Abstract
To explore the initial stages of amyloid β peptide (Aβ42) deposition on membranes, we have studied the interaction of Aβ42 in the monomeric form with lipid monolayers and with bilayers in either the liquid-disordered or the liquid-ordered (Lo) state, containing negatively charged phospholipids. Molecular dynamics (MD) simulations of the system have been performed, as well as experimental measurements. For bilayers in the Lo state, in the absence of the negatively charged lipids, interaction is weak and it cannot be detected by isothermal calorimetry. However, in the presence of phosphatidic acid, or of cardiolipin, interaction is detected by different methods and in all cases interaction is strongest with lower (2.5–5 mol %) than higher (10–20 mol %) proportions of negatively charged phospholipids. Liquid-disordered bilayers consistently allowed a higher Aβ42 binding than Lo ones. Thioflavin T assays and infrared spectroscopy confirmed a higher proportion of β-sheet formation under conditions when higher peptide binding was measured. The experimental results were supported by MD simulations. We used 100 ns MD to examine interactions between Aβ42 and three different 512 lipid bilayers consisting of palmitoylsphingomyelin, dimyristoyl phosphatidic acid, and cholesterol in three different proportions. MD pictures are different for the low- and high-charge bilayers, in the former case the peptide is bound through many contact points to the bilayer, whereas for the bilayer containing 20 mol % anionic phospholipid only a small fragment of the peptide appears to be bound. The MD results indicate that the binding and fibril formation on the membrane surface depends on the composition of the bilayer, and is the result of a subtle balance of many inter- and intramolecular interactions between the Aβ42 and membrane.
Introduction
Alzheimer's disease (AD) is a late-onset neurological disorder with progressive loss of memory and cognitive abilities as a result of excessive neurodegeneration (1). AD is characterized by extracellular aggregates of β-amyloid (Aβ) peptides known as amyloid plaques (2). The Aβ peptide is derived from the sequential processing of the amyloid precursor protein (APP) by β- and γ-secretases. Nonamyloidogenic processing of APP involves α-secretase that cleaves APP inside the Aβ region precluding Aβ formation (3). Lateral organization of membranes (4) and subcellular localization (5) of the substrate and the secretases has been documented to regulate Aβ generation. Previous work suggests that β-secretase associates with liquid-ordered (Lo) microdomains in the membrane (6,7) and that integrity of those (raft) domains is required for β-cleavage of APP to occur. Cleavage by α-secretase, in contrast, would occur outside the Lo microdomains (4).
Aβ peptide is released from cells in a soluble form, and progressively undergoes aggregation forming oligomers, multimers, and fibrils, ending with deposition of extracellular plaques (8). Oligomers have been indicated as the most toxic Aβ species (9), appearing likely before plaque deposition at an early stage of AD pathology. However, protofibrillar and fibrillar aggregates were also shown to be toxic (10,11). The structure of the Aβ peptide, along its passage from monomer to small aggregate to fibril, has been a subject of great interest for both experimental (12,13) and computational (14,15) researchers. Some workers have proposed (16) that free radical formation would be an essential factor in amyloidogenic Aβ aggregation. A recent study shows that lysophosphatidylcholine micelles enhance the formation of Aβ42 fibrils (17).
The toxicity of Aβ peptide appears to require conversion of the monomeric form to an aggregated fibrillar species. Although it is clear that the first stages of Aβ aggregation are essential to the progress of AD, it has not yet been determined exactly what factors influence this initial conversion from monomer to oligomer. Recent work (12,13) has demonstrated that cell membranes may play a significant catalytic role in increasing Aβ aggregation rates. Experimental work (12–16) has shown that Aβ, when interacting with lipid vesicles of various compositions, especially anionic lipids, will aggregate at a much faster rate than in solution. Further results have shown that this interaction between Aβ and lipids will induce a structural conversion (12–16) from a disordered peptide into a peptide dominated by β-structure. Various reports (18,19) show that anionic lipids promote fibril elongation. However, a quantitative description of the interaction of essentially monomeric Aβ peptide with phospholipid membranes is not yet available.
Computational methods have been extensively used to describe the conformational behavior of Aβ peptides in aqueous solution. They have provided valuable insight into the structure of Aβ in different steps of aggregation. However, only a few studies (20,21) investigated the Aβ42 structure bound on the bilayer surface, whereas others (22,23) studied the stability and structure of Aβ inserted into membrane. Davies and Berkowitz (20,21) examined Aβ42-phospholipid bilayer interactions using molecular dynamics (MD) simulations. The results showed that Aβ42 is attracted to the lipid surface during simulations regardless of the Aβ42 charge or the membrane charge. In their second study (21), the authors used replica exchange MD and observed that Aβ42 exhibits significant conformational flexibility on the membrane surface.
In this work both biophysical and computational techniques have been used to elucidate the molecular and structural details of the initial stages of Aβ monomer-membrane interaction. In MD simulations, we have investigated the structure of Aβ42 close to the surface of heterogeneous bilayers consisting of palmitoylsphingomyelin (PSM), dimyristoyl phosphatidic acid (DMPA), and cholesterol (Chol) in different proportions. In experiments, we have used large unilamellar vesicles (LUVs) and monolayers composed of lipids giving rise (in bilayers) to Lo phases, because β-secretase cleavage is believed to occur mainly in Lo domains (6,7), and occasionally we have used lipids giving rise to liquid-disordered (Ld) phases, predominant in cell membranes. Negatively charged phospholipids have been added to the lipid compositions to confirm the role of these lipids in bilayer-peptide interaction. Isothermal titration calorimetry (ITC), Langmuir monolayer experiments, thioflavin T (ThT) assays, infrared (IR) spectroscopy, and MD simulations have been performed, with good agreement between the computational and experimental results.
Experimental Procedures
Due to space limitations experimental procedures are described in detail in the Supporting Material.
MD simulations
Initial coordinates for Aβ42 were based on the NMR structure (24) determined in aqueous solutions of fluorinated alcohols. This three-dimensional NMR structure shows two helical regions encompassing residues 8–25 and 28–38, connected by a typical type I β-turn (pdb code 1IYT). The Aβ42 structure has been constructed in the Maestro interface of the Schrödinger program (Schrödinger LLC, New York, NY). The structure was submitted to the protein preparation utility, which included bond order correction, hydrogen addition, and subsequent optimization in the OPLS-AA force field (26). Aβ42 was modeled as a zwitterionic structure, with ionized C-, and N-terminus of the peptide. The force field parameters of Aβ42 were based on GROMOS 96 (27).
Three models of the 512 lipid bilayer consisting of PSM, DMPA, and Chol in three different proportions were used for our simulations (more details in the Supporting Material data).
Experimental studies
Materials
Aβ42 was supplied by Mario Negri Institute (Milan, Italy), a partner in the European Union Project (FP7/2007-2013), 1,1,1,3,3,3-hexafluoro-2-propanol and ThT were purchased from Sigma (St. Louis, MO). Brain sphingomyelin (SM), DMPA, bovine heart cardiolipin (CL), and cholesterol were purchased from Avanti Polar Lipids (Alabaster, AL). Egg phosphatidyl choline (PC) was from Lipid Products (South Nutfield, United Kingdom).
Aβ42 sample preparation
Aβ42 stock solution was prepared by dissolving the peptide at 1 mg/ml in 1,1,1,3,3,3-hexafluoro-2-propanol to render monomeric Aβ42. The volatile solvent was removed under vacuum in a Speed Vac (ThermoSavant, Holbrook, NY). The predominant monomeric form at least in the first 6 h after preparation was checked by ThT fluorescence (28) (Fig. S1 in the Supporting Material). ITC experiments took 5 h on average; other measurements were completed in <1 h.
Preparation of LUV
LUVs were prepared as described (29).
ITC
ITC was performed as described in detail by Arnulphi et al. (30).
Lipid monolayer measurements
Monolayers at the air-water interface in a Langmuir balance were studied at 22°C as indicated (31).
ThT assay
ThT fluorescence was performed as described (28).
IR spectroscopy measurements
See Methods in the Supporting Material.
Results
MD simulations
To characterize in more detail the conformational behavior of Aβ42 at the bilayer surface, an analysis of the Aβ42 secondary structure along the MD simulation trajectory was performed. The profile of secondary changes of Aβ42 and several snapshots extracted from the A–C trajectories are shown in Fig. 1. Fig. S2 illustrates the orientation and structure of the most frequently occurring conformations of Aβ42 during the simulations. The A model (PSM/Chol/DMPA = 50:50:0) provides reference data on a role of negatively charged phospholipids on conformational dynamics of Aβ42. In this model, shown in Fig. 1, during the 50 ns simulation the initial helical conformation of the Aβ42 segments does not undergo significant changes. The 10–20, and to some extent 5–10 regions, remain helical for the time of the simulation and β-sheet structures are not observed. Therefore, we did not extend simulation time to 100 ns. A conserved pattern of secondary structure is shown, indicating a strong intramolecular hydrogen bond network in the protein. In the B model, PSM/Chol/DMPA = 47.5:47.5:5.0, the secondary structure motif changes to mostly unordered coil conformation after 25 ns simulation, but after that Aβ42 is able to adopt conformations containing more ordered secondary structure. Remarkably, after 85 ns the segment of residues 10–18 starts to develop a β-sheet structure, which is stable to the end of the simulation. The simulation snapshots shown in Fig. S2 were clustered based on the root mean-square of Aβ42 backbone residual distances. The results revealed that the single most common conformation occurs in 28.7% of the trajectory. The second most common structure cluster contained only 1.6% of the snapshots. Fig S2 shows that the prevailing cluster of Aβ42 is tightly bound on the membrane surface in the B model. The peptide secondary structure resembles surprisingly the β-hairpin conformation of the Aβ42 fibril. The connection of the Aβ-surface association and the formation of the β-sheet structures that could promote aggregation has been indicated previously (21). On the contrary, in model C with the bilayer composition PSM/Chol/DMPA = 40:40:20, after the initial loss of the helical structure and beginning at 20 ns simulation, the segment of residues 12–20 builds up a 5-helical conformation that is stable for the rest of the 100 ns simulation trajectory. The residues 30–35 adopted an α-helical conformation in the trajectory interval 25–70 ns. Clustering analysis revealed the existence of a single most prevailing Aβ42 structure, shown in Fig S2, with an occurrence of 46.6% of the trajectory, the second most common structure has an occurrence of 2.6%.
Figure 1.
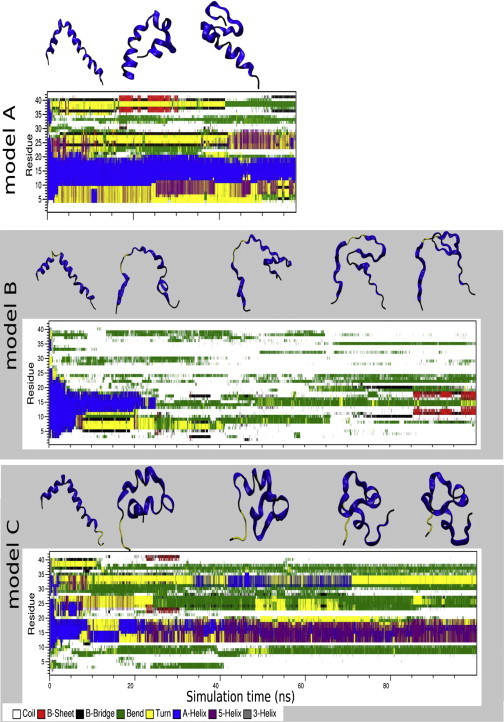
Time evolution of the Aβ42 peptide secondary structure in models A (SM/Chol/DMPA = 50:50:0), B (47.5:47.5:5.0), and C (40:40:20). The secondary structures were calculated by the DSSP program (50). The snapshot structures of Aβ42 were taken from trajectories at 20-ns intervals.
The conformational transitions of Aβ42 are also reflected in values of the radius of gyration as a result of interactions with the phospholipid membrane. See Fig. 2 C, which shows the calculated radius of gyration of Aβ42 during the simulation in all three models. In model B the radius of gyration varies between 1.4 and 2 nm during the simulation, whereas in model A and C, it is much lower and more uniform at ∼1 nm. These differences between the models indicate a different conformational behavior of the Aβ42 peptide.
Figure 2.
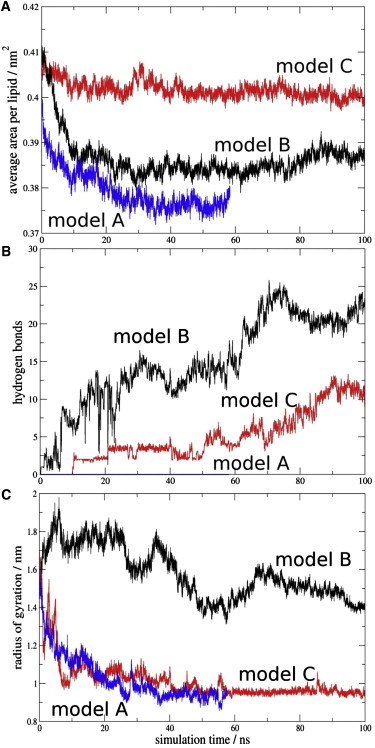
Time evolution of the MD simulations in the model A (SM/Chol/DMPA = 50:50:0), B (47,5:47.5:5.0), and C (40:40:20). For the sake of clarity, the data were averaged every 20 ps. (A) Average area per lipid molecule. (B) Hydrogen bonds between Aβ42 and membrane phospholipids. The proton donor-acceptor distance threshold was 0.35 nm and the angle was 30°. (C) Radius of gyration of the Aβ42 peptide in all models.
One of the most common parameters describing the lipidic bilayer properties is the area per lipid headgroup. Here, we have calculated the average area per lipid headgroup (Alipid) for fully hydrated 512 lipid bilayers during the MD simulations simply as the product of the box dimensions in the x axis and y axis, divided by the lipid number in a single leaflet (Fig. 2 A.). After the initial 10 ns, the Alipid in all simulations varies within 0.05 nm2, indicating that the bilayer model is sufficiently equilibrated. In model A, the Alipid is lower compared to the B model, and the Alipid of the model C is ∼0.2 nm2 higher than in the model B. The results from our three bilayer models averaged over the last 50 ns of simulation give 0.379, 0.386, and 0.401nm2 for the A, B, and C models, respectively. This was expected, because the ratio of the cholesterol molecules in model A and B is larger than in model C. The averaged values are clearly lower than the calculated values of the individual constituents in our system. For the 1-palmitoyl-2-oleoyl phosphatidic acid, similar to DMPA, values ranging from 0.485 to 0.511 nm2 were reported (32). For PSM, values from 0.565 to 0.59 nm2 were obtained by Mombelli et al. (33), and for Chol the experimentally obtained Alipid is 0.39 nm2 (32). However, in a mixture with phospholipids the condensation effect of cholesterol is well known (34). They observed an average Alipid of 0.39 nm2 in a 1:1 mixture of Chol and dipalmitoylphosphatidylcholine (DPPC), which corresponds well with our models.
Among many of the inter- and intramolecular interactions between Aβ42 and the membrane surface, hydrogen bonds play an important role and may be involved in the formation of Aβ42 fibrils. Therefore, the number of hydrogen bonds between Aβ42 amino acids and membrane lipids, representing interactions of Aβ42 with the phospholipid membrane surface was calculated as a function of time (Fig. 2 B). The evolution of the hydrogen bond contacts shows clearly more interactions in model B, gradually evolving during the simulations, with up to 25 interactions after 70 ns. In the case of model C, the number of interactions is lower, with up to 12 hydrogen bond contacts after 90 ns simulation. On the contrary, no hydrogen bonds have been observed in model A for the time of the simulation. We note that all simulations started from a structure with no hydrogen bond interactions between Aβ42 and the membrane. To analyze the contribution of individual Aβ42 amino acids to the hydrogen bonding network with the membrane the relative occurrence of hydrogen bonds for each residue has been calculated (Fig. 3). The B model is characterized by multiple interactions between Aβ42 and PSM, whereas the hydrogen bonds to Chol are scarce and those to DMPA, negligible. The peptide region mostly involved in interactions is the hydrophobic segment (residues 29–42). The hydrophilic part that contributes to the interactions involves residues Asp-1, Ala-2, Arg-5, and residues 24–28. Of importance, the prevailing binding residue is Lys-28, with multiple interactions with PSM and some to Chol. The crucial role of Lys-28 in binding to bilayer phospholipids has been suggested earlier (21,35), and our model is in agreement with those results. The number of interacting Aβ42 residues in model C is much smaller. As expected, due to the higher DMPA concentration, the hydrogen bonds to DMPA are more common than in model B. Here, the most occurring binding residue is Asp-1, interacting dominantly with PSM and to some extent also with DMPA. Hydrophilic residues Arg-5, Ser-26, Asn-27, and Lys-28 show interactions with DMPA, and less to PSM. It is noteworthy that Lys-28 is the second most involved binding residue in this model. The region of residues 29–42 do not interact with the membrane. Fig. S3 summarizes these results.
Figure 3.

Hydrogen bond occurrence between the residues of Aβ42 and membrane lipids in the models B and C. Distance and angle threshold between the hydrogen bond donor and acceptor was 0.35 nm and 30°, respectively. Relative occurrences for multivalent interactions are summed to individual residues.
Experimental measurements
Lipid monolayers at the air-water interface
The surface active properties of Aβ42 and its monomeric interaction with membrane lipids were first tested in a Langmuir balance at the air-water interface. In the absence of lipids, injection of Aβ42 into the aqueous phase causes an increase in surface pressure, the latter reaching equilibrium after ≈1 h (Fig. S4 A). This shows that Aβ42 is surface active, like many other peptides (31). The increase in surface pressure is dose-dependent and reaches a plateau at ∼10 mN/m, for Aβ concentrations slightly above 1 μM (Fig. S4 B). Thus, at these and higher concentrations the interface is saturated with adsorbed peptide and the peptide partitions between the interface and the bulk water (31). Previous studies have found plateau values in the 12–17 mN/m range with Aβ42 or Aβ40 (36–39), the origin of the variability being unclear at present.
Aβ insertion into lipid monolayers was assayed in a different set of experiments, in which a lipid monolayer was extended at the air-water interface and the peptide was injected into the aqueous subphase. The initial surface pressure of the lipid monolayer was fixed as desired, at values >10 mN/m to avoid simultaneous peptide insertion and peptide adsorption. Aβ insertion into the lipid monolayer at the interface causes a further increase in surface pressure Δπ (Fig. 4 A). As the initial pressure increases, Δπ decreases (Fig. 4 B) until a point is reached, at 30–33 mN/m for this system, at which peptide insertion is no longer possible. Fig. 4 B includes results for three different lipid compositions in the monolayer. In the 10–30 mN/m range, peptide insertion becomes easier in the order: SM/Chol (1:1) < SM/Chol/DMPA (40:40:20) < SM/Chol/DMPA (47.5:47.5:5), all figures given as mole ratios. However, the limiting initial pressure for all three lipid compositions is the same, close to 30 mN/m, and is supposed to be (with large fluctuations) the average surface pressure in the cell membranes. The same limiting initial pressure of 30 mN/m was found by Maltseva et al. (37,38) for Aβ40. This may mean that, under the cellular conditions, Aβ exists in equilibrium between the free and membrane-bound forms. The fact that insertion is facilitated by the presence of negatively charged lipids in the monolayer supports the role of electrostatic interactions in stabilizing Aβ42 insertion into lipid monolayers, in agreement with previous suggestions (37,39). A compression isotherm of the SM/Chol/DMPA (40/40/20) mixture indicated an average area/molecule of ≈ 0.4 nm2, in agreement with the MD calculations (isotherm not shown).
Figure 4.
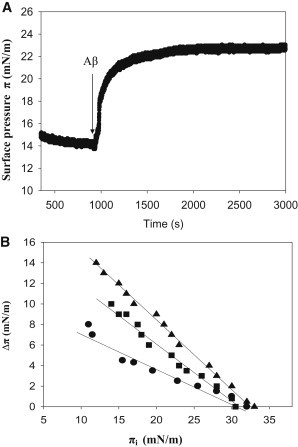
Changes in surface pressure of lipid monolayers, upon insertion of Aβ42 monomers at varying initial pressures. (A) A representative experiment, obtained with SM/Chol/DMPA (40/40/20) at 15 mN/m. (B) Equilibrium values. ●, SM/Chol (1:1), ▴, SM/Chol/DMPA (47.5/47.5/5), ■, SM/Chol/DMPA (40/40/20). Average values mean ± SE (n = 3). Aβ42 stock solution was 50 μM. Aβ42 final concentration in the trough was 1.22 μM.
The physical state of lipid bilayers containing DMPA
The SM/Chol/DMPA bilayers under study are in the Lo phase. Egg PC and PC/DMPA bilayers exist in the Ld phase (see the Supporting Material and Fig. S5).
Calorimetric studies
Small amounts of LUV were added to an Aβ solution and measurement of heat exchanges at varying lipid/peptide ratios allowed the calculation of ΔH, ΔS, and ΔG of the process. A typical experiment is shown in Fig. S6. When LUVs were composed of SM and Chol only, in the absence of negatively charged lipids, no measurable heats of interaction were observed. This may be related to the relatively poor insertion of Aβ into SM/Chol monolayers described previously, particularly at initial lateral pressures close to 30 mN/m, that are considered to represent the average lateral pressure in cell membranes (Fig. 4 B). Reliable measurements were obtained however with LUV containing DMPA. The experimental values are summarized in Table 1 (Nos. 1–4). The peptide association constant (Ka) decreases regularly (and correspondingly ΔG is made less negative) when DMPA concentration increases from 2.5 to 20 mol % (Fig. S7). This is in full agreement with the surface pressure measurements of peptide insertion (Fig. 4 B).
Table 1.
Thermodynamic parameters for the interaction of Aβ (1–42) with unilamellar vesicles of various lipid compositions
| No. 1 |
No. 2 |
No. 3 |
No. 4 |
No. 5 |
No. 6 |
No. 7 |
No. 8 |
No. 9 |
|
|---|---|---|---|---|---|---|---|---|---|
| SM/Ch/DMPA (48.75:48.75:2.5) | SM/Ch/DMPA (47.5:47.5:5) | SM/Ch/DMPA (45:45:10) | SM/Ch/DMPA (40:40:20) | SM/Ch/CL (47.5:47.5:5) | SM/Ch/CL (40:40:20) | PC | PC/DMPA (95:5) | PC/DMPA (80:20) | |
| Ka (M−1) | 7.8 ± 0.7 × 104 | 3.1 ± 0.9 × 104 | 1.1 ± 0.5 × 104 | 5.8 ± 0.8 × 103 | 1.6 ± 0.2 × 105 | 4.3 ± 0.5 × 104 | 1.4 ± 0.4 × 105 | 4.9 ± 0.36 × 105 | 4.0 ± 0.14 × 105 |
| Kd (μM) | 18.4 ± 0.5 | 32.0 ± 1.1 | 100.0 ± 1.5 | 172.0 ± 12.5 | 6.2 ± 1.2 | 25.0 ± 3.0 | 6.9 ± 1.1 | 2.0 ± 0.1 | 2.5 ± 0.1 |
| ΔH (Kcal/mol) | −2.3 ± 0.2 | −7.3 ± 0.05 | −1.2 ± 0.2 | −2.8 ± 0.19 | −12.6 ± 1.4 | −1.4 ± 0.8 | −2.9 ± 0.1 | −1.5 ± 0.1 | −2.5 ± 0.1 |
| ΔS (cal/mol) | 14.7 ± 0.6 | −3.0 ± 0.1 | 15.0 ± 1.3 | 8.1 ± 0.2 | −16.8 ± 0.4 | 16.4 ± 0.6 | 14.1 ± 0.6 | 21.1 ± 0.5 | 17.6 ± 0.5 |
| ΔG (Kcal/mol) | −6.9 ± 0.3 | −6.4 ± 0.1 | −5.8 ± 0.4 | −5.3 ± 0.01 | −11.9 ± 0.6 | −7.0 ± 0.7 | −7.3 ± 0.3 | −8.0 ± 0.5 | −7.9 ± 0.7 |
Average values mean ± SE (n = 3).
DMPA has been used throughout this work as a representative of the structurally simplest class of phospholipids with a net negative charge being in the fluid phase at 37°C. To discard that the observed increased binding was specific for DMPA, a structurally very different lipid, also negatively charged, namely CL was used (Table 1, Nos. 5, 6). The presence of CL favored Aβ42 interaction with bilayers even more than DMPA, and again the phenomenon was observed that the high CL concentration was less effective than the lower one in promoting peptide binding.
All the above mixtures give rise to bilayers in the Lo state. With the aim of testing the influence of membrane order on Aβ42 binding a number of experiments were performed with Ld bilayers (Table 1, Nos. 7–9). The results clearly show that these membranes bound Aβ42 monomers with higher activity than those in the Lo phase (Table 1, Nos. 1–6). Even pure PC bilayers bound the peptide. However, in this case the presence of DMPA increased only moderately, by two- to threefolds, the Ka and high DMPA concentrations did not appear to cause a large decrease in binding.
ThT assays
ThT fluorescence increases with β-sheet contents of peptides (40). In our case the test was applied to SM/Chol mixtures with or without DMPA. The results are summarized in Fig. 5. β-sheet formation increased with time, DMPA favored the process, and DMPA was more effective at 5 mol % than at 20 mol %, in good agreement with the calorimetric and surface pressure results. These data also corroborate the recent observation by Sani et al. (18) that negatively charged surfaces favored the Aβ peptide transition to an aggregated β-sheet conformation.
Figure 5.
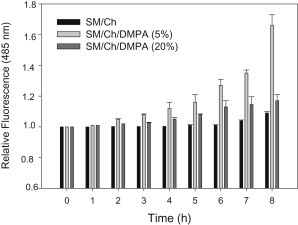
Time course of ThT fluorescence in the presence of Aβ42 in mixtures with LUV of varying lipid compositions. Lipid/protein mol ratio was 200:1, at 5 μM Aβ42. Average values mean ± SE (n = 3).
Secondary structure measurements
Formation of β-sheet structures upon binding of Aβ42 to bilayers was directly demonstrated by IR spectroscopy. The spectra were recorded under conditions similar to those in the ThT experiments, i.e., 1:200 peptide/lipid mol ratio. The spectral region corresponding to the amide I band of Aβ42 is shown in Fig. 6. An almost exclusively β-sheet structure was seen in all cases, as shown by the bands centered at 1626 cm−1 (41), however the intensity increased in the order: SM/Chol < SM/Chol/DMPA (20 mol %) < SM/Chol/DMPA (5 mol %), in agreement with all the results discussed previously.
Figure 6.
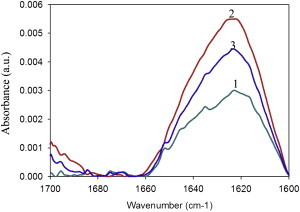
Difference IR spectra of Aβ42 in mixtures with multilamellar vesicles (MLV) of varying lipid compositions. Spectra of free peptide were subtracted from spectra of MLV-peptide mixtures. MLV composition was: 1, SM/Chol (1/1); 2, SM/Chol/DMPA (47.5/47.5/5); 3, SM/Chol/DMPA (40/40/20). Lipid/protein mol ratio was 200:1 at 80 μM Aβ42.
Discussion
The main results in this work include the thermodynamic parameters of Aβ42 monomer peptide binding to lipid bilayers in the fluid-ordered and fluid-disordered states, in the presence and absence of negatively charged lipids. Comparable experiments have been performed using MD methods. These, together with a number of other techniques, e.g., fluorescence and IR spectroscopy, and Langmuir through measurements, cooperate in providing a rather detailed picture of monomeric Aβ42-membrane interactions.
The binding studies have been interpreted in terms of a two-state model that suits best the experimental results. However, it should be kept in mind that the phenomena we observe include at least three states, namely free, bound, and adsorbed peptide, the bound-adsorbed transition being kinetically controlled.
Ordered versus disordered fluid bilayers
There has been some confusion on whether Lo domains (rafts) increase or decrease Aβ accumulation in membranes. These experiments have been usually carried out by increasing or decreasing cholesterol contents in bilayers thus increasing or decreasing Lo domain formation. In some cases Aβ accumulation has been related to the presence of cholesterol (42) and lipid rafts (4). Cholesterol-rich regions of hippocampal cell membranes have been associated with increased Aβ formation (43). However, other authors challenge the significance of the previous experiments, suggesting that APP was overexpressed in those models. In mice (44) and AD patients (45) inhibition of cholesterol synthesis or depletion of membrane cholesterol contents actually led to an increased deposition of Aβ aggregates. In our opinion, the previous studies (4,42–45) are in fact measuring the joint result of two different processes, namely APP hydrolysis by β- or γ-secretase yielding Aβ42, and Aβ deposition and aggregation. β-secretase appears to be located preferentially in Lo domains (4,46,47), thus Aβ is probably generated in those domains, but deposition and aggregation may occur in different domains. In this study Aβ binding to fluid-ordered bilayers was studied in particular detail, because of the association of β-secretase to that sort of domains. Particular care was taken to ensure that Aβ in the monomeric form was initially added to the membrane suspension, thus in our case only the process of peptide binding to membranes was considered, irrespective of the locus or rate of Aβ generation. Interestingly, the Aβ42 affinity for Ld membranes was higher than that for the Lo environment in which it is originally generated.
The role of anionic phospholipids
Data in the literature suggest that negatively charged phospholipids increase Aβ fibrillization (18,19). Electrostatic forces help to stabilize Aβ-lipid interactions (40,42). Our measurements (Table 1) confirm the previous observations, but both the calorimetric and MD results indicate that binding does not increase linearly with the amount of negative charges. Rather, a low dose of anionic phospholipids (2.5–5 mol % phosphatidic acid (PA) or CL) increase notoriously peptide binding, but higher doses have a smaller effect that MD pictures attribute to overall repelling electrostatic interactions: only Lys-28 appears to have a positive interaction with the anionic lipids (21). The latter authors observed a dose-dependent increase in Aβ binding to PC bilayers containing increasing proportions (0–50 mol %) of PA. The discrepancy may be because Chauhan et al. (19) used fluid-disordered bilayers in their experiments, in our hands high proportions of PA did not vary much the Aβ association constant to the fluid-disordered bilayers. It should also be noted that those authors measured Aβ binding with an indirect method, based on fluorescent probes. The available results may be summarized by stating that anionic phospholipids do increase Aβ binding both to fluid-ordered and to fluid-disordered bilayers, although the dose-effect pattern appears to be a complex one.
The view of molecular dynamics
The results obtained from 100 ns MD simulation of Aβ42 on the 512 lipid bilayers provide insights into interactions occurring between Aβ42 and the surface of the heterogeneous lipid bilayer. The Aβ42 conformational transitions observed during the MD simulations indicate that 100 ns is a sufficient simulation time. We are aware of the fact that 100 ns simulation is not sufficient to sample all available conformations of Aβ42. However, our results are consistent and in a reasonable agreement with the experimental data and, therefore, allow us to make some general explanations. The previous simulations of Aβ42 show that while in aqueous solution Aβ42 undergoes conformational flexibility (48), in near-homogeneous 128 lipid bilayers, represented by DPPC and dioleoyl phosphatidlyserine, the Aβ42 peptide adopts a more ordered secondary structure (20,21).
Analysis of the structure and interactions in model B, containing 5% DMPA (Fig. 1, Fig. S2) suggests that the Aβ42 peptide associates with the surface of the phospholipid membrane, forming multiple interactions mainly with the PSM polar head groups. The association results in a change of the Aβ42 secondary structure and an increase of the radius of gyration in model B compared to model C. Cluster analysis of the Aβ42 backbone structures during the simulation revealed that the most frequently occurring secondary structure conformation is similar to the β-hairpin conformer of Aβ42. This is in agreement with the mechanism of the accelerated surface Aβ42 fibril formation proposed by Bokvist et al. (35) and also with the MD study of Aβ42 in the presence of DPPC and dioleoyl phosphatidlyserine at different pH performed by Davies et al. (21). A different behavior of Aβ42 is observed for model C containing 20% DMPA. A distinctly lower number of interactions occur between Aβ42 and the phospholipids (Figs. 2 and 3). It was suggested (21) that Lys-28 may play a role in anchoring the Aβ42 to the bilayer. The interactions formed by Asp-1 and Lys-28 are strong enough to keep the peptide close to the surface, but in contrast to the B model, the peptide does not adsorb on it. We believe that unfavorable electrostatic interactions between the phospholipid head group of DMPA and Aβ42 inhibit binding of Aβ42 on the membrane. These results suggest that an inhibiting role of DMPA depends on charge density on bilayer surface (in other words on the molar ratio of DMPA). Whereas 5% of DMPA support binding of Aβ42, the percentage of 20% prevents strong binding. The intramolecular interactions in the peptide maintain a somewhat helical conformation with a markedly smaller radius of gyration than in the B model. In contrast to the B and C models, in the A model containing no negatively charged lipids, no interactions between the protein and the membrane surface were observed. The protein remains apart from the surface, with strong intramolecular hydrogen bonding network governing the conserved secondary structure. Overall, ours and previous simulation results (20,21) indicate that the binding and fibril formation on the membrane surface depends on the bilayer composition, and is the result of a subtle balance of many inter- and intramolecular interactions between Aβ42 and the membrane. On one hand, intramolecular hydrogen bonds (e.g., Asp-23-Lys-28) are essential for the formation of a secondary structure of Aβ42. However, an intermolecular hydrogen bond of Lys-28 is responsible for holding of Aβ42 on the bilayer surface. The previously mentioned intermolecular electrostatic interactions and hydrophobic interaction between the hydrophobic core of Aβ42 and bilayer contribute to this balance. In Fig. S3, the negatively charged DMPA lipids are color-coded red, and negatively charged residues of Aβ42 are in red, and positively charged residues in blue. A difference between the A, B, and C models can be clearly seen. At the beginning, long-range (electrostatic) interactions are important for bringing Aβ42 close to a bilayer. In the A model, these interactions are not available and Aβ42 remains apart from the bilayer. The situation is different in bilayer models containing DMPA. Attractive electrostatic interactions between negative charges of the DMPA phosphate group and positive charges of the Lys residues are responsible for bringing Aβ42 to the bilayer. Short-range (hydrogen bonding, hydrophobic) interactions then start to play a crucial role. In the B model, hydrophobic interactions between the bilayer and the C- and N-terminus of Aβ42 drive the conformational change of Aβ42 and its position on the bilayer surface. The hydrogen bond between Lys-28 and PSM contributes to this process. In the case of the C model, the electrostatic interactions are stronger compared to the B model, and as a result, Aβ42 is approaching locations with excessive negative charge density. However, repulsive electrostatic interactions of crowded DMPA negative charges with Asp negative charges of Aβ42 prevent Aβ42 from coming closer to the bilayer. As a consequence, the hydrogen bond between Lys-28 and phosphate of DMPA is holding Aβ42 close to a bilayer but hydrophobic interactions are too weak to initiate the transformation of the Aβ42 secondary structure.
Concluding remarks
The simulations and experiments described previously concur in showing that, for Aβ monomers interacting with lipid bilayers based on SM and Chol, which are in the Lo state, the presence of negatively charged phospholipids facilitates interaction with the Aβ peptide. However, low (2.5–5 mol %) DMPA or CL allows better interaction than 20 mol % of the same lipid. MD calculations predict for model B (5% DMPA) a larger number of interactions between peptide and bilayer than in model C (20% DMPA) or model A (no DMPA). Correspondingly, ThT fluorescence and IR spectroscopy show that β-sheet formation by Aβ42 occurs more readily at 5% than at 20% DMPA. Moreover, the MD methods indicate the development of a β-sheet structure by the peptide in bilayers containing 5% PA, whereas in the presence of 20 mol % PA the peptide retains a partially helical conformation. This may lead, under equilibrium conditions, to the situation of maximum β-sheet at low DMPA concentrations observed by ITC, Langmuir trough, and spectroscopic measurements (Fig. 7). It is also important that when Aβ42 interacts with bilayers in the Ld state, binding is stronger and less dependent on negatively charged lipids than with Lo bilayers. Our studies refer to Aβ42 binding to membranes irrespective of the place where the peptide has been generated. In principle, there would be no obstacle for Aβ42 being synthesized in Lo domains, yet binding preferentially and eventually giving rise to plaques, in the Ld regions that are predominant in cell membranes.
Figure 7.
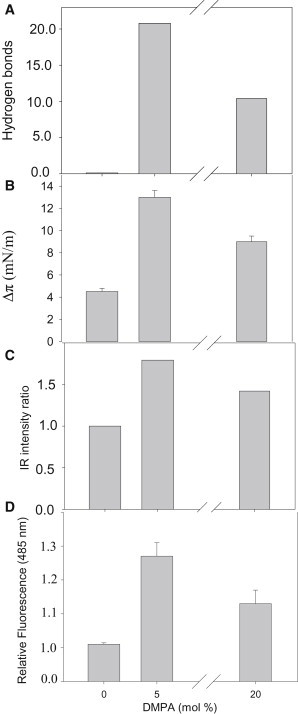
A comparison of data concerning Aβ42 binding to bilayers composed of SM/Chol (1:1 mol ratio), to which increasing amounts of anionic phospholipid (phosphatidic acid) had been added. (A) MD calculations of average H-bonds established between peptide and membrane for the last 20 ns. Data taken from Fig. 2B. (B) Langmuir balance. Increase in surface pressure of lipid monolayers (16 mN/m) of the compositions given previously due to the presence of Aβ42. Data are taken from Fig. 4B. (C) Relative change in intensity of the IR band centered at 1622 cm−1, assigned to antiparallel β-sheet vibration. Data taken from Fig. 6. (D) Changes in ThT fluorescence after 6 h incubation with Aβ42 and LUVs. Data taken from Fig. 5.
Acknowledgments
This research has received funding from the European Community's Seventh Framework Programme (FP7/2007-2013) under grant agreement No. 212043. This work was also supported by the Scientific Grant Agency of the Ministry of Education of Slovak Republic and Slovak Academy of Sciences (project VEGA-02/0101/11) and by the Spanish Ministerio de Ciencia e Innovación (BFU 2007-62062).
Footnotes
In memory of Professor J. M. Macarulla (1932–2012).
Supporting Material
References
- 1.Ross C.A., Poirier M.A. Protein aggregation and neurodegenerative disease. Nat. Med. 2004;10(Suppl):S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 2.Selkoe D.J. Alzheimer's disease: genes, proteins, and therapy. Physiol. Rev. 2001;81:741–766. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- 3.Kojro E., Gimpl G., Fahrenholz F. Low cholesterol stimulates the nonamyloidogenic pathway by its effect on the alpha -secretase ADAM 10. Proc. Natl. Acad. Sci. USA. 2001;98:5815–5820. doi: 10.1073/pnas.081612998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ehehalt R., Keller P., Simons K. Amyloidogenic processing of the Alzheimer β-amyloid precursor protein depends on lipid rafts. J. Cell Biol. 2003;160:113–123. doi: 10.1083/jcb.200207113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haass C., Lemere C.A., Selkoe D.J. The Swedish mutation causes early-onset Alzheimer's disease by β-secretase cleavage within the secretory pathway. Nat. Med. 1995;1:1291–1296. doi: 10.1038/nm1295-1291. [DOI] [PubMed] [Google Scholar]
- 6.Simons K., Vaz W.L. Model systems, lipid rafts, and cell membranes. Annu. Rev. Biophys. Biomol. Struct. 2004;33:269–295. doi: 10.1146/annurev.biophys.32.110601.141803. [DOI] [PubMed] [Google Scholar]
- 7.Rajendran L., Simons K. Lipid rafts and membrane dynamics. J. Cell Sci. 2005;118:1099–1102. doi: 10.1242/jcs.01681. [DOI] [PubMed] [Google Scholar]
- 8.Nussbaum R.L., Ellis C.E. Alzheimer's disease and Parkinson's disease. N. Engl. J. Med. 2003;348:1356–1364. doi: 10.1056/NEJM2003ra020003. [DOI] [PubMed] [Google Scholar]
- 9.Haass C., Selkoe D.J. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer's amyloid β-peptide. Nat. Rev. Mol. Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 10.Crouch P.J., Harding S.M., Masters C.L. Mechanisms of A β mediated neurodegeneration in Alzheimer's disease. Int. J. Biochem. Cell Biol. 2008;40:181–198. doi: 10.1016/j.biocel.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 11.Cappai R., Barnham K.J. Delineating the mechanism of Alzheimer's disease A β peptide neurotoxicity. Neurochem. Res. 2008;33:526–532. doi: 10.1007/s11064-007-9469-8. [DOI] [PubMed] [Google Scholar]
- 12.Matsuzaki K. Physicochemical interactions of amyloid β-peptide with lipid bilayers. Biochim. Biophys. Acta. 2007;1768:1935–1942. doi: 10.1016/j.bbamem.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 13.Terzi E., Hölzemann G., Seelig J. Self-association of β-amyloid peptide (1-40) in solution and binding to lipid membranes. J. Mol. Biol. 1995;252:633–642. doi: 10.1006/jmbi.1995.0525. [DOI] [PubMed] [Google Scholar]
- 14.Anand P., Nandel F.S., Hansmann U.H.E. The Alzheimer β-amyloid (Aβ (1-39)) dimer in an implicit solvent. J. Chem. Phys. 2008;129:195102. doi: 10.1063/1.3021062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu C., Murray M.M., Bowers M.T. The structure of Aβ42 C-terminal fragments probed by a combined experimental and theoretical study. J. Mol. Biol. 2009;387:492–501. doi: 10.1016/j.jmb.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shoval H., Weiner L., Lichtenberg D. Polyphenol-induced dissociation of various amyloid fibrils results in a methionine-independent formation of ROS. Biochim. Biophys. Acta. 2008;1784:1570–1577. doi: 10.1016/j.bbapap.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 17.Sheikh A.M., Nagai A. Lysophosphatidylcholine modulates fibril formation of amyloid β peptide. FEBS J. 2011;278:634–642. doi: 10.1111/j.1742-4658.2010.07984.x. [DOI] [PubMed] [Google Scholar]
- 18.Sani M.A., Gehman J.D., Separovic F. Lipid matrix plays a role in Aβ fibril kinetics and morphology. FEBS Lett. 2011;585:749–754. doi: 10.1016/j.febslet.2011.02.011. [DOI] [PubMed] [Google Scholar]
- 19.Chauhan A., Ray I., Chauhan V.P.S. Interaction of amyloid β-protein with anionic phospholipids: possible involvement of Lys-28 and C-terminus aliphatic amino acids. Neurochem. Res. 2000;25:423–429. doi: 10.1023/a:1007509608440. [DOI] [PubMed] [Google Scholar]
- 20.Davis C.H., Berkowitz M.L. Interaction between amyloid-β (1–42) peptide and phospholipid bilayers: a molecular dynamics study. Biophys. J. 2009;96:785–797. doi: 10.1016/j.bpj.2008.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davis C.H., Berkowitz M.L. Structure of the amyloid-β (1–42) monomer absorbed to model phospholipid bilayers: a molecular dynamics study. J. Phys. Chem. B. 2009;113:14480–14486. doi: 10.1021/jp905889z. [DOI] [PubMed] [Google Scholar]
- 22.Lemkul J.A., Bevan D.R. A comparative molecular dynamics analysis of the amyloid β-peptide in a lipid bilayer. Arch. Biochem. Biophys. 2008;470:54–63. doi: 10.1016/j.abb.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 23.Lemkul J.A., Bevan D.R. Perturbation of membranes by the amyloid β-peptide—a molecular dynamics study. FEBS J. 2009;276:3060–3075. doi: 10.1111/j.1742-4658.2009.07024.x. [DOI] [PubMed] [Google Scholar]
- 24.Crescenzi O., Tomaselli S., Picone D. Solution structure of the Alzheimer amyloid β-peptide (1–42) in an apolar microenvironment. Similarity with a virus fusion domain. Eur. J. Biochem. 2002;269:5642–5648. doi: 10.1046/j.1432-1033.2002.03271.x. [DOI] [PubMed] [Google Scholar]
- 25.Reference deleted in proof.
- 26.Berger O., Edholm O., Jähnig F. Molecular dynamics simulations of a fluid bilayer of dipalmitoylphosphatidylcholine at full hydration, constant pressure, and constant temperature. Biophys. J. 1997;72:2002–2013. doi: 10.1016/S0006-3495(97)78845-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scott W., Hunenberger P., van Gunsteren W. The GROMOS biomolecular simulation program package. J. Phys. Chem. 1999;103:3596–3607. [Google Scholar]
- 28.Nilsson M.R. Techniques to study amyloid fibril formation in vitro. Methods. 2004;34:151–160. doi: 10.1016/j.ymeth.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 29.Mayer L.D., Hope M.J., Cullis P.R. Vesicles of variable sizes produced by a rapid extrusion procedure. Biochim. Biophys. Acta. 1986;858:161–168. doi: 10.1016/0005-2736(86)90302-0. [DOI] [PubMed] [Google Scholar]
- 30.Arnulphi C., Sot J., Goñi F.M. Triton X-100 partitioning into sphingomyelin bilayers at subsolubilizing detergent concentrations: effect of lipid phase and a comparison with dipalmitoylphosphatidylcholine. Biophys. J. 2007;93:3504–3514. doi: 10.1529/biophysj.107.104463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sánchez-Magraner L., Cortajarena A.L., Ostolaza H. Membrane insertion of Escherichia coli alpha-hemolysin is independent from membrane lysis. J. Biol. Chem. 2006;281:5461–5467. doi: 10.1074/jbc.M512897200. [DOI] [PubMed] [Google Scholar]
- 32.Phillips M.C. The physical state of phospholipids and cholesterol in monolayers, bilayers and membranes. Prog. Surf. Membrane. Sci. 1972;5:139–222. [Google Scholar]
- 33.Mombelli E., Morris R., Fraternali F. Hydrogen-bonding propensities of sphingomyelin in solution and in a bilayer assembly: a molecular dynamics study. Biophys. J. 2003;84:1507–1517. doi: 10.1016/S0006-3495(03)74963-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edholm O., Nagle J.F. Areas of molecules in membranes consisting of mixtures. Biophys. J. 2005;89:1827–1832. doi: 10.1529/biophysj.105.064329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bokvist M., Lindström F., Gröbner G. Two types of Alzheimer's β-amyloid (1-40) peptide membrane interactions: aggregation preventing transmembrane anchoring versus accelerated surface fibril formation. J. Mol. Biol. 2004;335:1039–1049. doi: 10.1016/j.jmb.2003.11.046. [DOI] [PubMed] [Google Scholar]
- 36.Lin M.S., Chen X.B., Chen W.Y. Dynamic fluorescence imaging analysis to investigate the cholesterol recruitment in lipid monolayer during the interaction between β-amyloid (1-40) and lipid monolayers. Colloids Surf. B Biointerfaces. 2009;74:59–66. doi: 10.1016/j.colsurfb.2009.06.027. [DOI] [PubMed] [Google Scholar]
- 37.Maltseva E., Brezesinski G. Adsorption of amyloid β (1–40) peptide to phosphatidylethanolamine monolayers. ChemPhysChem. 2004;5:1185–1190. doi: 10.1002/cphc.200400045. [DOI] [PubMed] [Google Scholar]
- 38.Maltseva E., Kerth A., Brezesinski G. Adsorption of amyloid β (1–40) peptide at phospholipid monolayers. ChemBioChem. 2005;6:1817–1824. doi: 10.1002/cbic.200500116. [DOI] [PubMed] [Google Scholar]
- 39.Thakur G., Pao C., Leblanc R.M. Surface chemistry of lipid raft and amyloid Aβ (1-40) Langmuir monolayer. Colloids Surf. B Biointerfaces. 2011;87:369–377. doi: 10.1016/j.colsurfb.2011.05.047. [DOI] [PubMed] [Google Scholar]
- 40.Biancalana M., Koide S. Molecular mechanism of Thioflavin-T binding to amyloid fibrils. Biochim. Biophys. Acta. 2010;1804:1405–1412. doi: 10.1016/j.bbapap.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arrondo J.L., Muga A., Goñi F.M. Quantitative studies of the structure of proteins in solution by Fourier-transform infrared spectroscopy. Prog. Biophys. Mol. Biol. 1993;59:23–56. doi: 10.1016/0079-6107(93)90006-6. [DOI] [PubMed] [Google Scholar]
- 42.Simons K., Ehehalt R. Cholesterol, lipid rafts, and disease. J. Clin. Invest. 2002;110:597–603. doi: 10.1172/JCI16390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simons M., Keller P., Simons K. Cholesterol depletion inhibits the generation of β-amyloid in hippocampal neurons. Proc. Natl. Acad. Sci. USA. 1998;95:6460–6464. doi: 10.1073/pnas.95.11.6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park I.H., Hwang E.M., Mook-Jung I. Lovastatin enhances Aβ production and senile plaque deposition in female Tg2576 mice. Neurobiol. Aging. 2003;24:637–643. doi: 10.1016/s0197-4580(02)00155-0. [DOI] [PubMed] [Google Scholar]
- 45.Abad-Rodriguez J., Ledesma M.D., Dotti C.G. Neuronal membrane cholesterol loss enhances amyloid peptide generation. J. Cell Biol. 2004;167:953–960. doi: 10.1083/jcb.200404149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Riddell D.R., Christie G., Dingwall C. Compartmentalization of β-secretase (Asp2) into low-buoyant density, noncaveolar lipid rafts. Curr. Biol. 2001;11:1288–1293. doi: 10.1016/s0960-9822(01)00394-3. [DOI] [PubMed] [Google Scholar]
- 47.Kalvodova L., Kahya N., Simons K. Lipids as modulators of proteolytic activity of BACE: involvement of cholesterol, glycosphingolipids, and anionic phospholipids in vitro. J. Biol. Chem. 2005;280:36815–36823. doi: 10.1074/jbc.M504484200. [DOI] [PubMed] [Google Scholar]
- 48.Sgourakis N.G., Merced-Serrano M., Garcia A.E. Atomic-level characterization of the ensemble of the Aβ(1-42) monomer in water using unbiased molecular dynamics simulations and spectral algorithms. J. Mol. Biol. 2011;405:570–583. doi: 10.1016/j.jmb.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao Y., Truhlar D.G. Density functionals with broad applicability in chemistry. Acc. Chem. Res. 2008;41:157–167. doi: 10.1021/ar700111a. [DOI] [PubMed] [Google Scholar]
- 50.Hess B., Kutzner C., Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 51.Tieleman D.P., Berendsen H.J.C. Molecular dynamics simulations of fully hydrated dipalmitoylphosphatidylcholine bilayer with different macroscopic boundary conditions and parameters. J. Chem. Phys. 1996;105:4871–4880. [Google Scholar]
- 52.Cheng M.H., Liu L.T., Tang P. Molecular dynamics simulations of ternary membrane mixture: phosphatidylcholine, phosphatidic acid, and cholesterol. J. Phys. Chem. B. 2007;111:14186–14192. doi: 10.1021/jp075467b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Höltje M., Förster T., Höltje H.D. Molecular dynamics simulations of stratum corneum lipid models: fatty acids and cholesterol. Biochim. Biophys. Acta. 2001;1511:156–167. doi: 10.1016/s0005-2736(01)00270-x. [DOI] [PubMed] [Google Scholar]
- 54.Hess B., Bekker H., Fraaije J.J. A linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 55.Humphrey W., Dalke A., Schulten K.J. Visual molecular dynamics. J. Mol. Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–38. [DOI] [PubMed] [Google Scholar]
- 56.Goñi F.M., Alonso A., Thewalt J.L. Phase diagrams of lipid mixtures relevant to the study of membrane rafts. Biochim. Biophys. Acta. 2008;1781:665–684. doi: 10.1016/j.bbalip.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Montes L.R., Alonso A., Bagatolli L.A. Giant unilamellar vesicles electroformed from native membranes and organic lipid mixtures under physiological conditions. Biophys. J. 2007;93:3548–3554. doi: 10.1529/biophysj.107.116228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Parasassi T., De Stasio G., Gratton E. Quantitation of lipid phases in phospholipid vesicles by the generalized polarization of Laurdan fluorescence. Biophys. J. 1991;60:179–189. doi: 10.1016/S0006-3495(91)82041-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dahlgren K.N., Manelli A.M., Jr., LaDu M.J. Oligomeric and fibrillar species of amyloid-beta peptides differentially affect neuronal viability. J. Biol. Chem. 2002;277:32046–32053. doi: 10.1074/jbc.M201750200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


