DNA mechanics. Curr. Opin. Struct. Biol. 2000;10:279–285. doi: 10.1016/s0959-440x(00)00085-3. [DOI] [PubMed] [Google Scholar]
- 49.Rosa A., Everaers R. Structure and dynamics of interphase chromosomes. PLoS Comput. Biol. 2008;4:e1000153. doi: 10.1371/journal.pcbi.1000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dubochet J., Noll M. Nucleosome arcs and helices. Science. 1978;202:280–286. doi: 10.1126/science.694532. [DOI] [PubMed] [Google Scholar]
- 51.Mangenot S., Leforestier A., Livolant F. Phase diagram of nucleosome core particles. J. Mol. Biol. 2003;333:907–916. doi: 10.1016/j.jmb.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 52.Cherstvy A.G., Everaers R. Layering, bundling, and azimuthal orientations in dense phases of nucleosome core particles. J. Phys. Condens. Matter. 2006;18:11429–11442. [Google Scholar]
- 53.Leforestier A., Fudaley S., Livolant F. Spermidine-induced aggregation of nucleosome core particles: evidence for multiple liquid crystalline phases. J. Mol. Biol. 1999;290:481–494. doi: 10.1006/jmbi.1999.2895. [DOI] [PubMed] [Google Scholar]
- 54.Ausió J., Abbott D.W., Moore S.C. Histone variants and histone modifications: a structural perspective. Biochem. Cell Biol. 2001;79:693–708. [PubMed] [Google Scholar]
- 55.Zlatanova J., Bishop T.C., van Holde K. The nucleosome family: dynamic and growing. Structure. 2009;17:160–171. doi: 10.1016/j.str.2008.12.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


