Abstract
The title compound [systematic name: 9-ethyl-13-hydroxy-14-methyl-2-(3,4,5-trimethoxy-6-methyltetrahydro-2H-pyran-2-yloxy)-3,3a,5b,6,9,10,11,12,13,14,16a,16b-dodecahydro-1H-as-indaceno[3,2-d][1]oxacyclododecine-7,15(2H,5aH)-dione], C33H50O9, was obtained by hydrolysis of Spinosyn A. The fused cyclopentene ring adopts a twisted conformation, while the fused cyclohexene and cyclopentane rings are in envelope conformations with the same C atom at the flaps. In the crystal, molecules are linked by O—H⋯O and C—H⋯O hydrogen bonds into a layer parallel to the ab plane.
Related literature
For the insecticidal activity and research background of Spinosyn, see: Sparks et al. (2008 ▶); Thompson et al. (2000 ▶); Salgado et al. (1998 ▶). For the structure of Spinosyn A, see: Evans & Black (1993 ▶).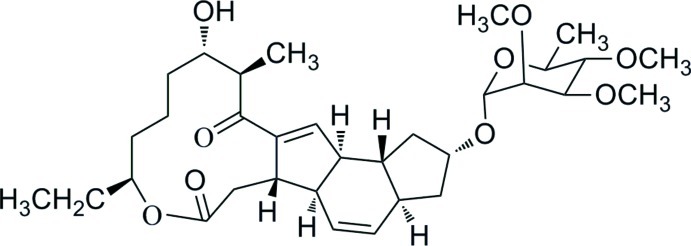
Experimental
Crystal data
C33H50O9
M r = 590.73
Orthorhombic,
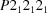
a = 8.7776 (15) Å
b = 8.7959 (15) Å
c = 41.737 (7) Å
V = 3222.4 (10) Å3
Z = 4
Mo Kα radiation
μ = 0.09 mm−1
T = 153 K
0.34 × 0.27 × 0.08 mm
Data collection
Rigaku AFC10/Saturn724+ diffractometer
25322 measured reflections
4876 independent reflections
4194 reflections with I > 2σ(I)
R int = 0.047
Refinement
R[F 2 > 2σ(F 2)] = 0.053
wR(F 2) = 0.134
S = 1.00
4876 reflections
389 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.75 e Å−3
Δρmin = −0.22 e Å−3
Data collection: CrystalClear (Rigaku, 2008 ▶); cell refinement: CrystalClear; data reduction: CrystalClear; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: CrystalStructure (Rigaku/MSC, 2009 ▶); software used to prepare material for publication: CrystalStructure.
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536812028851/is5144sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812028851/is5144Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O16—H16O⋯O18i | 0.92 (4) | 1.96 (3) | 2.840 (3) | 160 (3) |
| C3—H3B⋯O10ii | 0.99 | 2.47 | 3.310 (3) | 142 |
Symmetry codes: (i) 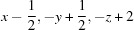 ; (ii)
; (ii) 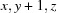 .
.
Acknowledgments
The authors thank Beijing Institute of Technology for the X-ray diffraction analysis.
supplementary crystallographic information
Comment
Spinosyns, a novel class of green pesticides, were characterized with high efficiency, fast degradation (Sparks et al., 2008), low toxicity (Thompson et al., 2000) and safety to environment (Salgado et al., 1998). Inspired by the high and broad insecticidal activity of Spinosyns and continuing our interest in its structure modification, we obtained pseudoaglycone of Spinosyn A from hydrolysis of the amino sugar forosamine. Here, we report the crystal structure of the title compound (Fig. 1).
Experimental
A solution of 2.0 g Spinosyn A (purchased from SHANGHAI HOHANCE GROUP, 98%) and 5eq H2SO4 was heated to 80 °C in ethanol (20 ml) for 2 h. The reaction mixture was cooled to room temperature and then filtered to give the title compound. The product was recrystallizated from ethanol to give colourless crystalline powder (m.p. 441–443 K).
Refinement
C-bound H atoms were included in a riding model approximation with C—H distances 0.95–1.00 Å, and with Uiso(H) = 1.2Ueq(C). The H atom of OH group was located in a difference Fourrier map and refined freely [O—H = 0.92 (4) Å]. The absolute configuration was determined according to the structure of Spinosyn A (Evans & Black, 1993).
Figures
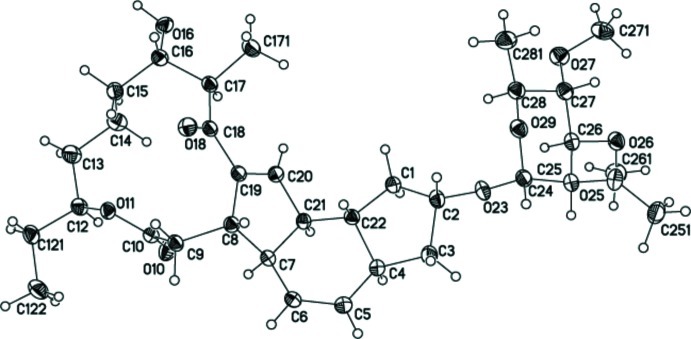
Fig. 1.
The molecular structure of the title compound with the atom numbering scheme. Displacement ellipsoids are drawn at the 30% probability level.
Crystal data
| C33H50O9 | F(000) = 1280 |
| Mr = 590.73 | Dx = 1.218 Mg m−3 |
| Orthorhombic, P212121 | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: P 2ac 2ab | Cell parameters from 11776 reflections |
| a = 8.7776 (15) Å | θ = 2.4–29.1° |
| b = 8.7959 (15) Å | µ = 0.09 mm−1 |
| c = 41.737 (7) Å | T = 153 K |
| V = 3222.4 (10) Å3 | Platelet, colorless |
| Z = 4 | 0.34 × 0.27 × 0.08 mm |
Data collection
| Rigaku AFC10/Saturn724+ diffractometer | 4194 reflections with I > 2σ(I) |
| Radiation source: Rotating Anode | Rint = 0.047 |
| Graphite monochromator | θmax = 29.1°, θmin = 2.4° |
| Detector resolution: 28.5714 pixels mm-1 | h = −10→12 |
| φ and ω scans | k = −11→12 |
| 25322 measured reflections | l = −57→43 |
| 4876 independent reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.053 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.134 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.00 | w = 1/[σ2(Fo2) + (0.0696P)2 + 0.586P] where P = (Fo2 + 2Fc2)/3 |
| 4876 reflections | (Δ/σ)max = 0.001 |
| 389 parameters | Δρmax = 0.75 e Å−3 |
| 0 restraints | Δρmin = −0.22 e Å−3 |
Special details
| Experimental. Spectral data: IR (KBr): 3517, 2968, 2930, 1624, 1650, 1606, 1457, 1379, cm-1; MS (MALDI-TOF) m/z: [M+Na]+ 613.3. |
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O10 | 0.5636 (3) | 0.0686 (2) | 0.89590 (4) | 0.0513 (6) | |
| O11 | 0.5456 (2) | 0.04001 (18) | 0.94906 (4) | 0.0318 (4) | |
| O16 | −0.0506 (2) | 0.1699 (2) | 0.96650 (5) | 0.0425 (5) | |
| O18 | 0.3702 (2) | 0.4082 (2) | 0.96960 (4) | 0.0375 (4) | |
| O23 | 0.2851 (2) | 0.94913 (19) | 0.81098 (4) | 0.0368 (4) | |
| O25 | 0.2547 (2) | 1.3408 (2) | 0.78898 (4) | 0.0412 (5) | |
| O26 | 0.0084 (3) | 1.2315 (2) | 0.75097 (5) | 0.0403 (4) | |
| O27 | −0.1883 (2) | 1.1350 (2) | 0.80154 (5) | 0.0455 (5) | |
| O29 | 0.1799 (2) | 1.1452 (2) | 0.84138 (4) | 0.0357 (4) | |
| C1 | 0.3003 (3) | 0.7119 (3) | 0.84098 (6) | 0.0350 (5) | |
| H1A | 0.2102 | 0.7109 | 0.8553 | 0.042* | |
| H1B | 0.2732 | 0.6649 | 0.8202 | 0.042* | |
| C2 | 0.3627 (3) | 0.8721 (3) | 0.83662 (6) | 0.0340 (5) | |
| H2 | 0.3465 | 0.9303 | 0.8569 | 0.041* | |
| C3 | 0.5355 (3) | 0.8560 (3) | 0.83072 (7) | 0.0386 (6) | |
| H3A | 0.5623 | 0.8882 | 0.8087 | 0.046* | |
| H3B | 0.5940 | 0.9182 | 0.8462 | 0.046* | |
| C4 | 0.5683 (3) | 0.6857 (3) | 0.83553 (6) | 0.0319 (5) | |
| H4 | 0.5598 | 0.6341 | 0.8143 | 0.038* | |
| C5 | 0.7166 (3) | 0.6402 (3) | 0.85043 (7) | 0.0386 (6) | |
| H5 | 0.8054 | 0.6996 | 0.8468 | 0.046* | |
| C6 | 0.7238 (3) | 0.5161 (3) | 0.86879 (6) | 0.0358 (5) | |
| H6 | 0.8196 | 0.4910 | 0.8779 | 0.043* | |
| C7 | 0.5914 (3) | 0.4131 (3) | 0.87605 (6) | 0.0313 (5) | |
| H7 | 0.6175 | 0.3089 | 0.8681 | 0.038* | |
| C8 | 0.5579 (3) | 0.4018 (3) | 0.91260 (6) | 0.0295 (5) | |
| H8 | 0.5880 | 0.5004 | 0.9227 | 0.035* | |
| C9 | 0.6420 (3) | 0.2733 (3) | 0.93019 (6) | 0.0307 (5) | |
| H9A | 0.7513 | 0.2766 | 0.9243 | 0.037* | |
| H9B | 0.6345 | 0.2907 | 0.9536 | 0.037* | |
| C10 | 0.5798 (3) | 0.1174 (3) | 0.92253 (6) | 0.0315 (5) | |
| C12 | 0.5017 (3) | −0.1199 (3) | 0.94565 (7) | 0.0357 (5) | |
| H12 | 0.4991 | −0.1471 | 0.9224 | 0.043* | |
| C13 | 0.3448 (4) | −0.1438 (3) | 0.95999 (9) | 0.0489 (7) | |
| H13A | 0.3546 | −0.1466 | 0.9836 | 0.059* | |
| H13B | 0.3063 | −0.2442 | 0.9530 | 0.059* | |
| C14 | 0.2249 (4) | −0.0210 (3) | 0.95098 (8) | 0.0449 (7) | |
| H14A | 0.2574 | 0.0328 | 0.9313 | 0.054* | |
| H14B | 0.1254 | −0.0701 | 0.9467 | 0.054* | |
| C15 | 0.2085 (3) | 0.0922 (3) | 0.97844 (7) | 0.0381 (6) | |
| H15A | 0.1735 | 0.0361 | 0.9976 | 0.046* | |
| H15B | 0.3107 | 0.1338 | 0.9834 | 0.046* | |
| C16 | 0.1004 (3) | 0.2250 (3) | 0.97290 (6) | 0.0355 (5) | |
| H16 | 0.0975 | 0.2888 | 0.9927 | 0.043* | |
| C17 | 0.1391 (3) | 0.3263 (3) | 0.94464 (6) | 0.0313 (5) | |
| H17 | 0.1169 | 0.2696 | 0.9244 | 0.038* | |
| C18 | 0.3045 (3) | 0.3737 (3) | 0.94445 (6) | 0.0293 (5) | |
| C19 | 0.3866 (3) | 0.3896 (3) | 0.91396 (6) | 0.0293 (5) | |
| C20 | 0.3231 (3) | 0.4125 (3) | 0.88526 (6) | 0.0311 (5) | |
| H20 | 0.2176 | 0.3998 | 0.8809 | 0.037* | |
| C21 | 0.4377 (3) | 0.4603 (3) | 0.86074 (6) | 0.0297 (5) | |
| H21 | 0.4207 | 0.4068 | 0.8399 | 0.036* | |
| C22 | 0.4360 (3) | 0.6331 (3) | 0.85635 (6) | 0.0292 (5) | |
| H22 | 0.4505 | 0.6794 | 0.8780 | 0.035* | |
| C24 | 0.2789 (3) | 1.1066 (3) | 0.81606 (6) | 0.0336 (5) | |
| H24 | 0.3835 | 1.1431 | 0.8216 | 0.040* | |
| C25 | 0.2305 (3) | 1.1817 (3) | 0.78489 (6) | 0.0342 (5) | |
| H25 | 0.2956 | 1.1435 | 0.7670 | 0.041* | |
| C26 | 0.0631 (3) | 1.1468 (3) | 0.77771 (6) | 0.0334 (5) | |
| H26 | 0.0529 | 1.0360 | 0.7729 | 0.040* | |
| C27 | −0.0359 (3) | 1.1844 (3) | 0.80648 (6) | 0.0351 (5) | |
| H27 | −0.0345 | 1.2965 | 0.8104 | 0.042* | |
| C28 | 0.0250 (3) | 1.1013 (3) | 0.83596 (6) | 0.0367 (5) | |
| H28 | 0.0209 | 0.9892 | 0.8320 | 0.044* | |
| C121 | 0.6240 (4) | −0.2127 (3) | 0.96221 (7) | 0.0422 (6) | |
| H12A | 0.6335 | −0.1777 | 0.9847 | 0.051* | |
| H12B | 0.5920 | −0.3206 | 0.9627 | 0.051* | |
| C122 | 0.7782 (4) | −0.2017 (4) | 0.94616 (9) | 0.0537 (8) | |
| H12C | 0.8121 | −0.0955 | 0.9461 | 0.064* | |
| H12D | 0.7703 | −0.2382 | 0.9240 | 0.064* | |
| H12E | 0.8519 | −0.2641 | 0.9579 | 0.064* | |
| C171 | 0.0413 (3) | 0.4720 (3) | 0.94530 (8) | 0.0438 (6) | |
| H17A | 0.0594 | 0.5308 | 0.9257 | 0.053* | |
| H17B | 0.0690 | 0.5333 | 0.9640 | 0.053* | |
| H17C | −0.0667 | 0.4444 | 0.9466 | 0.053* | |
| C251 | 0.3124 (4) | 1.4170 (4) | 0.76164 (8) | 0.0516 (8) | |
| H25A | 0.2365 | 1.4139 | 0.7444 | 0.062* | |
| H25B | 0.4061 | 1.3668 | 0.7545 | 0.062* | |
| H25C | 0.3347 | 1.5230 | 0.7671 | 0.062* | |
| C261 | 0.0420 (4) | 1.1631 (4) | 0.72113 (7) | 0.0528 (8) | |
| H26A | −0.0006 | 1.0601 | 0.7206 | 0.063* | |
| H26B | 0.1527 | 1.1580 | 0.7183 | 0.063* | |
| H26C | −0.0029 | 1.2237 | 0.7038 | 0.063* | |
| C271 | −0.2890 (4) | 1.2497 (4) | 0.79068 (8) | 0.0524 (8) | |
| H27A | −0.2558 | 1.2859 | 0.7696 | 0.063* | |
| H27B | −0.2886 | 1.3346 | 0.8059 | 0.063* | |
| H27C | −0.3922 | 1.2080 | 0.7890 | 0.063* | |
| C281 | −0.0613 (4) | 1.1369 (4) | 0.86623 (7) | 0.0502 (7) | |
| H28A | −0.0567 | 1.2464 | 0.8704 | 0.060* | |
| H28B | −0.0154 | 1.0817 | 0.8842 | 0.060* | |
| H28C | −0.1678 | 1.1056 | 0.8638 | 0.060* | |
| H16O | −0.083 (4) | 0.123 (4) | 0.9849 (8) | 0.057 (10)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O10 | 0.0789 (16) | 0.0422 (10) | 0.0329 (10) | −0.0082 (11) | 0.0023 (11) | −0.0019 (8) |
| O11 | 0.0363 (9) | 0.0259 (7) | 0.0333 (9) | 0.0026 (7) | 0.0007 (7) | 0.0025 (6) |
| O16 | 0.0279 (9) | 0.0581 (12) | 0.0415 (11) | −0.0038 (9) | 0.0016 (8) | 0.0115 (9) |
| O18 | 0.0358 (10) | 0.0461 (10) | 0.0307 (9) | −0.0012 (8) | −0.0039 (8) | −0.0016 (7) |
| O23 | 0.0478 (11) | 0.0299 (8) | 0.0327 (9) | 0.0108 (8) | −0.0055 (8) | 0.0000 (7) |
| O25 | 0.0510 (12) | 0.0312 (9) | 0.0415 (10) | −0.0039 (8) | −0.0036 (9) | 0.0063 (7) |
| O26 | 0.0445 (10) | 0.0424 (10) | 0.0339 (9) | 0.0078 (8) | −0.0048 (8) | 0.0037 (8) |
| O27 | 0.0357 (11) | 0.0481 (11) | 0.0527 (12) | −0.0028 (9) | −0.0035 (9) | −0.0016 (9) |
| O29 | 0.0401 (10) | 0.0347 (9) | 0.0324 (9) | 0.0062 (8) | −0.0021 (8) | −0.0028 (7) |
| C1 | 0.0333 (13) | 0.0378 (13) | 0.0340 (13) | 0.0042 (10) | −0.0005 (11) | 0.0060 (10) |
| C2 | 0.0408 (14) | 0.0318 (12) | 0.0295 (12) | 0.0066 (10) | −0.0033 (11) | 0.0012 (9) |
| C3 | 0.0407 (15) | 0.0314 (12) | 0.0438 (14) | 0.0002 (11) | −0.0017 (12) | 0.0052 (10) |
| C4 | 0.0322 (12) | 0.0308 (11) | 0.0328 (12) | 0.0011 (10) | 0.0000 (10) | 0.0028 (9) |
| C5 | 0.0310 (13) | 0.0403 (13) | 0.0445 (14) | −0.0004 (11) | 0.0022 (11) | 0.0069 (11) |
| C6 | 0.0256 (12) | 0.0418 (13) | 0.0399 (13) | 0.0040 (10) | −0.0017 (10) | 0.0072 (10) |
| C7 | 0.0297 (12) | 0.0309 (11) | 0.0332 (12) | 0.0047 (9) | −0.0002 (10) | 0.0033 (9) |
| C8 | 0.0288 (11) | 0.0290 (10) | 0.0307 (11) | −0.0001 (9) | −0.0013 (10) | 0.0007 (9) |
| C9 | 0.0269 (11) | 0.0314 (11) | 0.0339 (12) | 0.0021 (9) | −0.0040 (10) | 0.0027 (9) |
| C10 | 0.0292 (12) | 0.0329 (11) | 0.0324 (12) | 0.0035 (10) | 0.0013 (10) | 0.0007 (9) |
| C12 | 0.0391 (14) | 0.0241 (11) | 0.0439 (14) | 0.0040 (10) | 0.0016 (11) | −0.0024 (10) |
| C13 | 0.0447 (16) | 0.0284 (12) | 0.074 (2) | 0.0017 (12) | 0.0100 (15) | 0.0042 (13) |
| C14 | 0.0382 (15) | 0.0390 (14) | 0.0574 (18) | −0.0019 (12) | 0.0014 (13) | −0.0047 (12) |
| C15 | 0.0345 (13) | 0.0362 (12) | 0.0435 (14) | 0.0004 (11) | −0.0013 (12) | 0.0068 (10) |
| C16 | 0.0293 (12) | 0.0416 (13) | 0.0356 (13) | 0.0024 (10) | −0.0020 (10) | 0.0018 (10) |
| C17 | 0.0272 (12) | 0.0341 (12) | 0.0328 (12) | 0.0036 (9) | −0.0035 (10) | 0.0010 (10) |
| C18 | 0.0272 (11) | 0.0306 (11) | 0.0302 (11) | 0.0042 (9) | −0.0023 (9) | 0.0012 (9) |
| C19 | 0.0276 (11) | 0.0286 (11) | 0.0316 (11) | 0.0025 (9) | −0.0009 (9) | 0.0009 (9) |
| C20 | 0.0293 (12) | 0.0294 (11) | 0.0346 (12) | −0.0019 (9) | −0.0017 (10) | 0.0007 (9) |
| C21 | 0.0306 (12) | 0.0305 (10) | 0.0280 (11) | 0.0007 (10) | −0.0025 (9) | 0.0017 (9) |
| C22 | 0.0289 (11) | 0.0314 (11) | 0.0274 (11) | 0.0026 (9) | −0.0010 (9) | −0.0003 (8) |
| C24 | 0.0348 (13) | 0.0322 (12) | 0.0338 (12) | 0.0057 (10) | −0.0004 (10) | 0.0015 (9) |
| C25 | 0.0376 (14) | 0.0284 (11) | 0.0366 (13) | 0.0025 (10) | −0.0015 (11) | 0.0026 (9) |
| C26 | 0.0389 (13) | 0.0292 (11) | 0.0321 (12) | 0.0028 (10) | −0.0035 (11) | 0.0003 (9) |
| C27 | 0.0334 (13) | 0.0336 (12) | 0.0383 (13) | 0.0011 (10) | −0.0042 (11) | −0.0012 (10) |
| C28 | 0.0393 (14) | 0.0359 (12) | 0.0350 (13) | 0.0037 (11) | 0.0009 (11) | −0.0007 (10) |
| C121 | 0.0507 (17) | 0.0298 (12) | 0.0460 (15) | 0.0121 (11) | 0.0019 (13) | 0.0022 (11) |
| C122 | 0.0465 (18) | 0.0461 (16) | 0.069 (2) | 0.0170 (14) | 0.0014 (16) | 0.0023 (15) |
| C171 | 0.0347 (14) | 0.0420 (14) | 0.0547 (16) | 0.0071 (12) | −0.0002 (13) | 0.0040 (13) |
| C251 | 0.0525 (19) | 0.0458 (16) | 0.0565 (18) | −0.0060 (14) | −0.0004 (15) | 0.0164 (13) |
| C261 | 0.060 (2) | 0.0646 (19) | 0.0341 (14) | 0.0067 (17) | −0.0037 (14) | −0.0034 (13) |
| C271 | 0.0355 (15) | 0.069 (2) | 0.0528 (18) | 0.0098 (15) | −0.0034 (14) | −0.0046 (15) |
| C281 | 0.0521 (18) | 0.0603 (18) | 0.0383 (15) | 0.0103 (16) | 0.0090 (14) | −0.0005 (13) |
Geometric parameters (Å, º)
| O10—C10 | 1.200 (3) | C14—H14A | 0.9900 |
| O11—C10 | 1.334 (3) | C14—H14B | 0.9900 |
| O11—C12 | 1.465 (3) | C15—C16 | 1.523 (4) |
| O16—C16 | 1.436 (3) | C15—H15A | 0.9900 |
| O16—H16O | 0.92 (4) | C15—H15B | 0.9900 |
| O18—C18 | 1.236 (3) | C16—C17 | 1.517 (3) |
| O23—C24 | 1.402 (3) | C16—H16 | 1.0000 |
| O23—C2 | 1.438 (3) | C17—C18 | 1.510 (3) |
| O25—C251 | 1.417 (3) | C17—C171 | 1.543 (4) |
| O25—C25 | 1.425 (3) | C17—H17 | 1.0000 |
| O26—C261 | 1.414 (3) | C18—C19 | 1.469 (3) |
| O26—C26 | 1.425 (3) | C19—C20 | 1.336 (3) |
| O27—C271 | 1.416 (4) | C20—C21 | 1.496 (3) |
| O27—C27 | 1.421 (3) | C20—H20 | 0.9500 |
| O29—C24 | 1.410 (3) | C21—C22 | 1.531 (3) |
| O29—C28 | 1.432 (3) | C21—H21 | 1.0000 |
| C1—C22 | 1.520 (3) | C22—H22 | 1.0000 |
| C1—C2 | 1.523 (4) | C24—C25 | 1.519 (3) |
| C1—H1A | 0.9900 | C24—H24 | 1.0000 |
| C1—H1B | 0.9900 | C25—C26 | 1.531 (4) |
| C2—C3 | 1.543 (4) | C25—H25 | 1.0000 |
| C2—H2 | 1.0000 | C26—C27 | 1.518 (4) |
| C3—C4 | 1.538 (3) | C26—H26 | 1.0000 |
| C3—H3A | 0.9900 | C27—C28 | 1.527 (4) |
| C3—H3B | 0.9900 | C27—H27 | 1.0000 |
| C4—C5 | 1.497 (4) | C28—C281 | 1.506 (4) |
| C4—C22 | 1.522 (3) | C28—H28 | 1.0000 |
| C4—H4 | 1.0000 | C121—C122 | 1.513 (5) |
| C5—C6 | 1.335 (4) | C121—H12A | 0.9900 |
| C5—H5 | 0.9500 | C121—H12B | 0.9900 |
| C6—C7 | 1.505 (4) | C122—H12C | 0.9800 |
| C6—H6 | 0.9500 | C122—H12D | 0.9800 |
| C7—C21 | 1.550 (3) | C122—H12E | 0.9800 |
| C7—C8 | 1.557 (3) | C171—H17A | 0.9800 |
| C7—H7 | 1.0000 | C171—H17B | 0.9800 |
| C8—C19 | 1.509 (3) | C171—H17C | 0.9800 |
| C8—C9 | 1.537 (3) | C251—H25A | 0.9800 |
| C8—H8 | 1.0000 | C251—H25B | 0.9800 |
| C9—C10 | 1.511 (3) | C251—H25C | 0.9800 |
| C9—H9A | 0.9900 | C261—H26A | 0.9800 |
| C9—H9B | 0.9900 | C261—H26B | 0.9800 |
| C12—C121 | 1.516 (4) | C261—H26C | 0.9800 |
| C12—C13 | 1.516 (4) | C271—H27A | 0.9800 |
| C12—H12 | 1.0000 | C271—H27B | 0.9800 |
| C13—C14 | 1.554 (4) | C271—H27C | 0.9800 |
| C13—H13A | 0.9900 | C281—H28A | 0.9800 |
| C13—H13B | 0.9900 | C281—H28B | 0.9800 |
| C14—C15 | 1.525 (4) | C281—H28C | 0.9800 |
| C10—O11—C12 | 117.9 (2) | O18—C18—C19 | 118.9 (2) |
| C16—O16—H16O | 106 (2) | O18—C18—C17 | 120.8 (2) |
| C24—O23—C2 | 111.83 (19) | C19—C18—C17 | 120.1 (2) |
| C251—O25—C25 | 114.9 (2) | C20—C19—C18 | 125.9 (2) |
| C261—O26—C26 | 113.4 (2) | C20—C19—C8 | 111.8 (2) |
| C271—O27—C27 | 114.6 (2) | C18—C19—C8 | 121.9 (2) |
| C24—O29—C28 | 113.7 (2) | C19—C20—C21 | 112.0 (2) |
| C22—C1—C2 | 101.0 (2) | C19—C20—H20 | 124.0 |
| C22—C1—H1A | 111.6 | C21—C20—H20 | 124.0 |
| C2—C1—H1A | 111.6 | C20—C21—C22 | 110.8 (2) |
| C22—C1—H1B | 111.6 | C20—C21—C7 | 103.17 (18) |
| C2—C1—H1B | 111.6 | C22—C21—C7 | 108.9 (2) |
| H1A—C1—H1B | 109.4 | C20—C21—H21 | 111.2 |
| O23—C2—C1 | 110.8 (2) | C22—C21—H21 | 111.2 |
| O23—C2—C3 | 112.9 (2) | C7—C21—H21 | 111.2 |
| C1—C2—C3 | 106.7 (2) | C1—C22—C4 | 102.61 (19) |
| O23—C2—H2 | 108.8 | C1—C22—C21 | 120.7 (2) |
| C1—C2—H2 | 108.8 | C4—C22—C21 | 111.3 (2) |
| C3—C2—H2 | 108.8 | C1—C22—H22 | 107.2 |
| C4—C3—C2 | 104.6 (2) | C4—C22—H22 | 107.2 |
| C4—C3—H3A | 110.8 | C21—C22—H22 | 107.2 |
| C2—C3—H3A | 110.8 | O23—C24—O29 | 112.0 (2) |
| C4—C3—H3B | 110.8 | O23—C24—C25 | 108.1 (2) |
| C2—C3—H3B | 110.8 | O29—C24—C25 | 111.4 (2) |
| H3A—C3—H3B | 108.9 | O23—C24—H24 | 108.4 |
| C5—C4—C22 | 110.2 (2) | O29—C24—H24 | 108.4 |
| C5—C4—C3 | 118.5 (2) | C25—C24—H24 | 108.4 |
| C22—C4—C3 | 103.2 (2) | O25—C25—C24 | 106.4 (2) |
| C5—C4—H4 | 108.2 | O25—C25—C26 | 111.3 (2) |
| C22—C4—H4 | 108.2 | C24—C25—C26 | 110.4 (2) |
| C3—C4—H4 | 108.2 | O25—C25—H25 | 109.5 |
| C6—C5—C4 | 119.9 (2) | C24—C25—H25 | 109.5 |
| C6—C5—H5 | 120.1 | C26—C25—H25 | 109.5 |
| C4—C5—H5 | 120.1 | O26—C26—C27 | 108.2 (2) |
| C5—C6—C7 | 124.9 (2) | O26—C26—C25 | 111.8 (2) |
| C5—C6—H6 | 117.6 | C27—C26—C25 | 110.5 (2) |
| C7—C6—H6 | 117.6 | O26—C26—H26 | 108.7 |
| C6—C7—C21 | 115.32 (19) | C27—C26—H26 | 108.7 |
| C6—C7—C8 | 112.4 (2) | C25—C26—H26 | 108.7 |
| C21—C7—C8 | 104.87 (19) | O27—C27—C26 | 110.9 (2) |
| C6—C7—H7 | 108.0 | O27—C27—C28 | 107.5 (2) |
| C21—C7—H7 | 108.0 | C26—C27—C28 | 109.4 (2) |
| C8—C7—H7 | 108.0 | O27—C27—H27 | 109.7 |
| C19—C8—C9 | 114.1 (2) | C26—C27—H27 | 109.7 |
| C19—C8—C7 | 103.3 (2) | C28—C27—H27 | 109.7 |
| C9—C8—C7 | 115.1 (2) | O29—C28—C281 | 106.8 (2) |
| C19—C8—H8 | 108.0 | O29—C28—C27 | 109.3 (2) |
| C9—C8—H8 | 108.0 | C281—C28—C27 | 113.6 (2) |
| C7—C8—H8 | 108.0 | O29—C28—H28 | 109.0 |
| C10—C9—C8 | 113.1 (2) | C281—C28—H28 | 109.0 |
| C10—C9—H9A | 109.0 | C27—C28—H28 | 109.0 |
| C8—C9—H9A | 109.0 | C122—C121—C12 | 113.4 (2) |
| C10—C9—H9B | 109.0 | C122—C121—H12A | 108.9 |
| C8—C9—H9B | 109.0 | C12—C121—H12A | 108.9 |
| H9A—C9—H9B | 107.8 | C122—C121—H12B | 108.9 |
| O10—C10—O11 | 124.0 (2) | C12—C121—H12B | 108.9 |
| O10—C10—C9 | 124.3 (2) | H12A—C121—H12B | 107.7 |
| O11—C10—C9 | 111.6 (2) | C121—C122—H12C | 109.5 |
| O11—C12—C121 | 106.6 (2) | C121—C122—H12D | 109.5 |
| O11—C12—C13 | 109.5 (2) | H12C—C122—H12D | 109.5 |
| C121—C12—C13 | 112.9 (2) | C121—C122—H12E | 109.5 |
| O11—C12—H12 | 109.3 | H12C—C122—H12E | 109.5 |
| C121—C12—H12 | 109.3 | H12D—C122—H12E | 109.5 |
| C13—C12—H12 | 109.3 | C17—C171—H17A | 109.5 |
| C12—C13—C14 | 115.1 (2) | C17—C171—H17B | 109.5 |
| C12—C13—H13A | 108.5 | H17A—C171—H17B | 109.5 |
| C14—C13—H13A | 108.5 | C17—C171—H17C | 109.5 |
| C12—C13—H13B | 108.5 | H17A—C171—H17C | 109.5 |
| C14—C13—H13B | 108.5 | H17B—C171—H17C | 109.5 |
| H13A—C13—H13B | 107.5 | O25—C251—H25A | 109.5 |
| C15—C14—C13 | 109.6 (3) | O25—C251—H25B | 109.5 |
| C15—C14—H14A | 109.8 | H25A—C251—H25B | 109.5 |
| C13—C14—H14A | 109.8 | O25—C251—H25C | 109.5 |
| C15—C14—H14B | 109.8 | H25A—C251—H25C | 109.5 |
| C13—C14—H14B | 109.8 | H25B—C251—H25C | 109.5 |
| H14A—C14—H14B | 108.2 | O26—C261—H26A | 109.5 |
| C16—C15—C14 | 116.5 (2) | O26—C261—H26B | 109.5 |
| C16—C15—H15A | 108.2 | H26A—C261—H26B | 109.5 |
| C14—C15—H15A | 108.2 | O26—C261—H26C | 109.5 |
| C16—C15—H15B | 108.2 | H26A—C261—H26C | 109.5 |
| C14—C15—H15B | 108.2 | H26B—C261—H26C | 109.5 |
| H15A—C15—H15B | 107.3 | O27—C271—H27A | 109.5 |
| O16—C16—C17 | 105.1 (2) | O27—C271—H27B | 109.5 |
| O16—C16—C15 | 110.1 (2) | H27A—C271—H27B | 109.5 |
| C17—C16—C15 | 115.4 (2) | O27—C271—H27C | 109.5 |
| O16—C16—H16 | 108.7 | H27A—C271—H27C | 109.5 |
| C17—C16—H16 | 108.7 | H27B—C271—H27C | 109.5 |
| C15—C16—H16 | 108.7 | C28—C281—H28A | 109.5 |
| C18—C17—C16 | 112.4 (2) | C28—C281—H28B | 109.5 |
| C18—C17—C171 | 107.8 (2) | H28A—C281—H28B | 109.5 |
| C16—C17—C171 | 110.4 (2) | C28—C281—H28C | 109.5 |
| C18—C17—H17 | 108.7 | H28A—C281—H28C | 109.5 |
| C16—C17—H17 | 108.7 | H28B—C281—H28C | 109.5 |
| C171—C17—H17 | 108.7 | ||
| C24—O23—C2—C1 | 149.8 (2) | C8—C19—C20—C21 | −6.2 (3) |
| C24—O23—C2—C3 | −90.6 (3) | C19—C20—C21—C22 | −98.5 (2) |
| C22—C1—C2—O23 | 155.1 (2) | C19—C20—C21—C7 | 17.9 (3) |
| C22—C1—C2—C3 | 31.7 (3) | C6—C7—C21—C20 | −146.0 (2) |
| O23—C2—C3—C4 | −127.8 (2) | C8—C7—C21—C20 | −21.8 (2) |
| C1—C2—C3—C4 | −5.8 (3) | C6—C7—C21—C22 | −28.2 (3) |
| C2—C3—C4—C5 | −144.5 (2) | C8—C7—C21—C22 | 96.0 (2) |
| C2—C3—C4—C22 | −22.5 (3) | C2—C1—C22—C4 | −46.0 (2) |
| C22—C4—C5—C6 | 29.0 (3) | C2—C1—C22—C21 | −170.4 (2) |
| C3—C4—C5—C6 | 147.4 (3) | C5—C4—C22—C1 | 170.4 (2) |
| C4—C5—C6—C7 | 0.3 (4) | C3—C4—C22—C1 | 43.0 (2) |
| C5—C6—C7—C21 | −0.5 (4) | C5—C4—C22—C21 | −59.1 (3) |
| C5—C6—C7—C8 | −120.7 (3) | C3—C4—C22—C21 | 173.4 (2) |
| C6—C7—C8—C19 | 144.5 (2) | C20—C21—C22—C1 | −68.8 (3) |
| C21—C7—C8—C19 | 18.5 (2) | C7—C21—C22—C1 | 178.4 (2) |
| C6—C7—C8—C9 | −90.5 (3) | C20—C21—C22—C4 | 171.0 (2) |
| C21—C7—C8—C9 | 143.5 (2) | C7—C21—C22—C4 | 58.2 (3) |
| C19—C8—C9—C10 | 46.5 (3) | C2—O23—C24—O29 | −70.2 (3) |
| C7—C8—C9—C10 | −72.7 (3) | C2—O23—C24—C25 | 166.7 (2) |
| C12—O11—C10—O10 | 7.1 (4) | C28—O29—C24—O23 | −61.6 (3) |
| C12—O11—C10—C9 | −172.6 (2) | C28—O29—C24—C25 | 59.7 (3) |
| C8—C9—C10—O10 | 53.0 (4) | C251—O25—C25—C24 | 141.0 (2) |
| C8—C9—C10—O11 | −127.4 (2) | C251—O25—C25—C26 | −98.7 (3) |
| C10—O11—C12—C121 | 115.1 (2) | O23—C24—C25—O25 | −168.6 (2) |
| C10—O11—C12—C13 | −122.4 (3) | O29—C24—C25—O25 | 67.8 (3) |
| O11—C12—C13—C14 | 44.8 (3) | O23—C24—C25—C26 | 70.4 (3) |
| C121—C12—C13—C14 | 163.4 (3) | O29—C24—C25—C26 | −53.1 (3) |
| C12—C13—C14—C15 | −100.1 (3) | C261—O26—C26—C27 | −154.1 (2) |
| C13—C14—C15—C16 | 177.2 (2) | C261—O26—C26—C25 | 83.9 (3) |
| C14—C15—C16—O16 | 59.5 (3) | O25—C25—C26—O26 | 54.3 (3) |
| C14—C15—C16—C17 | −59.3 (3) | C24—C25—C26—O26 | 172.3 (2) |
| O16—C16—C17—C18 | −170.3 (2) | O25—C25—C26—C27 | −66.4 (3) |
| C15—C16—C17—C18 | −48.8 (3) | C24—C25—C26—C27 | 51.6 (3) |
| O16—C16—C17—C171 | 69.2 (3) | C271—O27—C27—C26 | −97.4 (3) |
| C15—C16—C17—C171 | −169.3 (2) | C271—O27—C27—C28 | 143.0 (2) |
| C16—C17—C18—O18 | −40.1 (3) | O26—C26—C27—O27 | 64.4 (3) |
| C171—C17—C18—O18 | 81.8 (3) | C25—C26—C27—O27 | −172.8 (2) |
| C16—C17—C18—C19 | 144.2 (2) | O26—C26—C27—C28 | −177.2 (2) |
| C171—C17—C18—C19 | −93.9 (3) | C25—C26—C27—C28 | −54.4 (3) |
| O18—C18—C19—C20 | −153.7 (2) | C24—O29—C28—C281 | 174.7 (2) |
| C17—C18—C19—C20 | 22.1 (4) | C24—O29—C28—C27 | −62.0 (3) |
| O18—C18—C19—C8 | 18.1 (4) | O27—C27—C28—O29 | 178.69 (19) |
| C17—C18—C19—C8 | −166.1 (2) | C26—C27—C28—O29 | 58.1 (3) |
| C9—C8—C19—C20 | −133.9 (2) | O27—C27—C28—C281 | −62.2 (3) |
| C7—C8—C19—C20 | −8.3 (3) | C26—C27—C28—C281 | 177.3 (2) |
| C9—C8—C19—C18 | 53.3 (3) | O11—C12—C121—C122 | −65.3 (3) |
| C7—C8—C19—C18 | 178.9 (2) | C13—C12—C121—C122 | 174.4 (3) |
| C18—C19—C20—C21 | 166.3 (2) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O16—H16O···O18i | 0.92 (4) | 1.96 (3) | 2.840 (3) | 160 (3) |
| C3—H3B···O10ii | 0.99 | 2.47 | 3.310 (3) | 142 |
Symmetry codes: (i) x−1/2, −y+1/2, −z+2; (ii) x, y+1, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: IS5144).
References
- Evans, D. A. & Black, W. C. (1993). J. Am. Chem. Soc. 115, 4497–4513.
- Rigaku (2008). CrystalClear Rigaku Corporation, Tokyo, Japan.
- Rigaku/MSC (2009). CrystalStructure Rigaku/MSC, The Woodlands, Texas, USA.
- Salgado, V. L. (1998). Pestic. Biochem. Physiol. 60, 91–102.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sparks, T. C., Crouse, G. D., Dripps, J. E., Anzeveno, P., Martynow, J., DeAmicis, C. V. & Gifford, J. (2008). J. Comput. Aided Mol. Des. 22, 393–401. [DOI] [PubMed]
- Thompson, G. D., Dutton, R. & Sparks, T. C. (2000). Pest Manage. Sci. 56, 696–702.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536812028851/is5144sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812028851/is5144Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report