Abstract
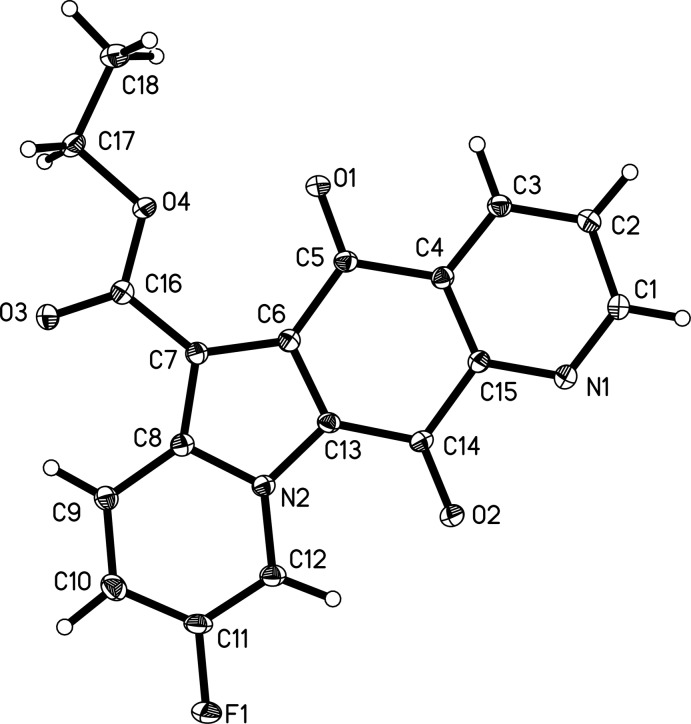
In the title molecule, C18H11FN2O4, the fused four- ring system is essentially planar, with an r.m.s. deviation of 0.032 Å. In the crystal, molecules are connected by π–π stacking interactions [centroid–centroid distances = 3.5684 (9) and 3.8247 (9) Å] into chains along [100].
Related literature
The title compound was obtained in an attempt to synthesize a Top1 (DNA topoisomerase IB) inhibitor. For general background to Top1, see: Pommier (2006 ▶). For the synthesis, see: Shen et al. (2008 ▶); Cheng et al. (2008 ▶). For a related structure, see: Wu et al. (2011 ▶). For the Top1 inhibitory activity of a related indolizinoquinoline-5,12-dione derivative, see: Wu et al. (2010 ▶).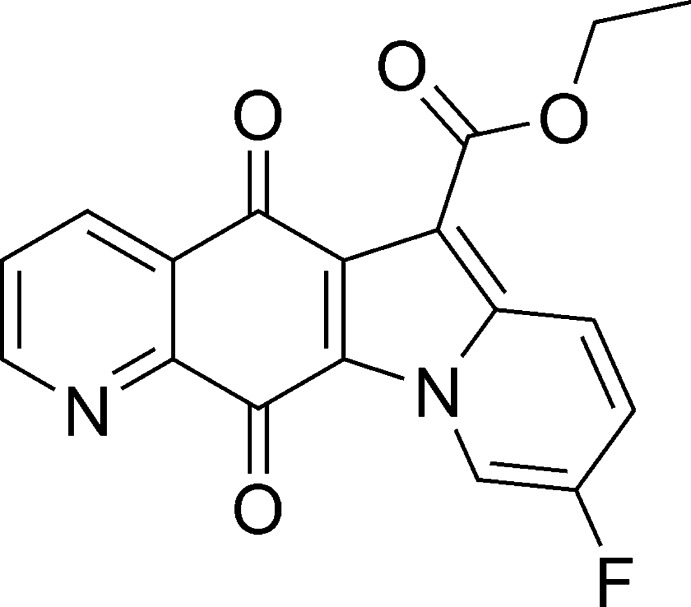
Experimental
Crystal data
C18H11FN2O4
M r = 338.29
Monoclinic,
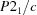
a = 6.85562 (10) Å
b = 12.12898 (16) Å
c = 17.0304 (2) Å
β = 94.2306 (13)°
V = 1412.25 (3) Å3
Z = 4
Cu Kα radiation
μ = 1.04 mm−1
T = 136 K
0.30 × 0.20 × 0.20 mm
Data collection
Agilent Xcalibur Onyx Nova diffractometer
Absorption correction: multi-scan (CrysAlis PRO; Agilent, 2011 ▶) T min = 0.550, T max = 1.000
5740 measured reflections
2723 independent reflections
2278 reflections with I > 2σ(I)
R int = 0.025
Refinement
R[F 2 > 2σ(F 2)] = 0.037
wR(F 2) = 0.101
S = 1.08
2723 reflections
270 parameters
All H-atom parameters refined
Δρmax = 0.30 e Å−3
Δρmin = −0.24 e Å−3
Data collection: CrysAlis PRO (Agilent, 2011 ▶); cell refinement: CrysAlis PRO; data reduction: CrysAlis RED (Agilent, 2011 ▶); program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: OLEX2 (Dolomanov et al., 2009 ▶); software used to prepare material for publication: publCIF (Westrip, 2010 ▶).
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S160053681202692X/lh5489sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S160053681202692X/lh5489Isup2.hkl
Supplementary material file. DOI: 10.1107/S160053681202692X/lh5489Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors gratefully acknowledge financial support from the National Natural Science Foundation of China (No. 30801425) and the Guangdong Natural Science Fund (No. 10151008901000022).
supplementary crystallographic information
Comment
Top1 is an essential nuclear enzyme, and can be used as a target to discover anticancer agents (Pommier, 2006). In our previous research, we found ethyl 7-fluoro-5,12-dioxo-5,12-dihydroindolizino[2,3-g] quinoline-6-carboxylate is a strong Top1 inhibitor with a different inhibitory mechanism from camptothecin, a well known Top1 inhibitor (Wu et al. 2010). In order to investigate the Top1 inhibitory activity of the 9-fluoro substituted isomer, the title compound was synthesized according to a modified literature method (Shen et al. 2008; Cheng et al. 2008; Wu et al. 2011) and its crystal structure was determined.
The asymmetric unit of the title compound is shown in figure 1. In the molecule the four fused aromatic rings system is approximately planar with an r.m.s. deviation = 0.032 Å. In the crystal, molecules are connected by π–π stacking interactions to form chains along [100]. Cg1···Cg1i = 3.5684 (9)Å and Cg1···Cg4ii = 3.8247 (9) Å, where Cg1 and Cg2 are the centroids of the N2/C8/C7/C6/C13 and C4/C5/C6/C13/C14/C15 rings [symmetry codes: (i) -x, 1-y, 1-z, (ii) 1-x, 1-y, 1-z].
Experimental
According to a modified literature method (Shen et al., 2008; Cheng et al., 2008; Wu et al., 2011), 12 equivalents of 3-fluoropyridine reacted with 6,7-dichloroquinoline-5,8-dione and ethyl acetoacetate to give the title compound as orange solid. Needle-shaped crystals suitable for X-ray analysis were obtained by slow evaporation of a solution of the title compound in chloroform-ethyl acetate (20/1, v/v).
Refinement
All H atoms were refined indpendently with isotropic displacement parameters.
Figures
Fig. 1.
The molecular structure of the title compound. The displacement ellipsoids are at the 30% probability level and H atoms are shown as small spheres of arbitrary radii.
Crystal data
| C18H11FN2O4 | F(000) = 696 |
| Mr = 338.29 | Dx = 1.591 Mg m−3 |
| Monoclinic, P21/c | Cu Kα radiation, λ = 1.5418 Å |
| Hall symbol: -P 2ybc | Cell parameters from 3543 reflections |
| a = 6.85562 (10) Å | θ = 2.6–72.7° |
| b = 12.12898 (16) Å | µ = 1.04 mm−1 |
| c = 17.0304 (2) Å | T = 136 K |
| β = 94.2306 (13)° | Needle, orange |
| V = 1412.25 (3) Å3 | 0.30 × 0.20 × 0.20 mm |
| Z = 4 |
Data collection
| Agilent Xcalibur Onyx Nova diffractometer | 2723 independent reflections |
| Radiation source: Nova (Cu) X-ray Source | 2278 reflections with I > 2σ(I) |
| Mirror monochromator | Rint = 0.025 |
| Detector resolution: 8.2417 pixels mm-1 | θmax = 72.9°, θmin = 5.2° |
| ω scans | h = −8→8 |
| Absorption correction: multi-scan (CrysAlis PRO; Agilent, 2011) | k = −14→10 |
| Tmin = 0.550, Tmax = 1.000 | l = −20→20 |
| 5740 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.037 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.101 | All H-atom parameters refined |
| S = 1.08 | w = 1/[σ2(Fo2) + (0.0396P)2 + 0.8533P] where P = (Fo2 + 2Fc2)/3 |
| 2723 reflections | (Δ/σ)max = 0.001 |
| 270 parameters | Δρmax = 0.30 e Å−3 |
| 0 restraints | Δρmin = −0.24 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| F1 | 0.02823 (18) | 0.90196 (9) | 0.34624 (7) | 0.0382 (3) | |
| O2 | 0.28727 (18) | 0.79052 (10) | 0.60332 (7) | 0.0266 (3) | |
| O3 | 0.1029 (2) | 0.36760 (11) | 0.33751 (7) | 0.0301 (3) | |
| O1 | 0.3911 (2) | 0.34756 (10) | 0.58977 (7) | 0.0307 (3) | |
| N1 | 0.4460 (2) | 0.68373 (12) | 0.73454 (9) | 0.0241 (3) | |
| O4 | 0.2439 (2) | 0.28907 (10) | 0.44611 (8) | 0.0291 (3) | |
| C4 | 0.4124 (2) | 0.51043 (14) | 0.66509 (10) | 0.0199 (4) | |
| N2 | 0.1785 (2) | 0.66666 (11) | 0.45980 (8) | 0.0195 (3) | |
| C13 | 0.2564 (2) | 0.62440 (14) | 0.53105 (10) | 0.0192 (4) | |
| C6 | 0.2735 (2) | 0.51065 (14) | 0.52314 (10) | 0.0193 (4) | |
| C16 | 0.1782 (2) | 0.37539 (14) | 0.40402 (10) | 0.0204 (4) | |
| C14 | 0.3084 (2) | 0.68974 (14) | 0.60029 (10) | 0.0199 (4) | |
| C7 | 0.2028 (2) | 0.48181 (14) | 0.44537 (10) | 0.0199 (4) | |
| C5 | 0.3582 (2) | 0.44629 (14) | 0.59152 (10) | 0.0202 (4) | |
| C15 | 0.3912 (2) | 0.62469 (14) | 0.66995 (10) | 0.0206 (4) | |
| C12 | 0.1396 (3) | 0.77509 (15) | 0.44053 (11) | 0.0238 (4) | |
| C1 | 0.5239 (3) | 0.62799 (15) | 0.79713 (11) | 0.0270 (4) | |
| C10 | 0.0304 (3) | 0.71363 (16) | 0.30895 (11) | 0.0286 (4) | |
| C3 | 0.4925 (2) | 0.45453 (15) | 0.73164 (10) | 0.0223 (4) | |
| C18 | 0.2975 (3) | 0.09848 (17) | 0.46821 (13) | 0.0328 (5) | |
| C8 | 0.1448 (2) | 0.58082 (14) | 0.40663 (10) | 0.0204 (4) | |
| C2 | 0.5502 (3) | 0.51463 (15) | 0.79815 (11) | 0.0253 (4) | |
| C9 | 0.0689 (2) | 0.60677 (15) | 0.32938 (11) | 0.0231 (4) | |
| C17 | 0.2236 (3) | 0.18208 (15) | 0.40799 (12) | 0.0265 (4) | |
| C11 | 0.0667 (3) | 0.79580 (15) | 0.36591 (11) | 0.0266 (4) | |
| H1 | 0.568 (3) | 0.6713 (18) | 0.8436 (12) | 0.028 (5)* | |
| H17A | 0.302 (3) | 0.1835 (16) | 0.3619 (12) | 0.023 (5)* | |
| H2 | 0.607 (3) | 0.4777 (17) | 0.8449 (12) | 0.026 (5)* | |
| H9 | 0.043 (3) | 0.5454 (19) | 0.2945 (13) | 0.033 (6)* | |
| H3 | 0.513 (3) | 0.3732 (18) | 0.7311 (12) | 0.031 (5)* | |
| H12 | 0.167 (3) | 0.8305 (18) | 0.4819 (13) | 0.032 (6)* | |
| H10 | −0.018 (3) | 0.7362 (18) | 0.2575 (13) | 0.034 (6)* | |
| H17B | 0.079 (3) | 0.1699 (18) | 0.3898 (12) | 0.033 (6)* | |
| H18A | 0.439 (4) | 0.117 (2) | 0.4875 (15) | 0.052 (7)* | |
| H18B | 0.217 (4) | 0.099 (2) | 0.5139 (15) | 0.045 (7)* | |
| H18C | 0.291 (3) | 0.022 (2) | 0.4457 (13) | 0.039 (6)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| F1 | 0.0529 (7) | 0.0221 (6) | 0.0372 (7) | 0.0044 (5) | −0.0126 (5) | 0.0072 (5) |
| O2 | 0.0321 (7) | 0.0185 (6) | 0.0281 (7) | 0.0017 (5) | −0.0049 (5) | −0.0011 (5) |
| O3 | 0.0403 (7) | 0.0246 (7) | 0.0237 (7) | −0.0013 (6) | −0.0091 (6) | −0.0024 (5) |
| O1 | 0.0468 (8) | 0.0173 (7) | 0.0265 (7) | 0.0026 (6) | −0.0071 (6) | 0.0008 (5) |
| N1 | 0.0267 (8) | 0.0217 (8) | 0.0235 (8) | 0.0003 (6) | −0.0008 (6) | −0.0025 (6) |
| O4 | 0.0408 (8) | 0.0168 (6) | 0.0278 (7) | 0.0042 (6) | −0.0096 (6) | −0.0034 (5) |
| C4 | 0.0181 (8) | 0.0186 (8) | 0.0229 (9) | −0.0005 (6) | 0.0003 (6) | −0.0009 (7) |
| N2 | 0.0178 (7) | 0.0175 (7) | 0.0229 (7) | 0.0000 (5) | −0.0005 (5) | 0.0003 (6) |
| C13 | 0.0175 (8) | 0.0178 (8) | 0.0218 (9) | 0.0005 (6) | −0.0015 (6) | 0.0025 (6) |
| C6 | 0.0169 (8) | 0.0202 (8) | 0.0208 (8) | 0.0004 (6) | 0.0009 (6) | −0.0008 (7) |
| C16 | 0.0165 (8) | 0.0220 (9) | 0.0227 (9) | −0.0015 (6) | 0.0012 (6) | −0.0008 (7) |
| C14 | 0.0183 (8) | 0.0160 (8) | 0.0252 (9) | −0.0001 (6) | 0.0010 (7) | −0.0008 (7) |
| C7 | 0.0173 (8) | 0.0196 (8) | 0.0223 (8) | −0.0005 (6) | −0.0016 (6) | −0.0004 (7) |
| C5 | 0.0201 (8) | 0.0179 (9) | 0.0225 (9) | −0.0007 (7) | 0.0012 (7) | 0.0019 (7) |
| C15 | 0.0212 (8) | 0.0181 (8) | 0.0219 (9) | −0.0023 (7) | −0.0010 (7) | −0.0002 (7) |
| C12 | 0.0235 (9) | 0.0192 (9) | 0.0284 (9) | 0.0015 (7) | −0.0011 (7) | 0.0025 (7) |
| C1 | 0.0330 (10) | 0.0241 (9) | 0.0231 (9) | −0.0005 (8) | −0.0046 (8) | −0.0025 (7) |
| C10 | 0.0277 (9) | 0.0320 (10) | 0.0251 (9) | 0.0004 (8) | −0.0049 (7) | 0.0037 (8) |
| C3 | 0.0230 (8) | 0.0194 (9) | 0.0243 (9) | −0.0006 (7) | 0.0003 (7) | 0.0020 (7) |
| C18 | 0.0386 (11) | 0.0218 (10) | 0.0375 (12) | 0.0030 (8) | −0.0018 (9) | 0.0016 (8) |
| C8 | 0.0174 (8) | 0.0199 (9) | 0.0237 (9) | −0.0011 (6) | −0.0001 (7) | −0.0002 (7) |
| C2 | 0.0297 (9) | 0.0226 (9) | 0.0229 (9) | 0.0005 (7) | −0.0027 (7) | 0.0028 (7) |
| C9 | 0.0219 (8) | 0.0235 (9) | 0.0234 (9) | −0.0002 (7) | −0.0023 (7) | 0.0005 (7) |
| C17 | 0.0309 (10) | 0.0186 (9) | 0.0293 (10) | 0.0002 (7) | −0.0023 (8) | −0.0053 (7) |
| C11 | 0.0286 (9) | 0.0183 (9) | 0.0320 (10) | 0.0021 (7) | −0.0029 (8) | 0.0069 (7) |
Geometric parameters (Å, º)
| F1—C11 | 1.352 (2) | C7—C8 | 1.413 (2) |
| O2—C14 | 1.232 (2) | C12—C11 | 1.354 (3) |
| O3—C16 | 1.213 (2) | C12—H12 | 0.98 (2) |
| O1—C5 | 1.219 (2) | C1—C2 | 1.387 (3) |
| N1—C15 | 1.343 (2) | C1—H1 | 0.98 (2) |
| N1—C1 | 1.339 (2) | C10—C9 | 1.363 (3) |
| O4—C16 | 1.329 (2) | C10—C11 | 1.400 (3) |
| O4—C17 | 1.453 (2) | C10—H10 | 0.95 (2) |
| C4—C5 | 1.498 (2) | C3—C2 | 1.380 (3) |
| C4—C15 | 1.397 (2) | C3—H3 | 1.00 (2) |
| C4—C3 | 1.398 (2) | C18—C17 | 1.503 (3) |
| N2—C13 | 1.387 (2) | C18—H18A | 1.02 (3) |
| N2—C12 | 1.377 (2) | C18—H18B | 0.99 (3) |
| N2—C8 | 1.388 (2) | C18—H18C | 1.00 (2) |
| C13—C6 | 1.392 (2) | C8—C9 | 1.414 (2) |
| C13—C14 | 1.443 (2) | C2—H2 | 0.97 (2) |
| C6—C7 | 1.420 (2) | C9—H9 | 0.96 (2) |
| C6—C5 | 1.484 (2) | C17—H17A | 0.98 (2) |
| C16—C7 | 1.474 (2) | C17—H17B | 1.02 (2) |
| C14—C15 | 1.501 (2) | ||
| C1—N1—C15 | 116.99 (15) | N1—C1—H1 | 117.0 (12) |
| C16—O4—C17 | 116.44 (13) | C2—C1—H1 | 119.3 (12) |
| C15—C4—C5 | 122.98 (15) | C9—C10—C11 | 118.60 (17) |
| C15—C4—C3 | 118.01 (16) | C9—C10—H10 | 123.7 (14) |
| C3—C4—C5 | 118.99 (15) | C11—C10—H10 | 117.7 (13) |
| C13—N2—C8 | 109.17 (14) | C4—C3—H3 | 121.3 (12) |
| C12—N2—C13 | 128.08 (15) | C2—C3—C4 | 118.75 (16) |
| C12—N2—C8 | 122.75 (15) | C2—C3—H3 | 119.9 (12) |
| N2—C13—C6 | 108.13 (14) | C17—C18—H18A | 109.7 (15) |
| N2—C13—C14 | 124.58 (15) | C17—C18—H18B | 110.7 (14) |
| C6—C13—C14 | 127.29 (15) | C17—C18—H18C | 111.0 (13) |
| C13—C6—C7 | 108.00 (15) | H18A—C18—H18B | 109 (2) |
| C13—C6—C5 | 118.44 (15) | H18A—C18—H18C | 109.6 (19) |
| C7—C6—C5 | 133.55 (16) | H18B—C18—H18C | 107.2 (19) |
| O3—C16—O4 | 123.11 (16) | N2—C8—C7 | 107.77 (14) |
| O3—C16—C7 | 122.68 (16) | N2—C8—C9 | 118.18 (15) |
| O4—C16—C7 | 114.21 (14) | C7—C8—C9 | 134.04 (16) |
| O2—C14—C13 | 123.79 (16) | C1—C2—H2 | 120.8 (12) |
| O2—C14—C15 | 121.81 (15) | C3—C2—C1 | 118.96 (17) |
| C13—C14—C15 | 114.40 (14) | C3—C2—H2 | 120.2 (12) |
| C6—C7—C16 | 132.87 (15) | C10—C9—C8 | 120.02 (17) |
| C8—C7—C6 | 106.93 (14) | C10—C9—H9 | 123.8 (13) |
| C8—C7—C16 | 120.18 (15) | C8—C9—H9 | 116.1 (13) |
| O1—C5—C4 | 119.70 (15) | O4—C17—C18 | 106.34 (15) |
| O1—C5—C6 | 124.04 (16) | O4—C17—H17A | 107.5 (12) |
| C6—C5—C4 | 116.21 (14) | O4—C17—H17B | 108.8 (12) |
| N1—C15—C4 | 123.62 (16) | C18—C17—H17A | 112.4 (11) |
| N1—C15—C14 | 115.71 (15) | C18—C17—H17B | 112.2 (12) |
| C4—C15—C14 | 120.65 (15) | H17A—C17—H17B | 109.3 (16) |
| N2—C12—H12 | 117.3 (13) | F1—C11—C12 | 117.46 (17) |
| C11—C12—N2 | 116.90 (17) | F1—C11—C10 | 118.99 (16) |
| C11—C12—H12 | 125.8 (13) | C12—C11—C10 | 123.55 (17) |
| N1—C1—C2 | 123.66 (17) | ||
| O2—C14—C15—N1 | −2.0 (2) | C14—C13—C6—C5 | −1.6 (3) |
| O2—C14—C15—C4 | 179.59 (17) | C7—C6—C5—O1 | 3.6 (3) |
| O3—C16—C7—C6 | 175.43 (17) | C7—C6—C5—C4 | −179.07 (17) |
| O3—C16—C7—C8 | −2.5 (3) | C7—C8—C9—C10 | −178.48 (19) |
| N1—C1—C2—C3 | −0.5 (3) | C5—C4—C15—N1 | −177.38 (16) |
| O4—C16—C7—C6 | −4.0 (3) | C5—C4—C15—C14 | 0.9 (3) |
| O4—C16—C7—C8 | 178.07 (15) | C5—C4—C3—C2 | 177.18 (16) |
| C4—C3—C2—C1 | 1.0 (3) | C5—C6—C7—C16 | 3.4 (3) |
| N2—C13—C6—C7 | −0.53 (19) | C5—C6—C7—C8 | −178.46 (17) |
| N2—C13—C6—C5 | 178.65 (14) | C15—N1—C1—C2 | 0.2 (3) |
| N2—C13—C14—O2 | 0.6 (3) | C15—C4—C5—O1 | 175.68 (17) |
| N2—C13—C14—C15 | −179.64 (15) | C15—C4—C5—C6 | −1.8 (2) |
| N2—C12—C11—F1 | −179.88 (16) | C15—C4—C3—C2 | −1.3 (2) |
| N2—C12—C11—C10 | 0.0 (3) | C12—N2—C13—C6 | −178.91 (16) |
| N2—C8—C9—C10 | −0.2 (3) | C12—N2—C13—C14 | 1.3 (3) |
| C13—N2—C12—C11 | 178.63 (17) | C12—N2—C8—C7 | 179.30 (15) |
| C13—N2—C8—C7 | 0.03 (18) | C12—N2—C8—C9 | 0.6 (2) |
| C13—N2—C8—C9 | −178.66 (15) | C1—N1—C15—C4 | −0.4 (3) |
| C13—C6—C7—C16 | −177.61 (17) | C1—N1—C15—C14 | −178.78 (16) |
| C13—C6—C7—C8 | 0.54 (19) | C3—C4—C5—O1 | −2.7 (2) |
| C13—C6—C5—O1 | −175.32 (17) | C3—C4—C5—C6 | 179.87 (15) |
| C13—C6—C5—C4 | 2.0 (2) | C3—C4—C15—N1 | 1.0 (3) |
| C13—C14—C15—N1 | 178.18 (15) | C3—C4—C15—C14 | 179.27 (15) |
| C13—C14—C15—C4 | −0.2 (2) | C8—N2—C13—C6 | 0.31 (19) |
| C6—C13—C14—O2 | −179.20 (17) | C8—N2—C13—C14 | −179.48 (16) |
| C6—C13—C14—C15 | 0.6 (3) | C8—N2—C12—C11 | −0.5 (3) |
| C6—C7—C8—N2 | −0.35 (18) | C9—C10—C11—F1 | −179.75 (17) |
| C6—C7—C8—C9 | 178.04 (18) | C9—C10—C11—C12 | 0.4 (3) |
| C16—O4—C17—C18 | −177.06 (16) | C17—O4—C16—O3 | 0.8 (3) |
| C16—C7—C8—N2 | 178.08 (15) | C17—O4—C16—C7 | −179.83 (15) |
| C16—C7—C8—C9 | −3.5 (3) | C11—C10—C9—C8 | −0.2 (3) |
| C14—C13—C6—C7 | 179.26 (16) |
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: LH5489).
References
- Agilent (2011). CrysAlis PRO and CrysAlis RED Agilent Technologies Ltd, Yarnton, Oxfordshire, England.
- Cheng, Y., An, L. K., Wu, N., Wang, X. D., Bu, X. Z., Huang, Z. S. & Gu, L. Q. (2008). Bioorg. Med. Chem. 16, 4617–4625. [DOI] [PubMed]
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341.
- Pommier, Y. (2006). Nat. Rev. Cancer, 6, 789–802. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Shen, D. Q., Cheng, Y., An, L. K., Bu, X. Z., Huang, Z. S. & Gu, L. Q. (2008). Chin. Chem. Lett. 19, 533–536.
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
- Wu, N., Wu, X. W., Agama, K., Pommier, Y., Du, J., Li, D., Gu, L. Q., Huang, Z. S. & An, L. K. (2010). Biochemistry, 49, 10131–10136. [DOI] [PMC free article] [PubMed]
- Wu, X. W., Wu, Z. P., Wang, L. X., Zhang, H. B., Chen, J. W., Zhang, W., Gu, L. Q., Huang, Z. S. & An, L. K. (2011). Eur. J. Med. Chem. 46, 4625–4633. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S160053681202692X/lh5489sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S160053681202692X/lh5489Isup2.hkl
Supplementary material file. DOI: 10.1107/S160053681202692X/lh5489Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report