Abstract
Calmodulin (CaM) expression is upregulated upon HIV-1 infection and interacts with proteins involved in viral processing, including the multifunctional HIV-1 MA protein. We present here the results of studies utilizing small-angle neutron scattering with contrast variation that, when considered in the light of earlier fluorescence and NMR data, show CaM binds MA in an extended open-clamp conformation via interactions with two tryptophans that are widely spaced in sequence and space. The interaction requires a disruption of the MA tertiary fold such that MA becomes highly extended in a long snakelike conformation. The CaM-MA interface is extensive, covering ∼70% of the length of the MA such that regions known to be important in MA interactions with critical binding partners would be impacted. The CaM conformation is semiextended and as such is distinct from the classical CaM-collapse about short α-helical targets. NMR data show that upon dissociation of the CaM-MA complex, either by the removal of Ca2+ or increasing ionic strength, MA reforms its native tertiary contacts. Thus, we observe a high level of structural plasticity in MA that may facilitate regulation of its activities via intracellular Ca2+-signaling during viral processing.
Introduction
Calmodulin (CaM) is a ubiquitous intracellular Ca2+ receptor that binds to hundreds of different target proteins and modulates their activities in response to Ca2+ signals (1, 2). This relatively small (148-amino-acid) and highly conserved eukaryotic protein thus plays a critical regulatory role in a diverse range of essential biological functions; e.g., metabolism, motility, memory, immune response, apoptosis. HIV-1 processing requires reprogramming the hosts cell transcriptome and proteome via interactions with multiple signaling pathways, including Ca2+-signaling (3), and CaM is known to interact with a number of viral proteins involved in HIV-1 replication: including Nef (4, 5), gp160 (5, 6, 7, 8), and MA (9, 10). The nature and purpose of these CaM interactions are not fully known, but the potential for their playing specific roles in the viral life cycle or in host defense mechanisms is of considerable interest. The MA protein was first identified as a potential CaM target from observations that CaM expression is upregulated in cells infected with HIV-1 (9). Furthermore, CaM and the Gag polyprotein (from which MA is derived) colocalize in these cells in a distribution that is distinct from the typical distribution of CaM in uninfected cells. Studies using peptides from MA showed that the N-terminal sequence of MA binds CaM in a Ca2+-dependent manner (10).
The 132-amino-acid MA protein (15 kDa) is one of several proteolytic products produced during viral packaging from the Gag-polyprotein that plays a pivotal role in HIV-1 processing (11). Before proteolytic cleavage, MA forms the first N-terminal domain of Gag and is posttranslationally modified at its amino-terminus by a myristoyl group (12). In this form, MA directs Gag to cytoplasmic regions of the plasma membrane enriched with negatively charged phosphatidylinositol-(4,5)-bisphosphate (PI(4,5)P2) phospholipids (13, 14, 15). The localization of Gag to the cell membrane is mediated by the exposure of the myristoyl group and is further enhanced by the interaction between basic N-terminal amino acids of MA and PI(4,5)P2 (16). This localization enables Gag to coordinate with a number of other host-cell proteins at the inner cell membrane to facilitate viral maturation and release. The cleaved MA protein exists as a distinct species within the mature HIV-1 virus and in the cytoplasm of recently infected cells and participates in a large number of cellular processes (17, 18). It interacts with DNA and RNA, forms part of the HIV-1 preintegration complex, and constitutes a key component of the viral matrix, forming an icosahedral arrangement of MA proteins that decorate the inner membrane of mature virons.
The structures of CaM and MA have been solved by NMR spectroscopy and x-ray crystallography. The CaM crystal structure (19) revealed a dumbbell-shaped structure with two globular lobes, each consisting of a pair of helix-loop-helix motifs that form a cup-shaped domain. The sides of the cup are formed by pairs of helices with the Ca2+-binding loops at the base. The inner surface is lined with hydrophobic residues that are exposed upon Ca2+ binding via concerted motions of the helices (20) to bind hydrophobic recognition sites in CaM's regulatory targets. The CaM Ca2+-binding lobes are connected by an extended α-helix that in solution has a flexible region that enables reorientation of the lobes so that CaM can bind to diverse target sequences (2, 21, 22, 23). The crystal (24) and NMR (25, 26) structures of the MA protein each show an N-terminal domain formed by four α-helices and three short β-strands that sequester the amino N-terminal linked myristoyl group (16). A fifth helix projects from the N-terminal domain, extending into a predominantly unstructured C-terminal tail.
We have shown previously that the Ca2+-dependent binding of CaM to MA induces a large conformational change in MA involving a dramatic loss of tertiary contacts in the unmyristoylated form (27) and Ghanam et al. (28) confirmed this same result for the myristoylated protein. This CaM-induced unfolding of MA could thus affect myristoylate exposure and/or the interaction between MA and PI(4,5)P2 at the inner cell membrane, which leads to the intriguing possibility that Gag localization to the membrane, or the number of the other processes MA is involved in, could be influenced by calcium signaling. In this regard, it is of interest to determine the overall conformation of MA within the CaM-MA complex and whether the CaM-induced unfolding of MA is reversed upon disassociation, i.e., is the unfolding of MA by CaM specific and reversible? This question is addressed here using small-angle neutron scattering (SANS), with deuterium labeling and contrast variation, to determine the shapes and disposition of CaM and MA within the CaM-MA complex and NMR spectroscopy to monitor the structural changes in MA upon dissociation from CaM.
Materials and Methods
Sample preparation for small-angle scattering
MA and CaM were expressed in Escherichia coli and purified as described previously in Chow et al. (27). Deuteration of CaM was achieved by growing cells in ModC1 minimal media (29) containing 90% D2O (v/v) with nondeuterated glycerol (40 g/L) as the carbon source to yield CaM with 73% of nonexchangeable hydrogens deuterated, as determined by partial trypsin digest MALDI-TOF mass spectrometry.
Complexes of CaM-MA were formed under conditions previously determined in Chow et al. (27) by mixing a 1:1 molar ratio of CaM and MA and dialyzing into buffers containing 50 mM MOPS pH 7.0, 5 mM CaCl2, 2 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP). A range of H2O/D2O ratios were used to yield samples for neutron scattering in 19%, 44%, 88%, and 100% v/v D2O with protein concentrations of 7.7, 7.0, 7.4, and 7.5 mg/mL, respectively. All protein concentrations were determined by UV absorbance (calculated molar extinction coefficients for CaM and MA were ε276 nm = 3030 M−1 cm−1 and ε280 nm = 17,085 M−1 cm−1, respectively (27)). The equilibrated dialysates were used as matched solvent blanks for the neutron scattering experiments. The final percentage of D2O for each equilibrated dialysate was checked by densitometry (DMA 5000 Density Meter; Anton-Paar, Ashland, VA). For each SANS measurement, 300 μL of protein solution or solvent blank was loaded into a 1-mm path-length, banjo-shaped cell (product No. 120-1 mm; HELLMA USA, Plainview, NY). Small-angle x-ray scattering (SAXS) measurements were made in a 2-mm glass capillary cell. Further details of sample preparation are provided in the Supporting Material.
Small-angle scattering data acquisition and reduction
Neutron scattering
SANS data were acquired using the SANS instrument Quokka at the Australian Nuclear Science and Technology Organisation (30, 31) using a wavelength, λ, of 4.94 Å (Δλ/λ of 6.65%) with the sample holder maintained in the range 10–20°C. Data were acquired for the attenuated direct beam sample and matched solvent transmissions and scattering, and empty cell and blocked beam with sample-to-detector positions suitable to give a measured q-range 0.007–0.3 Å−1.
Data for each sample measured at different detector positions were combined and reduced to I(q) versus q (q = (4πsinθ)/λ, 2θ is the scattering angle) using IGOR Pro 6.1 (WaveMetrics, Portland, OR) with Quokka data reduction macros installed (32). Detector sensitivity corrections were applied using the H2O or 19% D2O solvent scattering. Radially averaged I(q) versus q for each measurement was obtained (before combination of data from different detector setting) after subtracting the scattering due to the empty cell and blocked beam with corrections for sample and buffer transmissions. The data were put on an absolute scale by reference to the measured beam flux. The scattering due to the protein was obtained by subtraction of the solvent scattering from that of the protein-plus-solvent sample.
X-ray scattering
SAXS data were acquired using the Nanostar instrument (Bruker AXS, Madison, WI) at the Australian Nuclear Science and Technology Organisation. Samples were maintained at 10°C during measurement. The scattering data were reduced to I(q) versus q using the program “SAXS: Small Angle X-Ray Scattering System V4.1.29”, which circularly averages the raw data and corrects for detector sensitivity and nonlinearity. All data were placed on an absolute scale using the scattering of water as a standard (33) and the scattering due to the protein was obtained by subtraction of the solvent-plus-cell scattering from that of the protein-plus-solvent-plus cell.
Small-angle scattering data analysis and molecular modeling
All scattering data analysis and modeling used programs of the ATSAS program package (34). Guinier analyses used PRIMUS to yield estimates of the intensity at zero scattering angle, I(0), and radius of gyration, Rg. GNOM was used to determine P(r) versus r and the associated I(0) and Rg values. Determination of the molecular mass, Mr, of the CaM-MA complex in solution used the SAXS-determined zero-angle scattering I(0) value (0.2 cm−1) and the method of Orthaber et al. (33) (see the Supporting Material) with values for the protein contrast (3.084 × 1010 cm−2) and partial specific volume (0.727 cm3 g−1) determined using the amino-acid sequences and the program MULCh (35).
Ab initio shape modeling was performed using the combined SAXS and SANS data and the multiphase dummy-atom modeling program MONSA in the slow mode with P1 symmetry and default parameters. Rigid body modeling (detailed in the Supporting Material) used the program SASREF7. The programs CRYSOL or CRYSON were used to calculate SAXS or SANS profiles, respectively, from Protein Data Bank (PDB) structures.
Nuclear magnetic resonance spectroscopy
MA and CaM were expressed and purified as above, except CaM production was from H2O media and 15N-labeled MA was produced using M9 minimal growth medium with 15NH4Cl as the sole nitrogen source (27). 15N-MA and unlabeled CaM were dialyzed into a buffer containing 10 mM sodium phosphate (pH 5.5), 5 mM CaCl2 and 1 mM TCEP. The CaM-MA complex was formed as above by mixing a 1:1 molar ratio of 15N-MA and CaM with excess Ca2+ in the solvent. For the KCl titration experiments, a final concentration range from 0–500 mM was investigated, and for the EGTA titration, a final concentration range of 0–10 mM was investigated. For each titration of KCl or EGTA into solutions initially containing the CaM-MA complex, the sample was allowed to equilibrate for a minimum of 10 min after each addition before NMR spectra were recorded. Spectra for denatured 15N-labeled MA were collected after dialysis of the sample into the above buffer that additionally contained 6 M guanidinium HCl.
All two-dimensional 1H-15N-HSQC spectra were recorded at 298 K (25°C) using an Avance III 600- or 800-MHz spectrometer (Bruker) equipped with a triple resonance cryogenic TCI probe. NMR data were processed with the program TOPSPIN (Bruker) and analyzed with SPARKY (T. D. Goddard and D. G. Kneller, University of California, San Francisco). Previously published NMR 1H-15N chemical shift assignments for MA (BMRB #7250) were used to assign the 1H-15N HSQC spectrum of 15N-MA.
Results
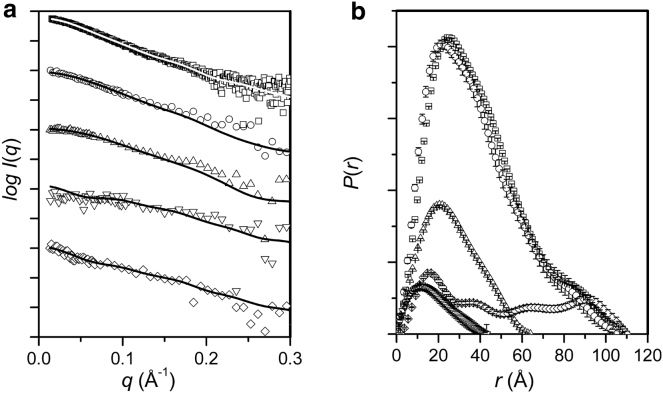
SANS data, I(q) versus q, for the CaM-MA complex in solutions prepared with 19%, 44%, 88%, and 100% v/v D2O along with SAXS data for the complex are shown in Fig. 1 a with corresponding, P(r) versus r profiles in Fig. 1 b. All the scattering data are for calcium-saturated CaM (73% deuterated) complexed with nondeuterated MA, which we denote here simply as CaM-MA.
Figure 1.
Scattering data for CaM-MA. (a) I(q) versus q and (b) P(r) versus r. Symbol key: SAXS data (□), and SANS data for CaM-MA in 19% D2O (○), 44% D2O (▵), 88% (▿), and 100% D2O (⋄). The solid lines in a are the fits of the MONSA-derived model in Fig. 3. The I(q) profiles have been offset on the vertical scale for clarity. The P(r) profiles are on a relative scale that accounts for the contrast of each scattering profile.
Guinier plots for each SANS and SAXS profile (see Fig. S1 a in the Supporting Material) show the expected linearity for a solution of monodisperse scattering particles and the SAXS data are consistent with the expected results for the complex. The molecular mass calculated from the SAXS data is 34,500 Da (see Materials and Methods), which is within experimental uncertainty (<10%) of that expected for the 1:1 CaM-MA complex based on amino-acid sequence (31,700 Da). The Rg and Dmax values determined from the SAXS data for the complex with deuterated CaM (31.4 Å and 110 Å, respectively, Table 1) are in reasonable agreement with those obtained previously for the complex formed with nondeuterated CaM (30.5 Å and 110 Å, respectively) (27). A plot of the square-root of I(0) as a function of % D2O in the solvent (Fig. S1 b) has the expected linear dependence with an intercept on the D2O axis that predicts the average scattering density for the complex is matched to that of the solvent at 75% D2O. This value is in excellent agreement with that expected from chemical and isotopic composition (74.6%, calculated based on the amino-acid sequence using MULCh (35).)
Table 1.
SAXS and SANS contrast series parameters for CaM-MA
| Δρ∗ (cm−2) | Rg† (Å) | Rg‡ (Å) | Dmax‡ (Å) | |
|---|---|---|---|---|
| SAXS | 3.084 × 1010 | 30.9 ± 0.2 | 31.4 ± 0.3 | 110 |
| SANS 19% D2O | 3.035 × 1010 | 29.7 ± 1.3 | 30.0 ± 0.8 | 105 |
| SANS 44% D2O | 1.700 × 1010 | 20.9 ± 1.3 | 20.7 ± 1.0 | 64 |
| SANS 88% D2O | −0.858 × 1010 | 13 ± 1 | 13.3 ± 0.9 | 43 |
| SANS 100% D2O | −1.414 × 1010 | 41 ± 6 | 41 ± 2 | 110 |
Values of Δρ calculated using MULCh (35).
Rg determined from Guinier analysis and data for which qRg < 1.3.
Rg and Dmax determined from P(r) analysis.
The P(r) profiles represent the probable distribution of pairwise distances between atoms within the scattering particle weighted by the product of the contrast values at their positions (r); hence, P(r) goes to zero at the maximum linear dimension of the particle, Dmax. In the SAXS experiment the complex is an approximately uniform scattering density object and the P(r) profile is as expected from our previous studies (27). On the other hand, the P(r) profiles calculated from the SANS data show the anticipated variations with percent D2O due to the changing contrast values that give rise to the large changes in effective I(0) and Rg values observed in the Guinier analyses (Table 1).
CaM within the CaM-MA complex adopts a semiextended conformation
In ∼44% D2O, the scattering density of MA in the CaM-MA complex was calculated to match the solvent and therefore was expected to not contribute significantly to the SANS data. If the solvent matching is sufficiently good, these data would yield the structural parameters for CaM within the complex. To test the effectiveness of the solvent matching, the molecular mass of the scattering particle for these data were determined using Kratky analysis. The volume and corresponding molecular mass, Mr, value obtained was 16,400 Da (using the correction factors from Fisher et al. (36) to account for the finite q-range measured), in excellent agreement with the expected 16,700 Da based on amino-acid sequence.
The Guinier- and P(r)-derived Rg values from the 44% D2O data (20.9 ± 1.3 Å and 20.7 ± 1.0 Å, respectively) are in excellent agreement with each other, and lie in between the values expected for the fully extended dumbbell shape of the crystal structure (23 Å, from CRYSOL calculation with PDB:1CLL) and the compact version of CaM when complexed with the classical helical binding target skeletal muscle myosin light chain kinase (skMLCK, Rg ∼17 Å from both SANS data (37) and the NMR structure (22)). The P(r) determined from the 44% D2O data (Fig. 1 b) also has an asymmetric shape with a peak at ∼20 Å and a Dmax of 64 Å, characteristic of a mostly, but not fully extended, dumbbell-shaped CaM.
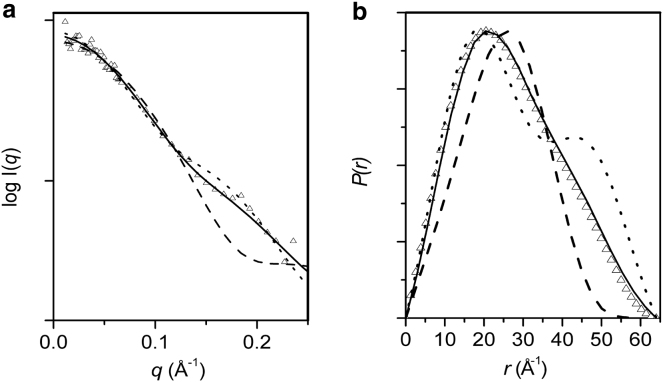
Our previous investigations (27) and those of Ghanam et al. (28) have shown that the tryptophans in MA, W16, and W36, interact directly with CaM in the hydrophobic clefts of each CaM lobe indicating bidentate-like binding of CaM to MA. Recently, the NMR structure of CaM bound to its target sequence in Munc13 has been determined (38, 39). The structure reveals that the hydrophobic cleft in each Ca2+-binding lobe of CaM binds one of two tryptophans that, like the CaM-binding tryptophans in MA, are widely separated in sequence (by 24 amino acids in Munc13 compared to 19 in MA.) This binding facilitates CaM adopting a semiextended conformation with the Munc13 peptide, although in this complex it appears that, as for uncomplexed CaM in solution, there is flexibility in CaM's interconnecting helix region. We compared the scattering profile for CaM in the CaM-MA complex with that of a semiextended CaM in its complex with the Munc13 peptide and that of the classical collapsed CaM binding mode induced by shorter helical target sequences. Note that the latter typically contain one tryptophan and an additional large hydrophobic residue spaced by 12 amino acids (as in skMLCK (22).
The Rg value of CaM in the complex with MA is within experimental error of that calculated for the semiextended CaM from the CaM-Munc13 peptide complex (20.7 ± 1.0 Å, coordinates taken from the first structure in the NMR ensemble in PDB:2KDU; compare to ∼21 Å, Table 1). The predicted scattering profile calculated from the atomic coordinates of the semiextended CaM also provides an overall superior fit to the 44% D2O data (χ = 0.5) when compared to either the fully extended CaM structure (PDB:1CLL, χ = 0.6) or the collapsed form observed on binding to a skMLCK peptide (PDB:2BBM (CaM), χ = 0.7) (Fig. 2 a). The χ-values for these fits are all <1, indicating the propagated statistical errors are an overestimate of the errors in the scattering data for fitting a smooth function. Nonetheless, consistent with the trends in χ, visual inspection of the fits affirms that the semiextended CaM is superior; discrepancies in the fits of the fully extended or collapsed conformations of CaM to the SANS data are evident in the mid-to-high q-region (>0.1 Å−1) that is most impacted by the spatial disposition of the globular lobes. Further, the expected differences are observed in the P(r) profiles for these different degrees of CaM extension (Fig. 2 b). It thus appears that the CaM lobes reorient in such a way that, when engaging with W16 and W36 of MA, the overall conformation of CaM adopts a semiextended binding mode very similar to that observed in the CaM-Munc13 ensemble.
Figure 2.
Comparisons of CaM in CaM-MA with CaM-peptide structures. (a) 44% D2O I(q) versus q for CaM-MA (▵) with CRYSON fits of the crystal structures of CaM (PDB:1CLL) (dotted line), and CaM taken from its structures in its complexes with Munc13 (PDB:2KDU) (solid line) and skMLCK (PDB:2BBM) (dashed line). (b) P(r) versus r for the experimental and modeled profiles shown in panel a using the same key.
The solvent-matching strategy that allowed for the analysis of the CaM structural parameters was not applicable to the MA component of the complex because the theoretical solvent match point for the 73% deuterated CaM was somewhat greater than 100% D2O. The alternate strategy of developing a shape model for the complex, however, provides information about the MA structure and its disposition with respect to CaM in the complex.
The semiextended CaM binds a highly extended MA in an open-clamp conformation
The multiphase bead modeling method (MONSA, see Materials and Methods) was used to simultaneously fit the full set of contrast variation data (SANS and SAXS) and produce a shape model for the CaM-MA complex (Fig. 3). The modeling calculations were repeated six times and fits to the experimental data indicate good agreement, within the propagated statistical errors (Fig. 1 a; χ-values for the fits shown are 1.0, 0.6, 1.0, 0.9, 1.0 for the 19%, 44%, 88%, 100% D2O SANS and SAXS data, respectively; Table S1 in the Supporting Material gives χ-values for fits from all six calculations, each of which yielded models with the same features). We note the relatively poor fit of the MONSA model to the lowest-q region (<0.03 Å−1) of the 88% D2O SANS data. This effect is attributable to the fact that the MONSA model assumes the CaM and MA components are uniform density objects. However, at contrast points close to the solvent match point of a component, internal density fluctuations become significant. For the 88% D2O data, the CAM-MA has very low contrast and the assumption of uniform density starts to break down.
Figure 3.
MONSA-derived shape model for CaM-MA.
CaM in the MONSA model has an Rg of 20 Å and Dmax equal to ∼64 Å in good agreement, within experimental error, with the values determined by Guinier or P(r) from the 44% D2O SANS data. For the MA in the MONSA model, the Rg value is 36 Å, which is significantly greater that the value for the complex as a whole, and its Dmax is equal to that of the complex and ∼70% greater than that for CaM. In comparison, the Rg and Dmax values for free MA in solution measured by SAXS (Rg = 19.9, Dmax = 70 Å) (30) indicate a much more compact structure consistent with the crystal (24) and NMR (25) structures that show a globular N-terminal domain with an unstructured or disordered C-terminal tail. This result is consistent with earlier evidence that CaM-MA complex formation is accompanied by a dramatic loss of tertiary contacts within MA (27, 28).
The MONSA model for the CaM-MA complex shows an extended complex with one dimension significantly longer that the other two. CaM has a semiextended dumbbell shape and binds toward one end of the relatively narrow, bent, and highly extended MA. There appear to be extensive contacts between CaM and MA as their longest dimensions are approximately aligned such that the full length of the semiextended CaM spans 70% of the length of MA.
Dissociation of the CaM-MA complex results in restoration of the native, uncomplexed MA structure
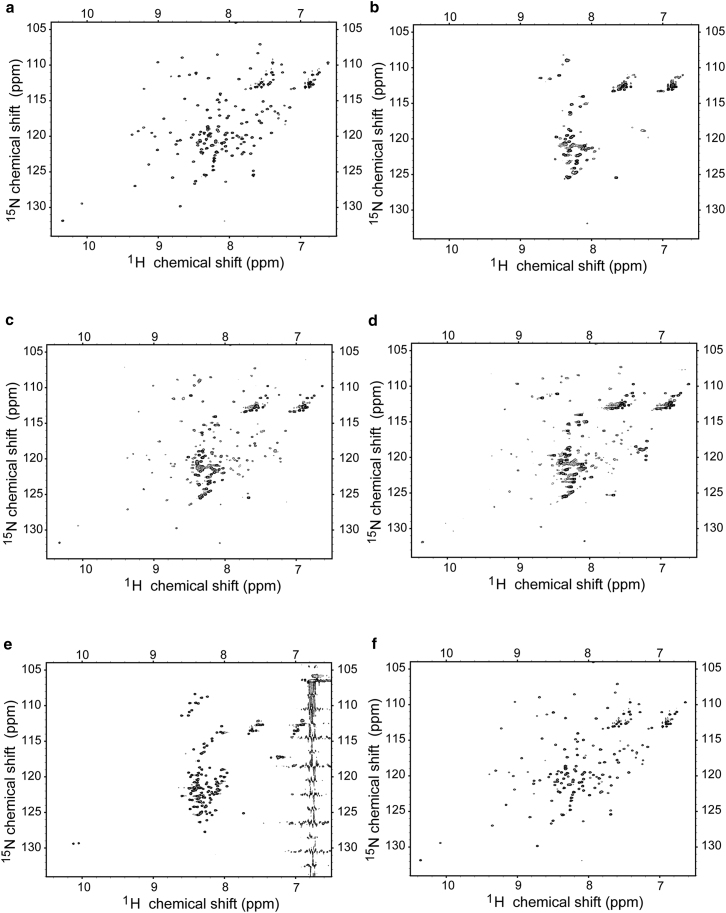
The two-dimensional 1H-15N-HSQC spectra recorded for 15N-MA free and in complex with unlabeled CaM are very similar to previously published spectra recorded for myristoylated (28) or unmyristoylated (25) MA. In particular, many peaks in the MA spectrum disappeared or moved to the region of the spectrum typically associated with less structured residues upon binding to CaM (Fig. 4, a and b), consistent with the loss of tertiary contacts that would be expected for MA to form the narrow and highly elongated structure as characterized in the SANS experiment on the complex. Our previous studies indicated that this loss of tertiary contacts is associated with only some loss of the predominantly helical structure in MA (circular dichroism data indicate free MA has 53 ± 3% helical structure compared to an estimated 41 ± 2% in the complex (27)).
Figure 4.
Two-dimensional 15N-1H-HSQC spectra from CaM-MA. (a) 15N-MA only; (b) 15N-MA-CaM complex in 10 mM NaPO4 (pH 5.5), 5 mM CaCl2, 1 mM TCEP; (c and d) the 15N-MA-CaM complex after titration with either KCl (to 500 mM) or EGTA (10 mM), respectively, showing many peaks corresponding to folded MA have been restored; (e) 15N-MA in 6 M GuHCl, and (f) 15N-MA after removal of 6 M GuHCl.
To test whether the unfolding of MA upon binding to CaM is reversible, the complex was titrated with either the Ca2+-chelator EGTA (0–10 mM) or by increasing ionic strength of the solution (KCl, 0–500 mM). EGTA is known to remove Ca2+ from CaM at μM concentrations with a resultant closure of the hydrophobic clefts in each of the globular lobes that are key to target recognition and binding. KCl additions were chosen, as we have reported previously that increasing salt concentrations decreases the strength of the CaM-MA interaction (27). With increasing KCl, there is a gradual return of the 15N-HSQC spectrum to that observed for free 15N-MA with a large proportion of peaks reappearing at 500 mM KCl (Fig. 4 c). A similar result was obtained upon addition of EGTA (Fig. 4 d).
To ascertain the degree of MA unfolding by CaM, a two-dimensional 1H-15N-HSQC spectrum of 15N-MA in 6 M guanidine hydrochloride (GuHCl) (Fig. 4 e) was compared to that recorded on 15N-MA complexed with CaM (no salt). The comparison reveals a distinct pattern of unfolding for these two conditions (Fig. 4, b and e). The GuHCl-induced unfolding appears to be more complete as the HSQC spectrum for this condition displays many more sharp peaks when compared with the spectrum for the CaM-MA complex. In both cases, all observable peaks show little dispersion and are located in the region of the spectrum associated with unstructured and solvent exposed residues. The spectral features are consistent with the fact that the previous circular dichroism data indicate that MA bound to CaM appears to retain a significant portion of its helical structure (27). Upon redialysis into a buffer without GuHCl, MA again was able to adopt its original conformation (Fig. 4 f).
As observed previously in Chow et al. (27), signals for the indole rings of both tryptophan residues (W16 and W36), disappear when MA is bound to CaM. Upon dissociation of MA from CaM, either by addition of KCl or EGTA, the peak corresponding to at least one tryptophan residue in MA (W16) returns to its original peak position. The NMR data indicate that in the presence of 500 mM KCl or 10 mM EGTA, some MA remains in complex with CaM. This result is consistent with previous tryptophan fluorescence titration results that indicated weak and similar binding constants in either 500 mM KCl or 10 mM EGTA (approximately mM compared to the Kd of ∼170 nM determined in the presence of saturating Ca2+ and low salt (27)).
Discussion and Conclusions
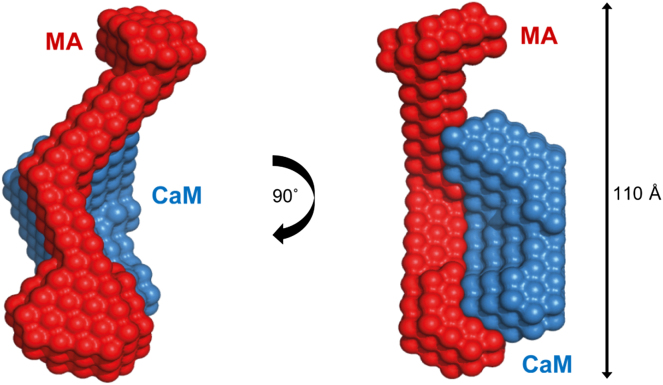
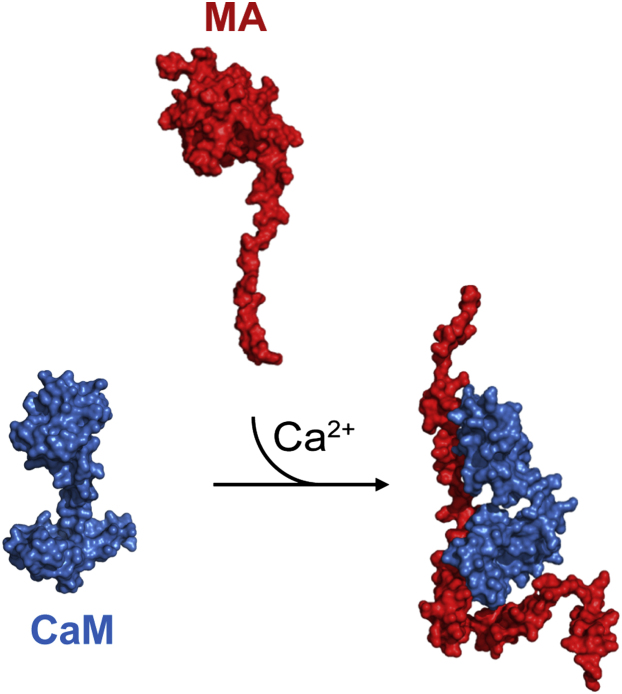
The contrast variation experiments described here take advantage of the different x-ray and neutron scattering properties of hydrogen and deuterium to reveal, to our knowledge for the first time, the shapes and dispositions of the CaM and MA components in the CaM-MA complex. A schematic representation of the CaM-MA complex derived from all of the available structural evidence is presented in Fig. 5 (see the Supporting Material and Fig. S2 for details of the construction of the schematic model). The scattering data show CaM adopts an overall semiextended conformation that can be described as an open-clamp that structurally disrupts the tertiary fold of MA as it undergoes a dramatic structural extension due to the loss of its native tertiary contacts. The MA protein within the CaM-MA complex appears as a long snakelike shape with no evident globular domain that bears any similarity to that observed in its uncomplexed state.
Figure 5.
Schematic of CaM-MA interaction. In the presence of Ca2+ and at low ionic strength, CaM binds MA in an open-clamp, semiextended bidentate conformation resulting in a dramatic extension of MA. Release of MA, which can be induced either by increasing ionic strength or loss of Ca2+, results in reformation of the native, free MA structure.
It appears that MA retains a significant proportion of its helical structure upon CaM binding, which would be in line with CaM-binding sequences generally. Furthermore, based on our NMR data, modulations in Ca2+ concentration and/or ionic strength alter the degree of CaM-MA association and MA displays an inherent structural pliancy that facilitates the protein spontaneously refolding to its native structure upon disassociation from CaM. Consequently, these two small proteins constitute a structurally dynamic system that is sensitive to alterations in environmental conditions. By inference, the formation or disassociation of the CaM-MA complex could be modulated by Ca2+ signaling pathways within a cell.
In the structure of uncomplexed MA (25, 26), the tryptophans W16 and W36 are positioned in the first two α-helices (α1, α2) of the MA globular domain that are linked by a long, basic β-turn-β motif (residues 17–31) that results in an approximately perpendicular arrangement of α1 with respect to α2 with only a 10–15 Å spacing between the tryptophans (see Fig. S3). These two helices are key structural elements in forming the hydrophobic core of free MA (25, 26). The indole ring of W16 is deeply buried within the hydrophobic core that also accommodates the N-terminal myristoyl group (28). One face of the W36 indole ring packs against the hydrophobic core whereas the other face forms part of a highly conserved surface/solvent-exposed cleft that accepts PI(4,5)P2.
The basic β-turn-β motif also forms part of the PI(4,5)P2 binding pocket (16). The binding of CaM to MA via interactions with W16 and W36 is thus expected to significantly perturb the interactions that stabilize the MA hydrophobic core. The dramatic extension evident from the SANS data implies a concomitant reorientation of α1 relative to α2 either by distorting or completely disrupting the β-turn-β motif so that the tryptophans can interact with the hydrophobic clefts in the two separated CaM lobes (requiring an ∼40 Å separation distance for the tryptophans). The consequent impacts on the entire tertiary fold of MA, as observed in our NMR data, are dramatic. The disruption to the hydrophobic core and the topology of the W36/β-turn-β surface cleft are expected to have significant consequences on both myristoylate exposure and PI(4,5)P2 binding (the two key features of MA that facilitate Gag-localization to the plasma membrane). Intriguingly, the reverse process may also be affected, i.e., the release of MA from the matrix into the cytoplasm on viral fusion with the plasma membrane.
The amino-acid sequence between the first and second tryptophans of MA (that includes the positively charged β-turn-β element with the consensus sequence K(V/I)RLRP(X)GKK(X)Y(X)L) could be important for maintaining the extended conformation of CaM. This very short basic region of MA has been implicated in PI(4,5)P2 binding (16) as well as RNA (14) and DNA (40) binding, Gag processing (41), and Env incorporation (42, 43). The 19-amino-acid spacing between the two tryptophans in HIV-1 MA is fixed across HIV1, HIV2, and simian immunodeficiency virus although the first tryptophan at position 16 can be substituted for other hydrophobic residues including L, F, and Y. The physical constraint of having two widely spaced interaction sites for CaM binding, that to date has only been superseded by the 24-amino-acid spacer of Munc13, would in itself help CaM to clamp to MA in an extended conformation. It is interesting to note that SAXS (10) and NMR (44) data acquired from CaM-MA peptide complexes with sequence segments from MA that contain only one of the two CaM-binding tryptophans reveal two distinct CaM binding modes. Peptides containing W16 bind CaM in an extended conformation whereas peptides containing W36 bind CaM in its collapsed conformation similar to the classical binding mode originally observed for skMLCK (22).
In both cases, the tryptophan (either W16 or W36) interacts with the C-terminal lobe of CaM. We therefore hypothesize that the identity of amino acids directly upstream of the first CaM binding site in MA (position 16) is important in facilitating the extended CaM conformation that we observe in the CaM-MA complex. The inclusion of the K(V/I)RLRP(X)G consensus sequence of alternating hydrophobic and charged residues of the β-turn-β element upstream from W16 could allow MA to interact with the C-terminal domain of CaM in such a way as to correctly orient the N-terminal lobe for an interaction with the more-distant distant W36. At the same time a perturbation in the β-turn-β region that causes the angle between α1 and α2 to open could ultimately unravel MA as indicated by our scattering and NMR data.
There have been differing interpretations published concerning the nature of the stabilizing interactions in the CaM-MA complex. Our combined studies, as well as those of Izumi et al. (45), indicate that interactions with the CaM hydrophobic clefts and the tryptophans of MA as well as ionic interactions play a role in the interaction between the two proteins. Others have concluded that ionic interactions are not significant (28). For our SAXS and SANS studies, the CaM-MA complex could only be isolated in pure form (by size exclusion chromatography) and in sufficient quantities using very low ionic-strength solution conditions. Our pull-down assays (that are affected by successive rounds of washing) indicate that the complex has an increased propensity to dissociate at higher ionic strength. Our NMR data also show evidence for increased MA dissociation from CaM with increasing KCl concentrations, although even at 500 mM KCl there is some residual complex. Our combined observations, thus, are consistent with ionic interactions playing a significant role in the formation of the complex. It is interesting to speculate as to whether this dependence might have some biological relevance, given the changes in potassium and sodium levels over the course of the HIV infection cycle (46).
Acknowledgments
Deuterated CaM was produced using the facilities and expertise of the National Deuteration Facility at the Australian Nuclear Science and Technology Organisation, including Dr. Vanessa Lake's assistance with subcloning of CaM. The mass spectrometry data were obtained using the facilities of Sydney University Proteome Research Unit. We thank Dr. Ben Crossett for his assistance with the mass spectrometry data acquisition and analysis and Dr. Robert Knott for assistance with the SAXS data collection and reduction.
This work was supported by National Institutes of Health grant No. P50 GM082545.
Editor: Lois Pollack.
Footnotes
William A. Hamilton's present address is Instrument and Source Development Division, Oak Ridge National Laboratory, Oak Ridge, TN.
Cy M. Jeffries's present address is Bragg Institute, Australian Nuclear Science and Technology Organisation, New South Wales, Australia.
One table and three figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(12)00731-X.
Supporting Material
References
- 1.Chin D., Means A.R. Calmodulin: a prototypical calcium sensor. Trends Cell Biol. 2000;10:322–328. doi: 10.1016/s0962-8924(00)01800-6. [DOI] [PubMed] [Google Scholar]
- 2.Yamniuk A.P., Vogel H.J. Calmodulin's flexibility allows for promiscuity in its interactions with target proteins and peptides. Mol. Biotechnol. 2004;27:33–57. doi: 10.1385/MB:27:1:33. [DOI] [PubMed] [Google Scholar]
- 3.Brown J.N., Kohler J.J., et al. Goodenow M.M. HIV-1 activates macrophages independent of Toll-like receptors. PLoS ONE. 2008;3:e3664. doi: 10.1371/journal.pone.0003664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsubara M., Jing T., et al. Hayashi N. Myristoyl moiety of HIV Nef is involved in regulation of the interaction with calmodulin in vivo. Protein Sci. 2005;14:494–503. doi: 10.1110/ps.04969605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller M.A., Mietzner T.A., et al. Montelaro R.C. Identification of a calmodulin-binding and inhibitory peptide domain in the HIV-1 transmembrane glycoprotein. AIDS Res. Hum. Retroviruses. 1993;9:1057–1066. doi: 10.1089/aid.1993.9.1057. [DOI] [PubMed] [Google Scholar]
- 6.Micoli K.J., Pan G., et al. McDonald J.M. Requirement of calmodulin binding by HIV-1 gp160 for enhanced FAS-mediated apoptosis. J. Biol. Chem. 2000;275:1233–1240. doi: 10.1074/jbc.275.2.1233. [DOI] [PubMed] [Google Scholar]
- 7.Sham S.W., McDonald J.M., et al. Krishna N.R. Solution structure of a calmodulin-binding domain in the carboxy-terminal region of HIV type 1 gp160. AIDS Res. Hum. Retroviruses. 2008;24:607–616. doi: 10.1089/aid.2007.0202. [DOI] [PubMed] [Google Scholar]
- 8.Srinivas S.K., Srinivas R.V., et al. Segrest J.P. Cytosolic domain of the human immunodeficiency virus envelope glycoproteins binds to calmodulin and inhibits calmodulin-regulated proteins. J. Biol. Chem. 1993;268:22895–22899. [PubMed] [Google Scholar]
- 9.Radding W., Pan Z.Q., et al. McDonald J.M. Expression of HIV-1 envelope glycoprotein alters cellular calmodulin. Biochem. Biophys. Res. Commun. 1996;218:192–197. doi: 10.1006/bbrc.1996.0034. [DOI] [PubMed] [Google Scholar]
- 10.Radding W., Williams J.P., et al. McDonald J.M. Calmodulin and HIV type 1: interactions with Gag and Gag products. AIDS Res. Hum. Retroviruses. 2000;16:1519–1525. doi: 10.1089/088922200750006047. [DOI] [PubMed] [Google Scholar]
- 11.Ganser-Pornillos B.K., Yeager M., Sundquist W.I. The structural biology of HIV assembly. Curr. Opin. Struct. Biol. 2008;18:203–217. doi: 10.1016/j.sbi.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mervis R.J., Ahmad N., et al. Venkatesan S. The Gag gene products of human immunodeficiency virus type 1: alignment within the Gag open reading frame, identification of posttranslational modifications, and evidence for alternative Gag precursors. J. Virol. 1988;62:3993–4002. doi: 10.1128/jvi.62.11.3993-4002.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chukkapalli V., Hogue I.B., et al. Ono A. Interaction between the human immunodeficiency virus type 1 Gag matrix domain and phosphatidylinositol-(4,5)-bisphosphate is essential for efficient Gag membrane binding. J. Virol. 2008;82:2405–2417. doi: 10.1128/JVI.01614-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chukkapalli V., Oh S.J., Ono A. Opposing mechanisms involving RNA and lipids regulate HIV-1 Gag membrane binding through the highly basic region of the matrix domain. Proc. Natl. Acad. Sci. USA. 2010;107:1600–1605. doi: 10.1073/pnas.0908661107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ono A., Ablan S.D., et al. Freed E.O. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc. Natl. Acad. Sci. USA. 2004;101:14889–14894. doi: 10.1073/pnas.0405596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saad J.S., Miller J., et al. Summers M.F. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc. Natl. Acad. Sci. USA. 2006;103:11364–11369. doi: 10.1073/pnas.0602818103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bukrinskaya A. HIV-1 matrix protein: a mysterious regulator of the viral life cycle. Virus Res. 2007;124:1–11. doi: 10.1016/j.virusres.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 18.Fiorentini S., Marini E., et al. Caruso A. Functions of the HIV-1 matrix protein p17. New Microbiol. 2006;29:1–10. [PubMed] [Google Scholar]
- 19.Babu Y.S., Sack J.S., et al. Cook W.J. Three-dimensional structure of calmodulin. Nature. 1985;315:37–40. doi: 10.1038/315037a0. [DOI] [PubMed] [Google Scholar]
- 20.Vigil D., Gallagher S.C., et al. García A.E. Functional dynamics of the hydrophobic cleft in the N-domain of calmodulin. Biophys. J. 2001;80:2082–2092. doi: 10.1016/S0006-3495(01)76182-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoeflich K.P., Ikura M. Calmodulin in action: diversity in target recognition and activation mechanisms. Cell. 2002;108:739–742. doi: 10.1016/s0092-8674(02)00682-7. [DOI] [PubMed] [Google Scholar]
- 22.Ikura M., Clore G.M., et al. Bax A. Solution structure of a calmodulin-target peptide complex by multidimensional NMR. Science. 1992;256:632–638. doi: 10.1126/science.1585175. [DOI] [PubMed] [Google Scholar]
- 23.Meador W.E., Means A.R., Quiocho F.A. Modulation of calmodulin plasticity in molecular recognition on the basis of x-ray structures. Science. 1993;262:1718–1721. doi: 10.1126/science.8259515. [DOI] [PubMed] [Google Scholar]
- 24.Hill C.P., Worthylake D., et al. Sundquist W.I. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc. Natl. Acad. Sci. USA. 1996;93:3099–3104. doi: 10.1073/pnas.93.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Massiah M.A., Starich M.R., et al. Sundquist W.I. Three-dimensional structure of the human immunodeficiency virus type 1 matrix protein. J. Mol. Biol. 1994;244:198–223. doi: 10.1006/jmbi.1994.1719. [DOI] [PubMed] [Google Scholar]
- 26.Matthews S., Barlow P., et al. Campbell I. Refined solution structure of p17, the HIV matrix protein. Biochem. Soc. Trans. 1995;23:725–729. doi: 10.1042/bst0230725. [DOI] [PubMed] [Google Scholar]
- 27.Chow J.Y., Jeffries C.M., et al. Trewhella J. Calmodulin disrupts the structure of the HIV-1 MA protein. J. Mol. Biol. 2010;400:702–714. doi: 10.1016/j.jmb.2010.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghanam R.H., Fernandez T.F., et al. Saad J.S. Binding of calmodulin to the HIV-1 matrix protein triggers myristate exposure. J. Biol. Chem. 2010;285:41911–41920. doi: 10.1074/jbc.M110.179093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Middelberg A.P., O'Neill B.K., et al. Snoswell M.A. A novel technique for the measurement of disruption in high-pressure homogenization: studies on E. coli containing recombinant inclusion bodies. Biotechnol. Bioeng. 1991;38:363–370. doi: 10.1002/bit.260380406. [DOI] [PubMed] [Google Scholar]
- 30.Gilbert E.P., Schulz J.C., Noakes T.J. “QUOKKA”—the small-angle neutron scattering instrument at OPAL. Physica B. 2006;385–386:1180–1182. [Google Scholar]
- 31.Hamilton W.A., Garrett R., Gilbert E.P. QUOKKA: the small-angle neutron scattering instrument. Neutron News. 2009;20:24–28. [Google Scholar]
- 32.Kline S.R. Reduction and analysis of SANS and USANS data using IGOR Pro. J. Appl. Crystallogr. 2006;39:895–900. [Google Scholar]
- 33.Orthaber D., Bergmann A., Glatter O. SAXS experiments on absolute scale with Kratky systems using water as a secondary standard. J. Appl. Crystallogr. 2000;3:218–225. [Google Scholar]
- 34.Petoukhov M.V., Franke D., et al. Svergun D.I. New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 2012;45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whitten A.E., Cai S., Trewhella J. MULCh: ModULes for the analysis of small-angle neutron contrast variation from biomolecular assemblies. J. Appl. Crystallogr. 2008;41:222–226. [Google Scholar]
- 36.Fisher H., de Oliveira Neto M., et al. Craievich A.F. Determination of the molecular weight of proteins in solution from a single small-angle x-ray scattering measurement on a relative scale. J. Appl. Crystallogr. 2010;43:101–109. [Google Scholar]
- 37.Heidorn D.B., Seeger P.A., et al. Trewhella J. Changes in the structure of calmodulin induced by a peptide based on the calmodulin-binding domain of myosin light chain kinase. Biochemistry. 1989;28:6757–6764. doi: 10.1021/bi00442a032. [DOI] [PubMed] [Google Scholar]
- 38.Rodríguez-Castañeda F., Coudevylle N., et al. Griesinger C. 1H, 13C and 15N resonance assignments of the Calmodulin-Munc13-1 peptide complex. Biomol. NMR Assign. 2010;4:45–48. doi: 10.1007/s12104-009-9204-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodríguez-Castañeda F., Maestre-Martínez M., et al. Griesinger C. Modular architecture of Munc13/calmodulin complexes: dual regulation by Ca2+ and possible function in short-term synaptic plasticity. EMBO J. 2010;29:680–691. doi: 10.1038/emboj.2009.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cai M., Huang Y., et al. Clore G.M. Structural basis of the association of HIV-1 matrix protein with DNA. PLoS ONE. 2012;5:e15675. doi: 10.1371/journal.pone.0015675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fouchier R.A., Meyer B.E., et al. Malim M.H. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J. 1997;16:4531–4539. doi: 10.1093/emboj/16.15.4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhatia A.K., Campbell N., et al. Ratner L. Characterization of replication defects induced by mutations in the basic domain and C-terminus of HIV-1 matrix. Virology. 2007;369:47–54. doi: 10.1016/j.virol.2007.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Freed E.O., Englund G., Martin M.A. Role of the basic domain of human immunodeficiency virus type 1 matrix in macrophage infection. J. Virol. 1995;69:3949–3954. doi: 10.1128/jvi.69.6.3949-3954.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Samal A.B., Ghanam R.H., et al. Saad J.S. NMR, biophysical, and biochemical studies reveal the minimal Calmodulin binding domain of the HIV-1 matrix protein. J. Biol. Chem. 2011;286:33533–33543. doi: 10.1074/jbc.M111.273623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Izumi Y., Watanabe H., et al. Hayashi N. Solution x-ray scattering reveals a novel structure of calmodulin complexed with a binding domain peptide from the HIV-1 matrix protein p17. Biochemistry. 2008;47:7158–7166. doi: 10.1021/bi702416b. [DOI] [PubMed] [Google Scholar]
- 46.Voss T.G., Fermin C.D., et al. Garry R.F. Alteration of intracellular potassium and sodium concentrations correlates with induction of cytopathic effects by human immunodeficiency virus. J. Virol. 1996;70:5447–5454. doi: 10.1128/jvi.70.8.5447-5454.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.