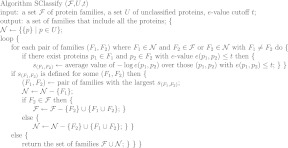Abstract
The goal of protein family classification is to group proteins into families so that proteins within the same family have common function or are related by ancestry. While supervised classification algorithms are available for this purpose, most of these approaches focus on assigning unclassified proteins to known families but do not allow for progressive construction of new families from proteins that cannot be assigned. Although unsupervised clustering algorithms are also available, they do not make use of information from known families. By computing similarities between proteins based on pairwise sequence comparisons, we develop supervised classification algorithms that achieve improved accuracy over previous approaches while allowing for construction of new families. We show that our algorithm has higher accuracy rate and lower mis-classification rate when compared to algorithms that are based on the use of multiple sequence alignments and hidden Markov models, and our algorithm performs well even on families with very few proteins and on families with low sequence similarity. A software program implementing the algorithm (SClassify) is available online (http://faculty.cse.tamu.edu/shsze/sclassify).
Key words: algorithms, gene clusters, protein families
1. Introduction
Depending on the definition of a family, proteins within gene families (or protein families) are usually homologous and have similar structure of conserved functional domains. The classification of proteins to families is an intrinsic part of comparative and evolutionary genomics. With the rapid increase in the amount of genomic data, the need for sensitive protein family classification methods continues to increase.
One strategy for protein family classification is through the use of unsupervised clustering algorithms, which take as input a large set of proteins, and perform all-against-all comparisons of sequence similarity using BLAST (Altschul et al., 1990), the Smith-Waterman local alignment algorithm (Smith and Waterman, 1981), or some other pairwise comparison algorithm. The pairwise scores are then used as a basis to apply a variety of clustering algorithms such as single linkage clustering. Other unsupervised approaches include the Markov clustering strategy (Enright et al., 2002), the density-based ordering method (Chen et al., 2006), and clustering algorithms based on the use of graph-theoretic properties (Kim and Lee, 2006). Unsupervised methods are useful for clustering a set of proteins, but they have two shortcomings: they do not assign proteins to existing families that can be found in databases such as Pfam (Bateman et al., 2000) and InterPro (Apweiler et al., 2000), and they do not make use of properties of known families, such as inter- and intra-family sequence diversity, to aid in the formation of new families.
Supervised classification algorithms can overcome some of these shortcomings. These methods include algorithms based on the use of profile hidden Markov models (Eddy, 1998), algorithms based on probabilistic suffix trees (Bejerano and Yona, 2001), algorithms based on the use of discriminative strategies such as support vector machines (Jaakkola et al., 2000; Liao and Noble, 2003), and algorithms based on sparse Markov transducers (Eskin et al., 2003). While most of these supervised techniques focus on the assignment of unclassified proteins to known families, they do not allow for progressive construction of new families from proteins that cannot be assigned.
We develop supervised classification algorithms that overcome the problems of existing supervised and unsupervised algorithms and achieve improved accuracy. By utilizing sequence similarity from pairwise comparisons, we show that our algorithm has higher accuracy rate and lower mis-classification rate when compared to algorithms that are based on the use of multiple sequence alignments and hidden Markov models. Our approach can assign proteins to existing families in databases and, by taking into account similarities between the unclassified proteins, can assign them to new families.
2. Methods
2.1. Classifying proteins to existing families
We first consider the problem of assigning an unclassified protein to known families. We assume that each protein belongs to at most one family, which consists of either individual protein domains or single domain proteins. The goal is to determine whether the unclassified protein belongs to one of the existing families. We consider the following algorithm: compute an e-value score from the given unclassified protein to each protein within the existing families by using a pairwise sequence comparison algorithm such as BLAST (Altschul et al., 1990) or the Smith-Waterman local alignment algorithm SSEARCH (Smith and Waterman, 1981), and assign the unclassified protein to the family with the highest average minus log e-value, where the average is taken over proteins in the family that have e-value below a cutoff (for the detailed algorithm see Fig. 1).
FIG. 1.
Algorithm SClassify when one unclassified protein is given.
Since sequences within the same family can have low similarity, we only consider proteins within a family that are of sufficiently high similarity to the unclassified protein during the computation of average scores. This avoids the problem of getting consistently low average scores, and thus not being able to assign any new proteins to these families. Note that the unclassified protein is assigned to at most one family that has the highest average score; thus, the e-value cutoff itself is not the only factor that decides the family assignment. When the unclassified protein is not sufficiently similar to any of the existing families, it is not assigned to any family. The worst case time complexity of the algorithm is O(fs), where f is the total number of proteins within the existing families, and s is the time to perform one pairwise comparison. The memory requirement of the algorithm is proportional to the total size of the existing families.
2.2. Constructing new families from unclassified proteins
We consider the problem when a set of more than one unclassified protein is given, and the goal is either to assign each unclassified protein to an existing family or to construct new families if necessary. We treat each unclassified protein initially as a family by itself, and iteratively merge a family that contains unclassified proteins either into an existing family or with another family that contains unclassified proteins. During each iteration, we make sure that the average minus log e-value between the two merged families is the highest among all pairs of families considered, where the average is taken over pairs of proteins that have e-value below a cutoff. The algorithm terminates when no pairs of proteins have e-value below the cutoff within all pairs of families considered, and thus it is no longer possible to merge, with the families that have not been merged into existing families becoming new families (for the detailed algorithm see Fig. 2).
FIG. 2.
Algorithm SClassify when a set of more than one unclassified protein is given.
The algorithm takes advantage of similarity between unclassified proteins but will not merge existing families together; thus, there is no need to perform pairwise comparisons between proteins in existing families. There are u(f + u) pairwise comparisons to perform, where f is the total number of proteins within the existing families, and u is the number of unclassified proteins. There are a total of O(u) iterations, with O(u(n + u)) average scores to compare within each iteration, and O(n + u) average scores to update after each merge, where n is the total number of existing families. Since each update takes constant time, the worst case time complexity of the algorithm is O(u(f + u)s + u2(n + u)), where s is the time to perform one pairwise comparison. The memory requirement of the algorithm is proportional to the number of scores that need to be stored, which is u(f + u).
3. Results
3.1. Data sets
We apply our algorithm to a few large-scale data sets, including curated families from the Pfam database (Bateman et al., 2000), protein families from the SCOP database (Murzin et al., 1995), full length prokaryotic sequences from the ProtClustDB database (Klimke et al., 2009), and curated proteins from the Swiss-Prot subset of the UniProt database (Apweiler et al., 2004). To compare the performance of our algorithm to slower algorithms, we use families within individual species from Pfam, including Arabidopsis thaliana, Caenorhabditis elegans, Drosophila melanogaster, Escherichia coli, Homo sapiens, Mus musculus, and Saccharomyces cerevisiae, with the proteins that are within each species forming a data set.
While the sequences from Pfam and SCOP are short sequences that correspond to protein domains, the sequences from ProtClustDB and UniProt are full length sequences that correspond to entire proteins (Table 1). We remove individual proteins that do not belong to any family within each of the data sets, while allowing proteins from different species to be within the same family in the data sets that contain multiple species. Except for UniProt, each remaining protein domain or full length sequence belongs to one family. We remove the very small percentage of proteins (less than 1%) that belong to more than one family from UniProt.
Table 1.
Data Sets for Performance Evaluation
| Data set | Family | Protein | avg_pro | avg_len |
|---|---|---|---|---|
| Pfam | 9318 | 2286710 | 245.4 | 151.1 |
| A. thaliana | 1962 | 36387 | 18.6 | 114.7 |
| C. elegans | 1164 | 15971 | 13.7 | 137.4 |
| D. melanogaster | 1294 | 13664 | 10.6 | 128.6 |
| E. coli | 404 | 2177 | 5.4 | 159.7 |
| H. sapiens | 1596 | 23051 | 14.4 | 102.2 |
| M. musculus | 1516 | 21891 | 14.4 | 110.8 |
| S. cerevisiae | 814 | 4526 | 5.6 | 157.2 |
| SCOP | 2234 | 15045 | 6.7 | 169.8 |
| ProtClustDB | 6521 | 356615 | 54.7 | 348.1 |
| UniProt | 8213 | 448469 | 54.6 | 343.7 |
For each set, family is the number of families, protein is the total number of proteins in all families, avg_pro is the average number of proteins within a family, and avg_len is the average length of proteins in amino acids.
3.2. Choice of parameters
We evaluate the accuracy of our algorithm by employing the 10-fold cross validation procedure over different e-value cutoffs. We randomly subdivide a given set of proteins into ten subsets, and take each subset as a testing data set while using the remaining nine subsets as a training data set. For each testing data set, we take each protein as an unclassified protein and apply our algorithm for classifying one protein against families that include proteins in the training data set. We define the accuracy rate to be the fraction of proteins that are classified to the correct family.
Since it is not likely that the 10-fold cross validation procedure groups many complete families into individual subsets, we further evaluate the mis-classification of our algorithm by removing proteins from one entire family at a time, and taking each protein as an unclassified protein while applying our algorithm for classifying one protein against all the other families. We define the mis-classification rate to be the fraction of proteins that are incorrectly classified to some other family. Since this procedure retains the largest number of already classified families during each test, it corresponds to the most difficult scenario that one can use for evaluating mis-classification. This procedure complements the above 10-fold cross validation procedure by evaluating the reliability of our algorithm on unclassified proteins that should not belong to any existing families.
Figure 3 shows the performance of our algorithm SClassify over different e-value cutoffs by using SSEARCH and BLAST to compute pairwise scores. There is a large performance difference between data sets that contain sequences corresponding to protein domains, including Pfam and SCOP, and data sets that contain full length sequences, including ProtClustDB and UniProt. To simultaneously achieve high accuracy rate and low mis-classification rate, a high e-value cutoff is needed for data sets that contain sequences corresponding to protein domains, and a low e-value cutoff is needed for data sets that contain full length sequences to avoid mis-classification.
FIG. 3.
Accuracy rate and mis-classification rate of SClassify over different SSEARCH e-value cutoffs and over different BLAST e-value cutoffs. SSEARCH and BLAST are used respectively to compute pairwise scores. Solid lines represent accuracy rates, while dotted lines represent mis-classification rates. More tests are performed for e-value cutoffs between 1 and 1e–5.
When SSEARCH is used to compute pairwise scores, we choose an e-value cutoff of 0.1 for data sets that contain sequences corresponding to protein domains, including Pfam and SCOP, which achieves an accuracy rate of at least 89% and a mis-classification rate of at most 9%. We choose a SSEARCH e-value cutoff of 1e–30 for data sets that contain full length sequences, including ProtClustDB and UniProt, which achieves an accuracy rate of at least 90% and a mis-classification rate of at most 13%. For these choices, the minimum accuracy rate is about the same as one minus the maximum mis-classification rate.
We use a similar strategy to obtain appropriate e-value cutoffs for BLAST, except that we consider a BLAST e-value to be above the cutoff if no hits are obtained between a protein pair. We choose a BLAST e-value cutoff of 0.1 for data sets that contain sequences corresponding to protein domains, including Pfam and SCOP, which achieves an accuracy rate of at least 85% and a mis-classification rate of at most 6%, resulting in a decrease of the minimum accuracy rate by 4% when using BLAST instead of SSEARCH. Such a decrease in performance is expected since SSEARCH computes optimal alignments, while BLAST employs a heuristic, and the similarity scores from SSEARCH are more accurate than the ones from BLAST. We choose a BLAST e-value cutoff of 1e–30 for data sets that contain full length sequences, including ProtClustDB and UniProt, which achieves an accuracy rate of at least 89% and a mis-classification rate of at most 13%, resulting in a similar minimum accuracy rate and maximum mis-classification rate as before, although the actual accuracy rate on the ProtClustDB data set decreases by 3% when using BLAST instead of SSEARCH. A caution is that, although the same e-value cutoffs are chosen for SSEARCH and BLAST, the SSEARCH e-value and the BLAST e-value use different formulas and are not directly comparable.
3.3. Comparison with other supervised algorithms
We compare the performance of SClassify to HMMER (Eddy, 1998), which classifies proteins against existing families according to profile hidden Markov models, to LIBSVM (Fan et al., 2005), which performs supervised classification based on the use of support vector machines, and to SMT (Eskin et al., 2003), which is a supervised classification algorithm that uses sparse Markov transducers to train a given set of known families. We employ the 10-fold cross validation procedure as before, and use the same training and testing sets in each case.
For SClassify, we use the variant for classifying one protein. For HMMER, we construct a profile hidden Markov model for each family in the training set from a multiple sequence alignment obtained by ClustalW (Thompson et al., 1994). These alignments are constructed either from a subset of distinct sequences in each family with BLAST e-values above a cutoff, or from all sequences in each family. For each family, a subset of distinct sequences is obtained by starting from an empty subset and iteratively adding a sequence that has the largest possible minimum BLAST e-value against sequences in the current subset as long as this largest minimum is above the cutoff. For the Pfam database, we also use the profile hidden Markov model that was created for each curated family from a subset of curated seed sequences in Bateman et al. (2000). For a given e-value cutoff, we assign an unclassified protein to the family with the lowest e-value if such a family exists, otherwise the unclassified protein is not assigned to any family. For LIBSVM, we follow a similar strategy as in Liao and Noble (2003), and define a feature vector for a protein based on the set of minus log e-values from BLAST to each training protein, while allowing missing values that correspond to pairwise scores above the BLAST e-value cutoff 1. Each attribute of the feature vector is normalized by using the same scaling factor for the training and testing data, and the radial basis kernel is used. Various gamma and cost parameters are tested, and the result with the best performance is selected for each data set. For SMT, we use the classifier variant that allows wild cards in the sequence motifs, and assign an unclassified protein to the family with the highest average log probability of the proteins within the family without using a cutoff.
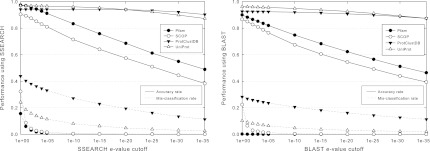
Figure 4 shows two boundary cases for HMMER in which the BLAST e-value cutoff to obtain a subset of distinct sequences in each family is set to a very high value 0.1, and in which all sequences in each family are used to obtain a multiple alignment. While the minimum mis-classification rate of HMMER is achieved in the first case, the accuracy rate is low. While the maximum accuracy rate of HMMER is achieved in the second case, the mis-classification rate is high. The performance of using other BLAST e-value cutoffs to obtain a subset of distinct sequences in each family is intermediate between the two. For Pfam, using the profile hidden Markov model that was created for each curated family from a subset of curated seed sequences in Bateman et al. (2000) simultaneously achieves high accuracy rate and low mis-classification rate. However, the accuracy rate of HMMER is significantly lower than the accuracy rate of SClassify at a fixed mis-classification rate, while the mis-classification rate of HMMER is significantly higher than the mis-classification rate of SClassify at a fixed accuracy rate (compare to Fig. 3). A caution is that the HMMER e-value is not directly comparable to the SSEARCH or BLAST e-values.
FIG. 4.
Accuracy rate and mis-classification rate of HMMER over different e-value cutoffs. Light bars and solid lines represent accuracy rates, while dark bars and dotted lines represent mis-classification rates. Bars denote performance on Pfam by applying HMMER on the profile hidden Markov model that was created for each curated family from a subset of curated seed sequences in Bateman et al. (2000). Lines denote performance by applying HMMER on the profile hidden Markov model that is constructed for each family in the training set from a multiple sequence alignment obtained by ClustalW. These alignments are constructed either from a subset of distinct sequences in each family with BLAST e-values above 0.1 (a BLAST e-value is considered to be above the cutoff if no hits are obtained) or from all sequences in each family. More tests are performed for e-value cutoffs between 1 and 1e–5.
Table 2 shows that SClassify has better accuracy than LIBSVM and much better accuracy than SMT on individual species from Pfam and on SCOP. When compared to our previous results, SClassify performs better on families that contain multiple species from Pfam, which may be due to the increased amount of information from the much larger number of proteins in multiple species.
Table 2.
Accuracy Rate Comparison of SClassify, LIBSVM, and SMT on Individual Species from Pfam and on SCOP
| Data set | SClassify (SSEARCH) | SClassify (BLAST) | LIBSVM | SMT |
|---|---|---|---|---|
| Pfam | ||||
| A. thaliana | 0.96 | 0.90 | 0.82 | 0.11 |
| C. elegans | 0.91 | 0.80 | 0.71 | 0.46 |
| D. melanogaster | 0.90 | 0.77 | 0.68 | 0.28 |
| E. coli | 0.90 | 0.82 | 0.61 | 0.40 |
| H. sapiens | 0.92 | 0.84 | 0.74 | 0.22 |
| M. musculus | 0.92 | 0.83 | 0.76 | 0.18 |
| S. cerevisiae | 0.81 | 0.72 | 0.52 | 0.38 |
| SCOP | 0.89 | 0.85 | 0.63 | 0.00 |
For SClassify, either SSEARCH or BLAST is used to compute pairwise scores.
3.4. Comparison with unsupervised algorithms
We compare the performance of SClassify to unsupervised clustering algorithms MCL (Enright et al., 2002), which uses the Markov cluster algorithm to classify a given set of proteins into families, and to BLASTClust (Altschul et al., 1990), which groups a given set of proteins into clusters based on computing pairwise scores from BLAST. In order to make the results from SClassify comparable to the clusters obtained from these unsupervised algorithms, we perform the 10-fold cross validation procedure as before and merge the family assignments of each protein from SClassify to obtain a set of clusters, with each protein that is not assigned to any existing family being in a new family by itself. This procedure roughly models the subdivision of a given set of proteins into clusters of proteins.
We evaluate the performance of each algorithm by checking whether each pair of proteins are correctly classified either to the same family or to different families. For each set of true clusters that are predefined in the original data set and each set of predicted clusters that are obtained from each algorithm, we compute the statistics TP, FP, and FN, which are the number of protein pairs that are within the same true cluster and within the same predicted cluster, the number of proteins pairs that are within the same predicted cluster but in different true clusters, and the number of protein pairs that are within the same true cluster but in different predicted clusters, respectively. From these statistics, we compute the precision PC = TP/(TP + FP), the recall RC = TP/(TP + FN), and the F-measure = 2 × (PC × RC)/(PC + RC). Note that the F-measure from SClassify roughly corresponds to the accuracy rate in our previous tests but not the mis-classification rate, and is highly correlated to the accuracy rate.
For SClassify, we use the variant for classifying one protein while using BLAST to compute pairwise scores. For MCL, we use the minus log e-value from BLAST as the edge weight, and test various inflation values while selecting the result with the best F-measure for each data set. For BLASTClust, we test various similarity thresholds and minimum length coverages while selecting the result with the best F-measure for each data set.
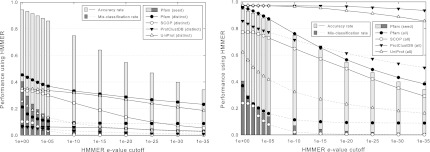
Figure 5 shows that, while MCL and BLASTClust are able to obtain high precision or high recall for some data sets, this is often at the expense of low recall or low precision respectively, and the overall F-measure is not high. By using information from known families, SClassify is able to obtain very good performance with respect to both precision and recall, which results in consistently high F-measure across all data sets.
FIG. 5.
Performance comparison of SClassify with unsupervised clustering algorithms MCL and BLASTClust. BLAST is used to compute pairwise scores. For each data set, PC is the precision, RC is the recall, and the lines denote the F-measure.
3.5. Classifying a set of proteins
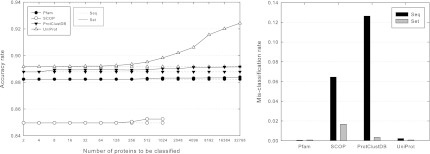
To evaluate the performance of SClassify when a set of unclassified proteins is given, we perform the 10-fold cross validation procedure as before. Instead of taking each protein within the testing data set as an unclassified protein individually, given a set size u, we subdivide the testing data set into subsets U that are roughly of the same size u and as evenly as possible. We apply the SClassify variant for classifying a set of proteins on each subset U independently against families that include proteins in the training data set. We compare the performance of this algorithm to the alternative strategy of applying the original algorithm for classifying one protein on each protein sequentially within each subset U according to a random order, in which each newly classified protein in U is retained in its assigned family before the next one in U is classified. To determine whether there are significant improvements in performance between these algorithms, we evaluate their accuracy rates over increasing values of u.
We further evaluate the mis-classification by removing proteins from one entire family at a time and setting U to be the removed family. Since it is possible that proteins in U are classified to different families, we take the largest new family that contains proteins in U to be the correct family for U after classification.
Figure 6 shows that significantly better accuracy rates can be obtained when u becomes large enough, which are especially evident on UniProt. Significantly lower mis-classification rates are also obtained, which are evident on SCOP and ProtClustDB.
FIG. 6.
Accuracy rate and mis-classification rate comparison of different variants of SClassify when a set of unclassified proteins is given. BLAST is used to compute pairwise scores. Given a set U of unclassified proteins, Seq denotes the original algorithm for classifying one protein on each protein sequentially in U according to a random order, while Set denotes the algorithm for classifying the proteins in U simultaneously by taking into account similarities between unclassified proteins.
4. Discussion
We have developed an algorithm SClassify that allows both accurate classification of proteins to existing families and progressive construction of new families. We have shown that there is a tradeoff between achieving high accuracy rate and low mis-classification rate. While we have used the default parameters in BLAST that limit the output to the top 250 alignments, removing this constraint increases the accuracy rate and the mis-classification rate simultaneously, and does not necessarily give better results.
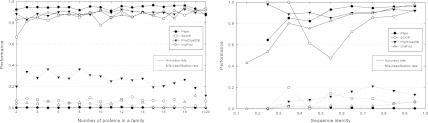
To investigate whether SClassify performs well on families with very few proteins and on families with low sequence similarity, we subdivide our results on the variant for classifying one protein into categories by grouping together all families that contain the same number of proteins and grouping together all families with average sequence identity between proteins within a specified range, where the sequence identity is obtained by dividing the number of exact matches by the average length of two aligned proteins. The performance of our algorithm remains high on families with very few proteins or on families with low sequence identity (Fig. 7).
FIG. 7.
Accuracy rate and mis-classification rate of SClassify within categories. Two categories are obtained by grouping together all families that contain the same number of proteins (each point denotes the performance on proteins within such families), and grouping together all families with average sequence identity between proteins within a specified range (each point denotes the performance on proteins within such families with average sequence identity within the range from l to r, where l is the label to the left of the point on the x-axis and r is the label to the right of the point on the x-axis). BLAST is used to compute pairwise scores. Solid lines represent accuracy rates, while dotted lines represent mis-classification rates. Missing points correspond to no proteins being assigned to the category.
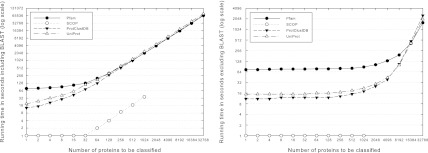
To evaluate the running time of SClassify on each data set, we apply SClassify against all existing protein families in each data set by using the variant for classifying a set of proteins. Although the worst case time complexity of SClassify is not linear, Figure 8 shows that the running time is roughly linear after the number of proteins to be classified becomes large enough and the computational overhead becomes relatively low. When BLAST is used to compute pairwise scores, it takes less than a day on one processor for the largest case with 32,768 proteins to classify. When the time to compute pairwise scores is excluded, SClassify is very fast and takes at most an hour in all the tests. If SSEARCH is used instead of BLAST, the total time to compute SSEARCH optimal alignments dominates, and the running time increases by about a factor of ten. The memory requirement is less than two gigabytes in all the tests with at most 32,768 proteins to classify.
FIG. 8.
Running time of SClassify on one processor while including and excluding the time to compute pairwise scores by BLAST. In each case, the average running time is given.
We have shown that for the purpose of protein family classification, it may not be necessary to consider more complicated models such as multiple sequence alignments and hidden Markov models, as it is possible to obtain better performance from the use of pairwise sequence comparisons alone. Since pairwise alignments may conflict with each other within a family, such multiple sequence representations will still be needed to define a consistent model for a family. These representations are especially important for determining conserved or critical residue positions within a family. In order to handle multiple domain proteins, domain prediction algorithms such as ADDA (Heger and Holm, 2003) can be used to determine domain boundaries before SClassify is applied.
To illustrate the use of SClassify in real life applications, we consider all 688,172 computationally predicted domains in Pfam, and apply SClassify to these domains against all existing curated families in Pfam (containing a total of over two million proteins, see Table 1), by using BLAST to compute pairwise scores. It takes about 60 processor-days to obtain all the BLAST scores, and about two weeks on one processor when the variant for classifying a set of proteins is used after all the BLAST scores are obtained.
When the variant for classifying a set of proteins is used on all the predicted domains, about 31% of these domains are assigned to some curated family. When the variant for classifying one protein is used sequentially according to a random order of predicted domains, about 27% of these domains are assigned to some curated family. These results are in contrast to the smaller number of predicted domains that are assigned (about 20%) when HMMER is applied with an e-value cutoff of 0.1 while using the profile hidden Markov model that was created for each curated family from a subset of curated seed sequences in Bateman et al. (2000). A caution is that the domain boundaries in the predictions may not be accurate and the assignments from different algorithms can be quite different from each other.
For the remaining domains that are not assigned to a curated family, both variants of SClassify are able to create new families. In both cases, about 63% of these new families contain only one domain. For the remaining new families that contain more than one domain, about 90% of them are completely contained within a computationally generated family in Pfam that contains these predicted domains. These results indicate that the new families generated by SClassify are largely consistent with the families generated by Pfam, except that a large number of them are split into smaller families by SClassify due to insufficient sequence similarities.
While most existing protein family classification algorithms are based on sequence similarity information from alignments, alignment-free approaches are also available (Ma and Chan, 2008). One future direction is to investigate whether it is possible to use these techniques to improve accuracy.
Acknowledgments
This work was supported by the Ministry of Science and Innovation, Spain (grant AGL2008-03177/AGR) and the National Science Foundation (grant MCB-0951120).
Disclosure Statement
No competing financial interests exist.
References
- Altschul S.F. Gish W. Miller W., et al. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Apweiler R. Attwood T.K. Bairoch A., et al. InterPro—an integrated documentation resource for protein families, domains and functional sites. Bioinformatics. 2000;16:1145–1150. doi: 10.1093/bioinformatics/16.12.1145. [DOI] [PubMed] [Google Scholar]
- Apweiler R. Bairoch A. Wu C.H., et al. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 2004;32:D115–D119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A. Birney E. Durbin R., et al. The Pfam Protein Families Database. Nucleic Acids Res. 2000;28:263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano G. Yona G. Variations on probabilistic suffix trees: statistical modeling and prediction of protein families. Bioinformatics. 2001;17:23–43. doi: 10.1093/bioinformatics/17.1.23. [DOI] [PubMed] [Google Scholar]
- Chen Y. Reilly K.D. Sprague A.P., et al. SEQOPTICS: a protein sequence clustering system. BMC Bioinform. 2006;7:S10. doi: 10.1186/1471-2105-7-S4-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S.R. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Enright A.J. Van Dongen S. Ouzounis C.A. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskin E. Noble W.S. Singer Y. Protein family classification using sparse Markov transducers. J. Comput. Biol. 2003;10:187–213. doi: 10.1089/106652703321825964. [DOI] [PubMed] [Google Scholar]
- Fan R.-E. Chen P.-H. Lin C.-J. Working set selection using second order information for training support vector machines. J. Mach. Learn. Res. 2005;6:1889–1918. [Google Scholar]
- Heger A. Holm L. Exhaustive enumeration of protein domain families. J. Mol. Biol. 2003;328:749–767. doi: 10.1016/s0022-2836(03)00269-9. [DOI] [PubMed] [Google Scholar]
- Jaakkola T. Diekhans M. Haussler D. A discriminative framework for detecting remote protein homologies. J. Comput. Biol. 2000;7:95–114. doi: 10.1089/10665270050081405. [DOI] [PubMed] [Google Scholar]
- Kim S. Lee J. BAG: a graph theoretic sequence clustering algorithm. Int. J. Data Mining Bioinform. 2006;1:178–200. doi: 10.1504/ijdmb.2006.010855. [DOI] [PubMed] [Google Scholar]
- Klimke W. Agarwala R. Badretdin A., et al. The National Center for Biotechnology Information's Protein Clusters Database. Nucleic Acids Res. 2009;37:D216–D223. doi: 10.1093/nar/gkn734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao L. Noble W.S. Combining pairwise sequence similarity and support vector machines for detecting remote protein evolutionary and structural relationships. J. Comput. Biol. 2003;10:857–868. doi: 10.1089/106652703322756113. [DOI] [PubMed] [Google Scholar]
- Ma P.C.H. Chan K.C.C. UPSEC: an algorithm for classifying unaligned protein sequences into functional families. J. Comput. Biol. 2008;15:431–443. doi: 10.1089/cmb.2007.0113. [DOI] [PubMed] [Google Scholar]
- Murzin A.G. Brenner S.E. Hubbard T., et al. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- Smith T.F. Waterman M.S. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- Thompson J.D. Higgins D.G. Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]