Abstract
Higher plants are sessile organisms that perceive environmental cues such as light and chemical signals and respond by changing their morphologies. Signaling pathways utilize a complex network of interactions to orchestrate biochemical and physiological responses such as flowering, fruit ripening, germination, photosynthetic regulation, and shoot or root development. In this session, the mechanisms of signaling systems that trigger plant responses to light and to the gaseous hormone, ethylene, were discussed. These signals are first sensed by a receptor and transmitted to the nucleus by a complex network. A signal may be transmitted to the nucleus by any of several systems including GTP binding proteins (G proteins), which change activity upon GTP binding; protein kinase cascades, which sequentially phosphorylate and activate a series of proteins; and membrane ion channels, which change ionic characteristics of the cells. The signal is manifested in the nucleus as a change in the activity of DNA-binding proteins, which are transcription factors that specifically interact and modulate the regulatory regions of genes. Thus, detection of an environmental signal is transmitted through a transduction pathway, and changes in transcription factor activity may coordinate changes in the expression of a portfolio of genes to direct new developmental programs.
Two major advances in plant biology have been critical for the analysis of signal transduction pathways. First, the ability to produce and analyze large numbers of mutant plants has allowed scientists to identify numerous alleles that are involved in signal transduction pathways. The Arabidopsis plant has desirable biological and genetic qualities such that tens of thousands of mutant plants can be screened to identify plants that have modified responses to a signal (1). Facile screens have been developed that precisely identify mutant plants that are either response-deficient, and fail to respond to a stimulus, or response-constitutive, and respond in the absence of the signal. Genetic analyses have been performed to elucidate both the order and interactions of pathway components, and suggest that a complex network of information input and interacting components are transmitted to the nucleus to direct changes in gene expression.
Second, biotechnological developments and genome project efforts in Arabidopsis have been critical for the success of the molecular genetic approach. Mutational techniques have been developed and include chemical mutagenesis and insertional mutagenesis, in which specific DNA sequences cause mutations by insertion into the genome, thereby “tagging” the mutated gene with a novel DNA sequence. In addition, this plant has a relatively small genome (≈120 million base pairs, 25 times smaller than humans) with only five chromosomes. The Arabidopsis genome has been cloned as very large fragments in yeast artificial chromosomes and bacterial artificial chromosomes with insert sizes ranging from 100,000 to 750,000 base pairs. These large genomic fragments are being ordered into contiguous regions (contigs) so that the genetic positions of markers becomes integrated with their physical positions in the contigs. Large numbers of cloned DNA sequences have been used to produce a physical map of the chromosomes such that the proximity of a new mutant allele to a unique DNA sequence can be established. This now allows new mutant alleles to be genetically mapped to a physically defined region of cloned DNA that greatly facilitates the isolation of the mutant gene; this process constitutes “map-based cloning.” Genetic complementation analysis in which the wild-type gene is introduced into the mutant plant can then be used to demonstrate genetic proof that the gene that causes the mutation has been identified. The complete sequence of the entire Arabidopsis genome is expected to be available by 2004 (1).
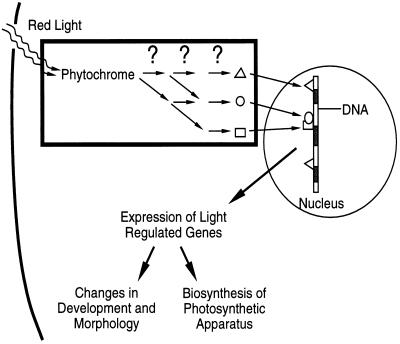
Joanne Chory, Associate Professor of the Salk Institute for Biological Studies, gave the first talk of the session, an overview of molecular genetic approaches that delineate the signal transduction pathway from light absorption by photoreceptors to the light-dependent developmental changes that prepare a plant for photosynthetic activity (photomorphogenesis) (2). Plants utilize at least three systems to perceive light: UV photoreceptors; blue light photoreceptors; and the red light photoreceptor known as phytochrome. Phytochrome senses the relative intensities of photosynthetically active red light (660 nm) and nonphotosynthetic near far-red light (730 nm), thus sensing information about light quality and the suitability for photosynthesis. Dark-grown plants develop as etiolated plants with characteristic features: absence of chlorophyll, suppression of cotyledon and leaf expansion, hypocotyl elongation, and the downward folding of cotyledons. Exposure of plants to red or blue light initiates developmental programs that prepare the plant to perform photosynthesis: chlorophyll and photosynthetic apparatus are produced, hypocotyl and stem elongation are suppressed, and the cotyledons and leaves unfold and expand (Fig. 1).
Figure 1.
The phytochrome signal transduction pathway. Signal perception by phytochrome, the light receptor, is transmitted to the nucleus by complex interactions shown inside the rectangle. Mutational analysis and molecular cloning have been used to identify and order the components of the signal transduction pathway.
Mutant screens for loss of the photomorphogenetic response have been used to identify plants that no longer respond to red, blue, or near far-red light, and these plants show characteristic features of etiolation when grown in the various wavelengths of light. These plants are effectively “blind” to the light! This approach has led to the identification of a number of Arabidopsis light-responsive mutants, including mutants in the biosynthesis of the phytochrome chromophore, two of the five phytochrome genes, and a blue light receptor gene. Individual mutations result in different degrees of deviation from normal photomorphogenesis; however, the combination of two light-responsive mutations generally demonstrate exaggerated effects. These results emphasize the importance of multiple signal inputs from various types and members of receptor families, and suggest that the systems are probably partially redundant and very complicated. Screening for mutants in light-response deficiency led to important advances in the understanding of sensory input information at the level of light receptors, but unfortunately has not led to much detailed information on the signal transduction pathways beyond the photoreceptors.
Mutant screens of Arabidopsis plants for photomorphogenetic response in the absence of light have been used to identify plants that constitutively show photomorphogenesis and are considered deetiolated (det) or constitutively photomorphogenetic (cop). These plants are delusional—behaving as though they have experienced light exposure in complete darkness! Six mutant genes have been cloned from this class of mutation by insertional mutagenesis or by map-based cloning. The COP1 protein has features of both transcription factors and G proteins, suggesting a general role for this protein in gene expression. The det2 gene has recently been characterized by the Chory lab as a steroid biosynthetic enzyme (steroid 5 alpha reductase), and has resulted in the rediscovery of a new class of phytohormones (3). Exogenous application of brassinolide to det2 mutant Arabidopsis plants produces the wild-type phenotype, suggesting that this chemical may represent a class of hormones with an important role in light-regulated development of higher plants.
Joseph Ecker, Associate Professor at the University of Pennsylvania, discussed how plants use various chemical signals to coordinate physiological processes. Plants use many different chemicals to send signals. Auxins, cytokinins, abscisic acid, gibberellic acid, and ethylene are “classical” phytohormones; however, a number of very recently discovered phytohormones include brassinolide (3); systemin, a peptide hormone; methyl jasmonate; and salicyclic acid.
The ethylene signal transduction pathway is ripe for analysis. The biochemical pathway and many of the genes for ethylene biosynthesis are well characterized. Exposure of dark-grown seedlings to ethylene leads to a set of clear morphological changes known as the “triple response.” These changes include (i) inhibition of hypocotyl and root elongation, (ii) radial swelling of the hypocotyl, and (iii) a diageotrophic (horizontal) growth habit. The triple response has been used to identify several types of Arabidopsis mutants (4). Ethylene-overproduction mutant plants produce high endogenous amounts of ethylene and consequently display the triple-response phenotype, even in the absence of exogenous ethylene. A more interesting phenotype is shown by the constitutive triple-response mutant (ctr1). These plants do not overproduce ethylene, but develop as though they have been exposed to high levels of ethylene gas. The ctr1 mutant is defective in the ethylene signal transduction pathway, apparently as a result of the loss of function of a negative regulator of ethylene response. The CTR1 gene has been cloned and encodes a protein kinase related to the Raf family.
Ethylene-insensitive mutants (ein, etr, ain) are ethylene response-deficient plants and represent a phenotype defined by numerous genes. These genes can be placed in a relative sequence of action by genetic analyses. One of these very early acting genes has now been cloned and reveals a most interesting surprise. The ETR1 polypeptide shows sequence similarity with bacterial and fungal histidine kinases such as SLN1, a component of the osmotic-stress transducing pathway in yeast (5). Interestingly, SLN1 acts through a phospho-relay system to activate a downstream protein kinase cascade. More recent biochemical studies have now shown that ETR1 has the capacity to generate ethylene binding sites when expressed in yeast, strongly supporting the notion that ETR1 encodes an ethylene receptor.
As additional molecular components are put in place in the light and hormone signal transduction pathways, the apparent complexity of the process grows, and the increased potential for controlling plant growth and development becomes evident. Ethylene coordinates the agronomically important process of fruit ripening, and prospects of controlling food production by modification of ripening processes have already been explored by Calgene (Davis, CA) with the Flavr-Savr tomato (6) and by scientists who have regulated ethylene production in tomato (7). Applications of signal transduction as well as basic understanding of plant developmental processes will have a very important role in future agricultural applications.
The discussion period brought out interesting academic and applied aspects of this work, including questions about the ethical, political, and environmental issues in biotechnology. A sample of two questions and the resulting discussion follows.
Question 1: How good does the antisense-ethylene tomato fruit taste? More generally what are the prospects for scientists to return the flavor to food that has been lost by modern breeding?
Joseph:
Breeding a tomato to satisfy the engineers is why they don’t taste good (laughter). The reason that tomatoes don’t taste good is not because there are not good varieties, but because of the engineering that is used to pick and handle them. In fact, this technology, for example, engineering an antisense ethylene gene, allows the fruit to basically develop to its full capacity on the vine in terms of all nonethylene-requiring processes. The way that it is done now is to not let the fruit develop completely, and then gas them with ethylene. The largest component of flavor is how long the fruit stays on the vine. There are other processes, unrelated to ethylene, that are required for fruit development, and if you pull the fruit off the vine these are not allowed to proceed.
Question 2: Both of you brought up the term redundancy with respect to gene families. Redundancy, strictly speaking, means that several members serve a common primary function, and if you believe that, can you make sense of it evolutionarily?
Joanne:
I can give you an example with phytochrome mutants. If you screen for mutants in white light, you find many phyB mutant plants that have really long hypocotyls, but we never found a phytochrome A mutant, and phytochrome A is a really abundant phytochrome. When we did a very specialized screen under a specific wavelength of light, we could find phyA mutants. Those mutants look absolutely normal under normal daylight. What a geneticist would say is that there is no phenotype under those conditions. If you look under the correct conditions, you can find a phenotype that specifically demonstrates that there is this special and unique function for this gene that it alone does.
Participant:
That’s another way to say that redundant is probably not the right word. Functional overlap is probably more precise.
References
- 1.Goodman H M, Ecker J R, Dean C. Proc Natl Acad Sci USA. 1995;92:10831–10835. doi: 10.1073/pnas.92.24.10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quail P H, Boylan M T, Parks B M, Short T W, Xu Y, Wagner D. Science. 1995;268:675–680. doi: 10.1126/science.7732376. [DOI] [PubMed] [Google Scholar]
- 3.Li J, Nagpal P, Vitart V, McMorris T C, Chory J. Science. 1996;272:398–401. doi: 10.1126/science.272.5260.398. [DOI] [PubMed] [Google Scholar]
- 4.Ecker J R. Science. 1995;268:667–675. doi: 10.1126/science.7732375. [DOI] [PubMed] [Google Scholar]
- 5.Chang C. Trends Biochem Sci. 1996;21:129–133. [PubMed] [Google Scholar]
- 6.Sheehy R E, Kramer M, Hiatt W R. Proc Natl Acad Sci USA. 1988;85:8805–8809. doi: 10.1073/pnas.85.23.8805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oeller P W, Lu M W, Taylor L P, Pike D A, Theologis A. Science. 1991;254:437–439. doi: 10.1126/science.1925603. [DOI] [PubMed] [Google Scholar]