Abstract
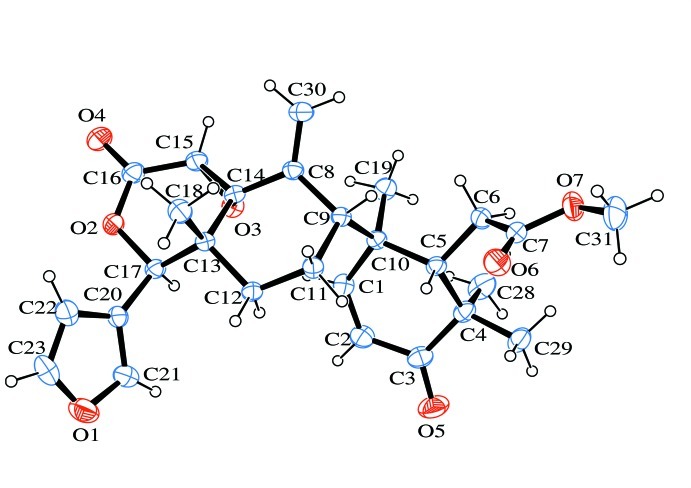
The title compound (systematic name: methyl 2-{(1R,2R)-2-[(1aS,4S,4aS,8aS)-4-(furan-3-yl)-4a-methyl-8-methylene-2-oxooctahydrooxireno[2,3-d]isochromen-7-yl]-2,6,6-trimethyl-5-oxocyclohex-3-en-1-yl}acetate), C27H32O7, was isolated from X. moluccensis seeds from Thailand. The conformations of the six-membered rings are distorted half-chair, chair and half-chair for the isolated cyclohexane, fused cyclohexane and lactone rings, respectively. In addition, the lactone ring bears in an equatorial orientation an essentially planar furan ring (r.m.s. deviation = 0.004 Å), which forms an angle of 63.87 (13)° with the mean plane defined by the ten atoms of the two fused six-membered rings (r.m.s. deviation = 0.213 Å). The absolute configuration was fixed on the basis of literature data.
Related literature
For general background to limonoids and their activities, see: Alvi et al. (1991 ▶); Yu et al. (2007 ▶); Li et al. (2009 ▶). For related structures, see: Chanin et al. (2010 ▶); Pudhom et al. (2009 ▶, 2010 ▶). For the bioactivity of limonoids, see: Koul et al. (2004 ▶); Endo et al. (2002 ▶); Nakagawa et al. (2001 ▶); Ravangpai et al. (2011 ▶). For puckering parameters, see: Cremer & Pople (1975 ▶).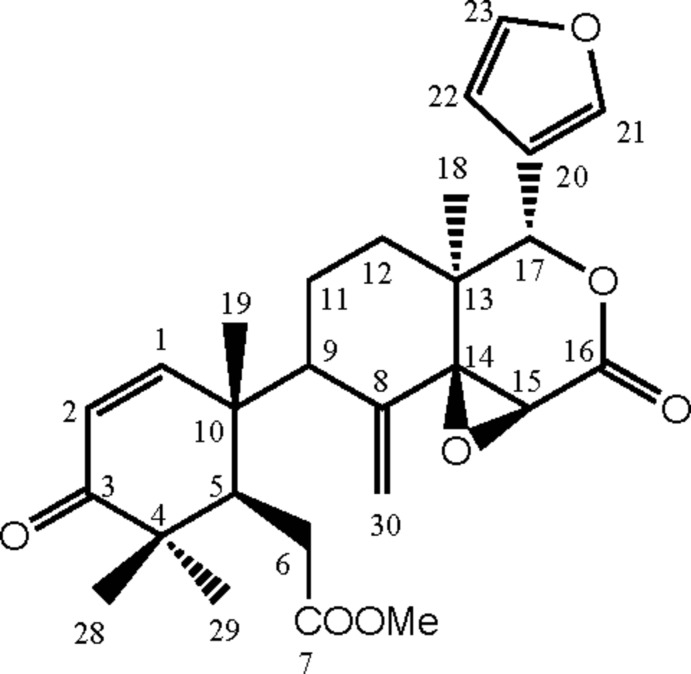
Experimental
Crystal data
C27H32O7
M r = 468.53
Orthorhombic,
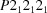
a = 8.8125 (5) Å
b = 12.5907 (7) Å
c = 21.9393 (11) Å
V = 2434.3 (2) Å3
Z = 4
Mo Kα radiation
μ = 0.09 mm−1
T = 296 K
0.48 × 0.40 × 0.36 mm
Data collection
Bruker SMART APEXII CCD area-detector diffractometer
13719 measured reflections
3132 independent reflections
2725 reflections with I > 2σ(I)
R int = 0.020
Refinement
R[F 2 > 2σ(F 2)] = 0.041
wR(F 2) = 0.117
S = 1.11
5520 reflections
312 parameters
H-atom parameters constrained
Δρmax = 0.68 e Å−3
Δρmin = −0.18 e Å−3
Data collection: APEX2 (Bruker, 2008 ▶); cell refinement: SAINT (Bruker, 2008 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 (Farrugia, 1997 ▶; software used to prepare material for publication: publCIF (Westrip, 2010 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536812027705/lr2061sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812027705/lr2061Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812027705/lr2061Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Acknowledgments
Financial support from the 90th Anniversary of Chulalongkorn University Fund (Ratchadaphisek Somphot Endowment Fund) and the Higher Education Research Promotion and National Research University Project of Thailand, Office of the Higher Education Commission (AS613A), is gratefully acknowledged. The authors are also grateful for research funding from the Thai Government Stimulus Package 2 (TKK2555), under the Project for Establishment of a Comprehensive Center for Innovative Food, Health Products and Agriculture.
supplementary crystallographic information
Comment
Limonoids are triterpene derivatives from a precursor with a 4,4,8-trimethyl-17-furanylsteroid skeleton. Limonoid examination of the Meliaceae family is of growing interest due to a range of biological activities, such as insect antifeedants and growth regulators, antibacterial, antifungal, antimalarial, anticancer, antiviral and anti-inflammatory activities (Koul et al., 2004; Endo et al., 2002; Nakagawa et al., 2001; Ravangpai, et al., 2011). The genus Xylocarpus (Meliaceae) has proved to be a rich source of an array of structurally diverse limonoids, including gedunin, andirobin, mexicanolide and phragmalin type limonoids, with a broad range of biological activities (Alvi et al., 1991; Yu et al., 2007; Li et al., 2009). We have recently reported the isolation and identification a number of limonoids from three Thai mangroves in this genus, X. granatum, X. moluccensis and X. rumphii (Chanin et al., 2010; Pudhom et al., 2009; Pudhom et al., 2010). Herein, we report the complete assignments of NMR and the crystal structure of the title compound isolated from X. moluccensis seeds.
In the molecular structure, the conformation of the six-membered rings are distorted half-chair, chair and half-chair for the isolated cyclohenane, fused cyclohexane and lactone ring respectively (Cremer & Pople, 1975). In addition, the lactone ring bears in equatorial orientation a planar furan ring (r.m.s. deviation= 0.004 Å) which form an angle of 63.87 (13)o with the mean square plane (r.m.s. deviation Å) defined by the ten atoms of the two fused six-membered rings.
Experimental
General Experiment Procedures. Melting point was measured using a Fisher-Johns melting point apparatus. NMR spectra were recorded with a Bruker AV400 (1H, 400 MHz; 13C, 100 MHz) spectrometer using tetramethylsilane as an internal standard. Mass spectra were obtained from a Bruker micrOTOF mass spectrometer.
Plant Material. Fruits of X. moluccensis were collected from Surat Thani province, Thailand, in January 2010. Plant materials were identified by Royal Forest Department, Bangkok, Thailand.
Extraction and Isolation of Andirobin (1). Air-dried powdered seeds of X. moluccensis (2 kg) were extracted with MeOH (5L x 2, each for two days) at room temperature. Extracts were pooled and the solvent were removed under reduced pressure. The combined MeOH extract was then suspended in water and partitioned with EtOAc. The EtOAc crude extract obtained (30 g) was chromatographed on a sililca gel column eluted with a gradient of acetone-hexane (from 1:9 to 1:0) to yield 12 fractions. Fraction 2 was further purified by silica gel column chromatography eluting with a 1:9 mixture of acetone-hexane and recrystallized from MeOH to afford the title compound (1, 25.0 mg).
Andirobin (1): colorless crystals; 1H NMR (400 MHz, CDCl3) d 7.34 (1H, s, H-23), 7.33 (1H, s, H-21), 7.07 (1H, d, J = 10.4 Hz, H-1), 6.27 (1H, s, H-22), 5.99 (1H, d, J = 10.4 Hz, H-2), 5.41 (1H, s, H-17), 5.30 (1H, s, H-30a), 5.20 (1H, s, H-30b), 3.97 (1H, s, H-15), 3.64 (3H, s, 7-COOCH3), 2.62 (1H, dd, J = 3.2, 6.8 Hz, H-5), 2.44 (1H, dd, J = 7.2, 17.2 Hz, H-6a), 2.39 (1H, d, J = 6.8 Hz, H-9), 2.28 (1H, dd, J = 3.2, 17.2 Hz, H-6 b), 1.90 (1H, m, H-11a), 1.73 (1H, m, H-11b), 1.60 (1H, m, H-12a), 1.16 (1H, m, H-12b), 1.04 (3H, s, 28-CH3), 1.01 (3H, s, 29-CH3), 0.90 (3H, s, 19-CH3), 0.87 (3H, s, 18-CH3);
13C NMR (100 MHz, CDCl3) d 203.7 (C=O, C-3), 174.3 (C=O, C-7), 166.7 (C=O, C-16), 153.5 (CH, C-1), 143.2 (CH, C-23), 140.9 (CH, C-21), 138.9 (C, C-8), 125.7 (CH, C-2), 122.3 (CH2, C-30), 119.8 (C, C-20), 109.7 (CH, C-22), 77.4 (CH, C-15), 67.8 (C, C-14), 55.5 (CH, C-17), 52.1 (CH3, 7-COOCH3), 48.8 (CH, C-9), 46.1 (C, C-4), 43.1 (C, C-10), 42.8 (CH, C-5), 38.6 (C, C-13), 31.5 (CH2, C-6), 29.5 (CH2, C-12), 22.7 (CH3, C-29), 22.5 (CH3, C-28), 21.3 (CH2, C-11), 20.2 (CH3, C-19), 14.6 (CH3, C-18).
Refinement
All H atoms were geometrically positioned and treated as riding atoms with distances C—H = 0.96 Å (CH3), 0.97 Å (CH2), 0.93 Å (CH), and Uiso(H) = 1.20 Ueq(C) for methylene and aromatic, 1.50 Ueq(C) for methyl. The absolute structure could not be determined from the X-ray analysis, but it was known from earlier work on related compounds (e.g. Alvi et al., 1991, Yu et al., 2007 and Li et al., 2009). 2388 Friedel pairs were therefore merged before the final refinement.
The maximum residual density ( 0.68 eÅ3) is larger than normally expected. However, the nearest atom to the corresponding minimum is O5 at 2.74 Å, which seems to indicate that de residual density can be associated to unmodeled disordered solvent molecules.
Figures
Fig. 1.
The molecular structure of the title compound with ellipsoids drawn at the 30% probability level.
Crystal data
| C27H32O7 | Z = 4 |
| Mr = 468.53 | F(000) = 1000 |
| Orthorhombic, P212121 | Dx = 1.278 Mg m−3 |
| Hall symbol: P 2ac 2ab | Mo Kα radiation, λ = 0.71073 Å |
| a = 8.8125 (5) Å | µ = 0.09 mm−1 |
| b = 12.5907 (7) Å | T = 296 K |
| c = 21.9393 (11) Å | Prism, colourless |
| V = 2434.3 (2) Å3 | 0.48 × 0.40 × 0.36 mm |
Data collection
| Bruker SMART APEXII CCD area-detector diffractometer | 2725 reflections with I > 2σ(I) |
| Radiation source: Mo Kα | Rint = 0.020 |
| Graphite monochromator | θmax = 27.5°, θmin = 1.9° |
| φ and ω scans | h = −11→7 |
| 13719 measured reflections | k = −15→15 |
| 3132 independent reflections | l = −28→19 |
Refinement
| Refinement on F2 | 0 restraints |
| Least-squares matrix: full | H-atom parameters constrained |
| R[F2 > 2σ(F2)] = 0.041 | w = 1/[σ2(Fo2) + (0.0714P)2 + 0.1995P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.117 | (Δ/σ)max = 0.014 |
| S = 1.11 | Δρmax = 0.68 e Å−3 |
| 5520 reflections | Δρmin = −0.18 e Å−3 |
| 312 parameters |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.8749 (3) | 0.19440 (18) | 0.08892 (10) | 0.0410 (5) | |
| H1 | 0.7868 | 0.1870 | 0.0660 | 0.049* | |
| C2 | 0.9926 (3) | 0.2397 (2) | 0.06213 (12) | 0.0504 (6) | |
| H2 | 0.9842 | 0.2570 | 0.0211 | 0.060* | |
| C3 | 1.1348 (3) | 0.2640 (2) | 0.09316 (12) | 0.0487 (6) | |
| C4 | 1.1303 (3) | 0.25998 (19) | 0.16248 (12) | 0.0427 (5) | |
| C5 | 1.0339 (2) | 0.16189 (17) | 0.18228 (10) | 0.0344 (4) | |
| H5 | 1.0890 | 0.1003 | 0.1664 | 0.041* | |
| C6 | 1.0335 (3) | 0.1469 (2) | 0.25150 (11) | 0.0417 (5) | |
| H6A | 1.0424 | 0.2159 | 0.2708 | 0.050* | |
| H6B | 0.9369 | 0.1166 | 0.2636 | 0.050* | |
| C7 | 1.1586 (3) | 0.07683 (19) | 0.27427 (10) | 0.0406 (5) | |
| C8 | 0.6587 (2) | 0.00557 (17) | 0.15184 (10) | 0.0352 (4) | |
| C9 | 0.8277 (2) | 0.03182 (16) | 0.15559 (9) | 0.0324 (4) | |
| H9 | 0.8598 | 0.0073 | 0.1960 | 0.039* | |
| C10 | 0.8716 (2) | 0.15387 (17) | 0.15342 (10) | 0.0338 (4) | |
| C11 | 0.9130 (2) | −0.04026 (19) | 0.10972 (10) | 0.0377 (5) | |
| H11A | 1.0170 | −0.0156 | 0.1068 | 0.045* | |
| H11B | 0.9156 | −0.1118 | 0.1261 | 0.045* | |
| C12 | 0.8468 (2) | −0.04512 (19) | 0.04580 (10) | 0.0364 (4) | |
| H12A | 0.9024 | −0.0975 | 0.0223 | 0.044* | |
| H12B | 0.8606 | 0.0233 | 0.0262 | 0.044* | |
| C13 | 0.6761 (2) | −0.07398 (16) | 0.04482 (9) | 0.0321 (4) | |
| C14 | 0.5934 (2) | −0.00153 (17) | 0.08910 (10) | 0.0338 (4) | |
| C15 | 0.4320 (2) | 0.0194 (2) | 0.07674 (11) | 0.0436 (5) | |
| H15 | 0.3670 | 0.0315 | 0.1123 | 0.052* | |
| C16 | 0.3595 (3) | −0.0313 (2) | 0.02300 (11) | 0.0466 (6) | |
| C17 | 0.6142 (2) | −0.05036 (19) | −0.01938 (10) | 0.0369 (5) | |
| H17 | 0.6351 | 0.0243 | −0.0288 | 0.044* | |
| C18 | 0.6499 (3) | −0.18975 (18) | 0.06402 (11) | 0.0447 (5) | |
| H18A | 0.5447 | −0.2074 | 0.0588 | 0.067* | |
| H18B | 0.6776 | −0.1984 | 0.1060 | 0.067* | |
| H18C | 0.7109 | −0.2358 | 0.0392 | 0.067* | |
| C19 | 0.7500 (3) | 0.21722 (19) | 0.18871 (12) | 0.0447 (6) | |
| H19A | 0.7865 | 0.2878 | 0.1964 | 0.067* | |
| H19B | 0.7291 | 0.1825 | 0.2267 | 0.067* | |
| H19C | 0.6587 | 0.2208 | 0.1649 | 0.067* | |
| C20 | 0.6791 (3) | −0.11713 (19) | −0.06933 (10) | 0.0394 (5) | |
| C21 | 0.7912 (3) | −0.0869 (2) | −0.10764 (12) | 0.0569 (7) | |
| H21 | 0.8380 | −0.0207 | −0.1070 | 0.068* | |
| C22 | 0.6448 (4) | −0.2222 (2) | −0.08653 (13) | 0.0603 (7) | |
| H22 | 0.5727 | −0.2659 | −0.0684 | 0.072* | |
| C23 | 0.7334 (4) | −0.2479 (3) | −0.13315 (12) | 0.0614 (8) | |
| H23 | 0.7318 | −0.3129 | −0.1533 | 0.074* | |
| C28 | 1.0646 (3) | 0.3679 (2) | 0.18309 (17) | 0.0640 (8) | |
| H28A | 1.0457 | 0.3662 | 0.2262 | 0.096* | |
| H28B | 0.9713 | 0.3813 | 0.1618 | 0.096* | |
| H28C | 1.1360 | 0.4233 | 0.1741 | 0.096* | |
| C29 | 1.2920 (3) | 0.2503 (2) | 0.18786 (14) | 0.0534 (6) | |
| H29A | 1.3322 | 0.1816 | 0.1780 | 0.080* | |
| H29B | 1.2898 | 0.2590 | 0.2313 | 0.080* | |
| H29C | 1.3549 | 0.3043 | 0.1701 | 0.080* | |
| C30 | 0.5775 (3) | −0.0189 (2) | 0.20065 (11) | 0.0503 (6) | |
| H30A | 0.4774 | −0.0411 | 0.1963 | 0.060* | |
| H30B | 0.6205 | −0.0139 | 0.2392 | 0.060* | |
| C31 | 1.3128 (4) | 0.0371 (3) | 0.35953 (15) | 0.0744 (9) | |
| H31A | 1.2729 | −0.0330 | 0.3658 | 0.112* | |
| H31B | 1.3411 | 0.0674 | 0.3980 | 0.112* | |
| H31C | 1.4004 | 0.0333 | 0.3336 | 0.112* | |
| O1 | 0.8261 (3) | −0.1661 (2) | −0.14718 (10) | 0.0712 (6) | |
| O2 | 0.44932 (17) | −0.06685 (16) | −0.02238 (8) | 0.0484 (4) | |
| O3 | 0.54407 (18) | 0.09958 (13) | 0.06392 (8) | 0.0427 (4) | |
| O4 | 0.22495 (19) | −0.0432 (2) | 0.01901 (10) | 0.0681 (6) | |
| O5 | 1.2467 (2) | 0.2941 (2) | 0.06570 (11) | 0.0795 (7) | |
| O6 | 1.2129 (2) | 0.00409 (15) | 0.24700 (8) | 0.0535 (4) | |
| O7 | 1.1985 (2) | 0.10284 (18) | 0.33109 (8) | 0.0591 (5) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0448 (12) | 0.0358 (11) | 0.0423 (12) | 0.0015 (10) | −0.0057 (10) | 0.0053 (9) |
| C2 | 0.0575 (14) | 0.0491 (14) | 0.0446 (13) | −0.0022 (12) | 0.0010 (11) | 0.0135 (11) |
| C3 | 0.0472 (13) | 0.0420 (12) | 0.0571 (15) | −0.0019 (11) | 0.0068 (12) | 0.0113 (11) |
| C4 | 0.0369 (10) | 0.0364 (11) | 0.0549 (14) | −0.0032 (9) | 0.0027 (10) | −0.0047 (10) |
| C5 | 0.0313 (9) | 0.0342 (10) | 0.0377 (11) | 0.0007 (9) | 0.0029 (9) | −0.0039 (9) |
| C6 | 0.0383 (11) | 0.0486 (13) | 0.0380 (11) | 0.0017 (10) | 0.0011 (10) | −0.0083 (10) |
| C7 | 0.0340 (10) | 0.0505 (13) | 0.0373 (11) | −0.0068 (10) | 0.0002 (9) | −0.0020 (10) |
| C8 | 0.0333 (9) | 0.0357 (10) | 0.0368 (10) | 0.0013 (9) | 0.0054 (9) | −0.0015 (9) |
| C9 | 0.0317 (9) | 0.0339 (10) | 0.0316 (9) | 0.0027 (8) | 0.0000 (8) | 0.0026 (8) |
| C10 | 0.0317 (9) | 0.0330 (10) | 0.0368 (10) | 0.0028 (8) | 0.0011 (9) | −0.0006 (8) |
| C11 | 0.0320 (9) | 0.0398 (11) | 0.0414 (11) | 0.0074 (9) | −0.0018 (9) | −0.0077 (10) |
| C12 | 0.0293 (9) | 0.0411 (11) | 0.0386 (10) | 0.0025 (9) | 0.0041 (9) | −0.0052 (9) |
| C13 | 0.0306 (9) | 0.0333 (10) | 0.0325 (10) | −0.0015 (8) | 0.0019 (8) | 0.0001 (8) |
| C14 | 0.0295 (9) | 0.0351 (10) | 0.0367 (10) | 0.0001 (8) | 0.0056 (8) | 0.0020 (9) |
| C15 | 0.0309 (10) | 0.0559 (14) | 0.0438 (12) | 0.0045 (10) | 0.0045 (9) | −0.0038 (11) |
| C16 | 0.0320 (10) | 0.0609 (15) | 0.0470 (13) | 0.0008 (11) | 0.0005 (10) | 0.0015 (11) |
| C17 | 0.0299 (9) | 0.0444 (11) | 0.0365 (10) | −0.0002 (9) | 0.0006 (8) | 0.0017 (10) |
| C18 | 0.0554 (13) | 0.0355 (11) | 0.0433 (12) | −0.0041 (11) | 0.0017 (11) | 0.0028 (9) |
| C19 | 0.0380 (11) | 0.0396 (12) | 0.0565 (14) | 0.0083 (10) | 0.0023 (11) | −0.0089 (11) |
| C20 | 0.0365 (10) | 0.0490 (12) | 0.0327 (10) | −0.0007 (10) | −0.0034 (9) | 0.0000 (10) |
| C21 | 0.0575 (15) | 0.0646 (16) | 0.0486 (14) | −0.0055 (14) | 0.0143 (13) | −0.0092 (13) |
| C22 | 0.0700 (18) | 0.0625 (16) | 0.0485 (14) | −0.0169 (15) | 0.0043 (14) | −0.0103 (13) |
| C23 | 0.079 (2) | 0.0610 (17) | 0.0440 (14) | 0.0081 (16) | −0.0053 (14) | −0.0161 (13) |
| C28 | 0.0568 (16) | 0.0386 (13) | 0.097 (2) | −0.0021 (12) | 0.0090 (16) | −0.0122 (14) |
| C29 | 0.0393 (11) | 0.0538 (15) | 0.0671 (16) | −0.0080 (12) | −0.0017 (12) | −0.0075 (13) |
| C30 | 0.0443 (12) | 0.0640 (16) | 0.0426 (12) | −0.0058 (12) | 0.0092 (11) | 0.0043 (12) |
| C31 | 0.0599 (16) | 0.102 (3) | 0.0610 (17) | 0.0075 (19) | −0.0247 (15) | −0.0005 (18) |
| O1 | 0.0636 (12) | 0.0953 (17) | 0.0548 (11) | 0.0116 (13) | 0.0136 (10) | −0.0144 (11) |
| O2 | 0.0303 (7) | 0.0707 (12) | 0.0441 (9) | −0.0009 (8) | −0.0025 (7) | −0.0084 (8) |
| O3 | 0.0379 (7) | 0.0408 (8) | 0.0493 (9) | 0.0077 (7) | −0.0028 (7) | 0.0008 (7) |
| O4 | 0.0308 (8) | 0.1065 (17) | 0.0671 (12) | −0.0061 (10) | 0.0017 (9) | −0.0101 (13) |
| O5 | 0.0559 (12) | 0.110 (2) | 0.0727 (13) | −0.0154 (12) | 0.0136 (11) | 0.0291 (14) |
| O6 | 0.0519 (10) | 0.0581 (11) | 0.0504 (10) | 0.0105 (9) | −0.0017 (8) | −0.0012 (9) |
| O7 | 0.0537 (10) | 0.0788 (13) | 0.0450 (10) | 0.0036 (10) | −0.0149 (8) | −0.0079 (9) |
Geometric parameters (Å, º)
| C1—C2 | 1.321 (3) | C14—C15 | 1.471 (3) |
| C1—C10 | 1.505 (3) | C15—O3 | 1.440 (3) |
| C1—H1 | 0.9300 | C15—C16 | 1.485 (3) |
| C2—C3 | 1.459 (4) | C15—H15 | 0.9800 |
| C2—H2 | 0.9300 | C16—O4 | 1.198 (3) |
| C3—O5 | 1.215 (3) | C16—O2 | 1.348 (3) |
| C3—C4 | 1.522 (4) | C17—O2 | 1.469 (2) |
| C4—C29 | 1.534 (3) | C17—C20 | 1.495 (3) |
| C4—C28 | 1.545 (3) | C17—H17 | 0.9800 |
| C4—C5 | 1.561 (3) | C18—H18A | 0.9600 |
| C5—C6 | 1.530 (3) | C18—H18B | 0.9600 |
| C5—C10 | 1.567 (3) | C18—H18C | 0.9600 |
| C5—H5 | 0.9800 | C19—H19A | 0.9600 |
| C6—C7 | 1.498 (3) | C19—H19B | 0.9600 |
| C6—H6A | 0.9700 | C19—H19C | 0.9600 |
| C6—H6B | 0.9700 | C20—C21 | 1.351 (4) |
| C7—O6 | 1.194 (3) | C20—C22 | 1.408 (4) |
| C7—O7 | 1.336 (3) | C21—O1 | 1.358 (3) |
| C8—C30 | 1.324 (3) | C21—H21 | 0.9300 |
| C8—C14 | 1.495 (3) | C22—C23 | 1.327 (4) |
| C8—C9 | 1.528 (3) | C22—H22 | 0.9300 |
| C9—C11 | 1.550 (3) | C23—O1 | 1.350 (4) |
| C9—C10 | 1.585 (3) | C23—H23 | 0.9300 |
| C9—H9 | 0.9800 | C28—H28A | 0.9600 |
| C10—C19 | 1.545 (3) | C28—H28B | 0.9600 |
| C11—C12 | 1.520 (3) | C28—H28C | 0.9600 |
| C11—H11A | 0.9700 | C29—H29A | 0.9600 |
| C11—H11B | 0.9700 | C29—H29B | 0.9600 |
| C12—C13 | 1.547 (3) | C29—H29C | 0.9600 |
| C12—H12A | 0.9700 | C30—H30A | 0.9300 |
| C12—H12B | 0.9700 | C30—H30B | 0.9300 |
| C13—C14 | 1.519 (3) | C31—O7 | 1.445 (4) |
| C13—C18 | 1.535 (3) | C31—H31A | 0.9600 |
| C13—C17 | 1.540 (3) | C31—H31B | 0.9600 |
| C14—O3 | 1.454 (3) | C31—H31C | 0.9600 |
| C2—C1—C10 | 125.4 (2) | C15—C14—C13 | 116.97 (19) |
| C2—C1—H1 | 117.3 | C8—C14—C13 | 116.10 (17) |
| C10—C1—H1 | 117.3 | O3—C15—C14 | 59.93 (13) |
| C1—C2—C3 | 123.9 (2) | O3—C15—C16 | 116.2 (2) |
| C1—C2—H2 | 118.1 | C14—C15—C16 | 119.0 (2) |
| C3—C2—H2 | 118.1 | O3—C15—H15 | 116.5 |
| O5—C3—C2 | 122.1 (2) | C14—C15—H15 | 116.5 |
| O5—C3—C4 | 121.8 (3) | C16—C15—H15 | 116.5 |
| C2—C3—C4 | 115.9 (2) | O4—C16—O2 | 119.0 (2) |
| C3—C4—C29 | 109.9 (2) | O4—C16—C15 | 122.5 (2) |
| C3—C4—C28 | 105.8 (2) | O2—C16—C15 | 118.41 (18) |
| C29—C4—C28 | 108.2 (2) | O2—C17—C20 | 105.46 (18) |
| C3—C4—C5 | 108.59 (19) | O2—C17—C13 | 111.37 (17) |
| C29—C4—C5 | 110.0 (2) | C20—C17—C13 | 115.22 (18) |
| C28—C4—C5 | 114.23 (19) | O2—C17—H17 | 108.2 |
| C6—C5—C4 | 112.02 (19) | C20—C17—H17 | 108.2 |
| C6—C5—C10 | 113.03 (17) | C13—C17—H17 | 108.2 |
| C4—C5—C10 | 115.81 (18) | C13—C18—H18A | 109.5 |
| C6—C5—H5 | 104.9 | C13—C18—H18B | 109.5 |
| C4—C5—H5 | 104.9 | H18A—C18—H18B | 109.5 |
| C10—C5—H5 | 104.9 | C13—C18—H18C | 109.5 |
| C7—C6—C5 | 113.70 (19) | H18A—C18—H18C | 109.5 |
| C7—C6—H6A | 108.8 | H18B—C18—H18C | 109.5 |
| C5—C6—H6A | 108.8 | C10—C19—H19A | 109.5 |
| C7—C6—H6B | 108.8 | C10—C19—H19B | 109.5 |
| C5—C6—H6B | 108.8 | H19A—C19—H19B | 109.5 |
| H6A—C6—H6B | 107.7 | C10—C19—H19C | 109.5 |
| O6—C7—O7 | 123.4 (2) | H19A—C19—H19C | 109.5 |
| O6—C7—C6 | 125.4 (2) | H19B—C19—H19C | 109.5 |
| O7—C7—C6 | 111.1 (2) | C21—C20—C22 | 104.8 (2) |
| C30—C8—C14 | 121.5 (2) | C21—C20—C17 | 125.2 (2) |
| C30—C8—C9 | 122.2 (2) | C22—C20—C17 | 130.0 (2) |
| C14—C8—C9 | 115.96 (17) | C20—C21—O1 | 110.9 (3) |
| C8—C9—C11 | 108.14 (17) | C20—C21—H21 | 124.6 |
| C8—C9—C10 | 116.50 (17) | O1—C21—H21 | 124.6 |
| C11—C9—C10 | 115.45 (17) | C23—C22—C20 | 108.0 (3) |
| C8—C9—H9 | 105.2 | C23—C22—H22 | 126.0 |
| C11—C9—H9 | 105.2 | C20—C22—H22 | 126.0 |
| C10—C9—H9 | 105.2 | C22—C23—O1 | 110.2 (3) |
| C1—C10—C19 | 108.03 (19) | C22—C23—H23 | 124.9 |
| C1—C10—C5 | 109.93 (18) | O1—C23—H23 | 124.9 |
| C19—C10—C5 | 113.42 (18) | C4—C28—H28A | 109.5 |
| C1—C10—C9 | 111.20 (17) | C4—C28—H28B | 109.5 |
| C19—C10—C9 | 108.42 (17) | H28A—C28—H28B | 109.5 |
| C5—C10—C9 | 105.86 (16) | C4—C28—H28C | 109.5 |
| C12—C11—C9 | 115.89 (17) | H28A—C28—H28C | 109.5 |
| C12—C11—H11A | 108.3 | H28B—C28—H28C | 109.5 |
| C9—C11—H11A | 108.3 | C4—C29—H29A | 109.5 |
| C12—C11—H11B | 108.3 | C4—C29—H29B | 109.5 |
| C9—C11—H11B | 108.3 | H29A—C29—H29B | 109.5 |
| H11A—C11—H11B | 107.4 | C4—C29—H29C | 109.5 |
| C11—C12—C13 | 113.28 (18) | H29A—C29—H29C | 109.5 |
| C11—C12—H12A | 108.9 | H29B—C29—H29C | 109.5 |
| C13—C12—H12A | 108.9 | C8—C30—H30A | 120.0 |
| C11—C12—H12B | 108.9 | C8—C30—H30B | 120.0 |
| C13—C12—H12B | 108.9 | H30A—C30—H30B | 120.0 |
| H12A—C12—H12B | 107.7 | O7—C31—H31A | 109.5 |
| C14—C13—C18 | 108.80 (18) | O7—C31—H31B | 109.5 |
| C14—C13—C17 | 107.38 (17) | H31A—C31—H31B | 109.5 |
| C18—C13—C17 | 112.39 (18) | O7—C31—H31C | 109.5 |
| C14—C13—C12 | 108.47 (17) | H31A—C31—H31C | 109.5 |
| C18—C13—C12 | 111.46 (19) | H31B—C31—H31C | 109.5 |
| C17—C13—C12 | 108.19 (17) | C23—O1—C21 | 106.1 (2) |
| O3—C14—C15 | 58.98 (15) | C16—O2—C17 | 120.06 (18) |
| O3—C14—C8 | 114.37 (18) | C15—O3—C14 | 61.09 (14) |
| C15—C14—C8 | 122.08 (19) | C7—O7—C31 | 116.5 (2) |
| O3—C14—C13 | 115.22 (17) | ||
| C10—C1—C2—C3 | −4.6 (4) | C9—C8—C14—C15 | −153.4 (2) |
| C1—C2—C3—O5 | 171.1 (3) | C30—C8—C14—C13 | −122.1 (2) |
| C1—C2—C3—C4 | −14.6 (4) | C9—C8—C14—C13 | 52.1 (3) |
| O5—C3—C4—C29 | −24.3 (4) | C18—C13—C14—O3 | −151.59 (18) |
| C2—C3—C4—C29 | 161.4 (2) | C17—C13—C14—O3 | −29.7 (2) |
| O5—C3—C4—C28 | 92.2 (3) | C12—C13—C14—O3 | 87.0 (2) |
| C2—C3—C4—C28 | −82.1 (3) | C18—C13—C14—C15 | −85.1 (2) |
| O5—C3—C4—C5 | −144.7 (3) | C17—C13—C14—C15 | 36.8 (3) |
| C2—C3—C4—C5 | 41.0 (3) | C12—C13—C14—C15 | 153.5 (2) |
| C3—C4—C5—C6 | 175.64 (19) | C18—C13—C14—C8 | 70.8 (2) |
| C29—C4—C5—C6 | 55.3 (3) | C17—C13—C14—C8 | −167.32 (18) |
| C28—C4—C5—C6 | −66.5 (3) | C12—C13—C14—C8 | −50.6 (2) |
| C3—C4—C5—C10 | −52.7 (2) | C8—C14—C15—O3 | 101.0 (2) |
| C29—C4—C5—C10 | −173.07 (19) | C13—C14—C15—O3 | −104.6 (2) |
| C28—C4—C5—C10 | 65.1 (3) | O3—C14—C15—C16 | 105.2 (2) |
| C4—C5—C6—C7 | −90.5 (2) | C8—C14—C15—C16 | −153.8 (2) |
| C10—C5—C6—C7 | 136.50 (19) | C13—C14—C15—C16 | 0.6 (3) |
| C5—C6—C7—O6 | −31.4 (3) | O3—C15—C16—O4 | −132.8 (3) |
| C5—C6—C7—O7 | 151.5 (2) | C14—C15—C16—O4 | 158.6 (3) |
| C30—C8—C9—C11 | 127.1 (2) | O3—C15—C16—O2 | 48.4 (3) |
| C14—C8—C9—C11 | −47.0 (2) | C14—C15—C16—O2 | −20.2 (4) |
| C30—C8—C9—C10 | −100.9 (3) | C14—C13—C17—O2 | −58.0 (2) |
| C14—C8—C9—C10 | 84.9 (2) | C18—C13—C17—O2 | 61.6 (2) |
| C2—C1—C10—C19 | 118.2 (3) | C12—C13—C17—O2 | −174.94 (18) |
| C2—C1—C10—C5 | −6.1 (3) | C14—C13—C17—C20 | −178.09 (18) |
| C2—C1—C10—C9 | −123.0 (3) | C18—C13—C17—C20 | −58.5 (3) |
| C6—C5—C10—C1 | 166.66 (19) | C12—C13—C17—C20 | 65.0 (2) |
| C4—C5—C10—C1 | 35.5 (2) | O2—C17—C20—C21 | 137.5 (3) |
| C6—C5—C10—C19 | 45.6 (3) | C13—C17—C20—C21 | −99.2 (3) |
| C4—C5—C10—C19 | −85.6 (2) | O2—C17—C20—C22 | −44.4 (3) |
| C6—C5—C10—C9 | −73.1 (2) | C13—C17—C20—C22 | 78.9 (3) |
| C4—C5—C10—C9 | 155.70 (18) | C22—C20—C21—O1 | 0.5 (3) |
| C8—C9—C10—C1 | −81.7 (2) | C17—C20—C21—O1 | 179.0 (2) |
| C11—C9—C10—C1 | 46.8 (2) | C21—C20—C22—C23 | −0.9 (3) |
| C8—C9—C10—C19 | 36.9 (3) | C17—C20—C22—C23 | −179.3 (3) |
| C11—C9—C10—C19 | 165.38 (18) | C20—C22—C23—O1 | 0.9 (4) |
| C8—C9—C10—C5 | 158.90 (17) | C22—C23—O1—C21 | −0.6 (4) |
| C11—C9—C10—C5 | −72.6 (2) | C20—C21—O1—C23 | 0.0 (3) |
| C8—C9—C11—C12 | 48.3 (3) | O4—C16—O2—C17 | 178.2 (3) |
| C10—C9—C11—C12 | −84.2 (2) | C15—C16—O2—C17 | −3.0 (4) |
| C9—C11—C12—C13 | −53.0 (3) | C20—C17—O2—C16 | 169.0 (2) |
| C11—C12—C13—C14 | 50.4 (2) | C13—C17—O2—C16 | 43.4 (3) |
| C11—C12—C13—C18 | −69.3 (2) | C16—C15—O3—C14 | −109.9 (2) |
| C11—C12—C13—C17 | 166.60 (19) | C8—C14—O3—C15 | −114.1 (2) |
| C30—C8—C14—O3 | 99.9 (3) | C13—C14—O3—C15 | 107.6 (2) |
| C9—C8—C14—O3 | −85.9 (2) | O6—C7—O7—C31 | −0.4 (4) |
| C30—C8—C14—C15 | 32.5 (3) | C6—C7—O7—C31 | 176.7 (2) |
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: LR2061).
References
- Alvi, K. A., Crews, P., Aalbergsberg, B., Prasad, R., Simpson, J. & Weavers, R. T. (1991). Tetrahedron, 47, 8943–8948.
- Bruker (2008). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Chanin, S., Nuanyai, T., Teerawatananond, T., Pengpreecha, S., Muangsin, N. & Pudhom, K. (2010). J. Nat. Prod. 73, 1456–1459. [DOI] [PubMed]
- Cremer, D. & Pople, J. A. (1975). J. Am. Chem. Soc. 97, 1354–1358.
- Endo, T., Kita, M., Shimada, T., Moriguchi, T., Hidaki, T., Matsumoto, R., Hasegawa, S. & Omura, M. (2002). Plant Biotechnol. 19, 397–403.
- Farrugia, L. J. (1997). J. Appl. Cryst. 30, 565.
- Koul, O., Sing, G., Singh, R., Daniewski, W. M. & Berlozecki, S. (2004). J. Biosci. 29, 409–416. [DOI] [PubMed]
- Li, M.-Y., Yang, X.-B., Pan, J.-Y., Feng, G., Xiao, Q., Sinkkonen, J., Satyanandamurty, T. & Wu, J. (2009). J. Nat. Prod. 72, 2110–2114. [DOI] [PubMed]
- Nakagawa, H., Duan, H. & Takaishi, Y. (2001). Chem. Pharm. Bull. 49, 649–651. [DOI] [PubMed]
- Pudhom, K., Sommit, D., Nuclear, P., Ngamrojanavanich, N. & Petsom, A. (2009). J. Nat. Prod. 72, 2188–2191. [DOI] [PubMed]
- Pudhom, K., Sommit, D., Nuclear, P., Ngamrojanavanich, N. & Petsom, A. (2010). J. Nat. Prod. 73, 263–269. [DOI] [PubMed]
- Ravangpai, W., Sommit, D., Teerawatananond, T., Sinpranee, N., Palaga, T., Pengpreecha, S., Muangsin, N. & Pudhom, K. (2011). Bioorg. Med. Chem. Lett 21, 4485–4489. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
- Yu, S., Wang, X.-N., Fan, C.-Q., Lin, L.-P., Ding, J. & Yue, J.-M. (2007). J. Nat. Prod. 70, 682–685. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536812027705/lr2061sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812027705/lr2061Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812027705/lr2061Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report