The influenza virus is constantly changing; immunity to one season’s strains does not translate into immunity in subsequent years. As a consequence, the vaccine is reformulated each year. However, several recent studies provide hope that one day a ‘universal’ influenza vaccine might be developed - one that protects against strains circulating not only in one season, but against all strains of influenza.
These studies examined neutralizing antibodies that block the infectivity of the virus, from people previously vaccinated or infected with influenza virus. The research pinpoints a handful of neutralizing antibodies that, surprisingly, are directed against a broad range of influenza strains. These observations from ‘the bedside’ are provoking basic research into how to develop a vaccine to induce such antibodies and harness them so they effectively protect against a range of strains. Ultimately, a new vaccine strategy may emerge1–3.
Vaccines are the cornerstone of prevention. The principle underlying licensed influenza virus vaccines is the induction of an immune response to two proteins, hemagglutinin (HA), the protein responsible for attachment to the receptor (sialic acid) and fusion of the viral membrane to the endosome during viral entry and neuraminidase (NA), the protein that cleaves sialic acid from the virion and cell membranes, releasing progeny virions. These two proteins are the main targets of protective immunity.
There are 16 HA and 9 NA subtypes among influenza A viruses and the 16 HA genes fall into two phylogenetic groups (I and II)4. Influenza viruses have succeeded as pathogens because of their ability to escape neutralization by antibodies elicited during previous infection or vaccination. This escape occurs annually by point mutations around the conserved receptor binding pocket of the HA (antigenic drift) or much more rarely, by the introduction of a novel HA subtype to which the population lacks immunity resulting in a pandemic (antigenic shift). Annual re-formulations of the vaccine are needed in order to keep pace with antigenic drift in the HA, and a completely new vaccine is needed in the event of antigenic shift. Despite the annual influenza vaccine campaigns, influenza accounts for up to 200,000 hospitalizations and 36,000 excess deaths annually in the United States5,6 and three to five million cases of severe illness and 250 000 to 500 000 deaths worldwide7.
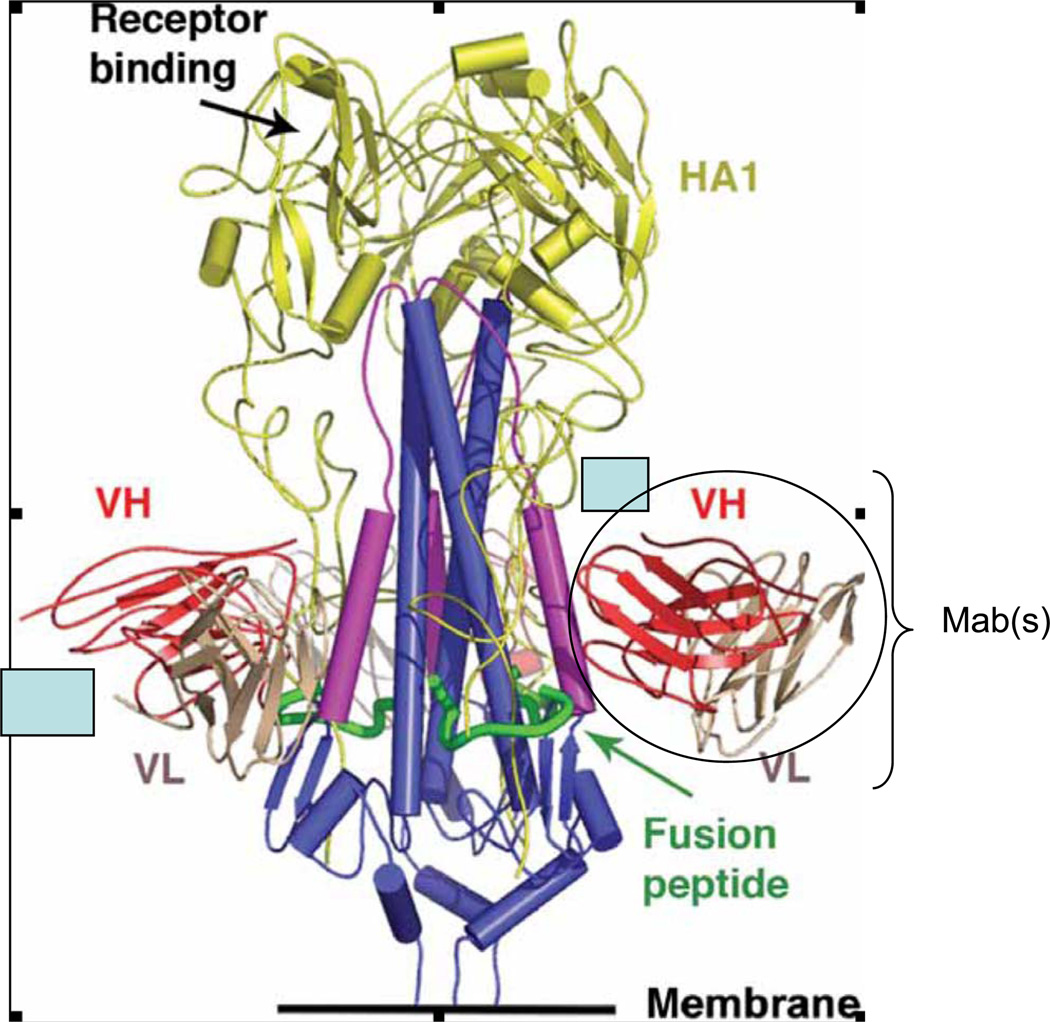
During influenza infection and following vaccination, neutralizing antibodies typically develop against epitopes on the globular head of the HA, a protein that looks a bit like a ‘lollipop’ with a stem and a head (Figure 1). Neutralizing antibodies are generally specific for antigenically related viruses and do not cross-react with other HA subtypes, though T cell responses cross-react across subtypes8,9.
FIGURE.
Schematic of an HA trimer identifying the fusion peptide, stem, globular head and conserved epitope based on Figure 4 in Sui et al. and Figure 1 of Ekiert et al.
Recent studies identified antibodies that broadly neutralize HAs of different subtypes. Gioia et al.10 examined antibodies from individuals immunized with inactivated seasonal influenza virus vaccines and observed neutralizing antibodies and enhanced T-cell reactivity against H5N1 ‘bird flu’ viruses. Three independent studies scanned combinatorial antibody libraries in which immunoglobulin light and heavy chains were amplified by PCR from survivors of H5N1 infection11 or recipients of seasonal influenza vaccine12, or a pooled non-immune human antibody phage display library2. All three of these studies yielded human monoclonal antibodies (MAb) that neutralized HAs from group I. Structural studies with two of the MAbs established that binding to the HA was mediated exclusively by the immunoglobulin heavy chain2,3 and the MAbs bound to a conserved epitope in the stem region of the HA containing the fusion peptide (Figure 1). The fusion peptide is a hydrophobic portion of the HA that is exposed when the HA undergoes a conformational change at low pH in the endosome; the fusion peptide inserts into the endosomal membrane and pulls the endosomal and virion membranes together, causing them to fuse. This portion of the HA is broadly conserved across several HA subtypes from group I. Based on where this region mapped, and on in vitro findings of fusion inhibition, the authors proposed a specific mechanism for how the antibodies neutralized the viruses. It seems the MAbs lock the fusion peptide in place, preventing the structural reorganization that is required for membrane fusion2. The MAbs were also able to prevent infection with several influenza subtypes in mice.
The human MAbs recognize an epitope in a region that was previously identified using a murine MAb that neutralized H1, H2, H5 and H6 HAs13,14. Why weren’t such cross-reactive neutralizing human antibodies identified earlier? There are two likely explanations: First, the dominant antibody response to the influenza HA is strain specific; most of the antibodies are directed at epitopes on the globular head of the HA and cross-reactive antibodies directed at the stem of the HA are rare. Second, these rare cross-reactive antibodies were identified using new technology that makes it possible to identify antibodies that cannot be found easily by other means. The MAbs were identified using combinatorial antibody libraries, which reflect the entire immunologic repertoire of an individual, from IgM+ memory B cells or from bone marrow RNA. One can increase the probability of finding antibodies of interest by screening large libraries.
These findings represent a significant advance because they provide clear evidence that the human immune system can recognize and produce a neutralizing antibody response to a conserved epitope on the HA that is shared across several influenza subtypes. Most of the universal vaccine design approaches currently being pursued focus on generating cross-protective cellular immunity; most such vaccines provoke a response against conserved viral proteins such as the ion channel M2 or the nucleoprotein (NP). But whereas cellular immune responses are critical for viral clearance, neutralizing antibodies directed at the HA can prevent infection with influenza viruses. Therefore, a universal HA-based vaccine could be combined with an M2 or NP based vaccine to provide heterosubtypic protection based on both antibody and cellular immune mechanisms. Further research will be needed to determine how the conserved HA epitope can be engineered into a vaccine. A number of vaccine platforms that present the epitope including virus like particles, peptide vaccines, or vectored vaccines can be explored and their success will be measured by the ability of a vaccine to elicit antibodies that resemble the MAbs in breadth of cross reactivity. Evaluation of such vaccines should include studies to determine whether high titers of such antibodies will drive the evolution of the HA to escape neutralization, as is common with antibodies directed at the immunodominant sites on the globular head of the HA. Structure-based explanations for the lack of binding of these newly identified MAbs to group II HAs have been proposed2,3. Identification of a similar conserved epitope for the group II HAs would be necessary to protect against the entire range of HA subtypes.
These findings raise research questions that also go beyond influenza. For instance, most of the cross-reactive neutralizing antibodies directed against the influenza viruses contained a particular antibody heavy chain sequence, the immunoglobulin VH 1–69 segment. Preferential use of the VH 1–69 germline is reported for antibodies against hepatitis C virus (HCV) and HIV and has been found in HCV associated lymphomas and antibodies associated with auto-immunity1. The selective VH gene usage in HIV, HCV, influenza reflect unusual properties of the epitopes; the basis and the consequences of the use of this germline by viruses of different families will be of interest to immunologists and virologists. Would a vaccine designed to elicit cross-reactive antibodies to the influenza HA increase the risk of autoimmunity? As Wang et al. suggest, it may also be possible to rationally design an antiviral drug to mimic this neutralizing antibody in order to block membrane fusion2.
ACKNOWLEDGEMENTS
This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, NIH).
Selected bibliography
- 1.Wang TT, Palese P. Universal epitopes of influenza virus hemagglutinins? Nat Struct Mol Biol. 2009;16:233–234. doi: 10.1038/nsmb.1574. [DOI] [PubMed] [Google Scholar]
- 2.Sui J, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol. 2009;16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ekiert DC, et al. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Russell RJ, et al. Structure of influenza hemagglutinin in complex with an inhibitor of membrane fusion. Proc Natl Acad Sci U S A. 2008;105:17736–17741. doi: 10.1073/pnas.0807142105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thompson WW, et al. Influenza-associated hospitalizations in the United States. JAMA. 2004;292:1333–1340. doi: 10.1001/jama.292.11.1333. [DOI] [PubMed] [Google Scholar]
- 6.Thompson WW, et al. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA. 2003;289:179–186. doi: 10.1001/jama.289.2.179. [DOI] [PubMed] [Google Scholar]
- 7.WHO. [October 16 2009]; http://www.who.int/mediacentre/factsheets/fs211/en/index.html.
- 8.Lee LY, et al. Memory T cells established by seasonal human influenza A infection cross-react with avian influenza A (H5N1) in healthy individuals. J Clin Invest. 2008;118:3478–3490. doi: 10.1172/JCI32460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roti M, et al. Healthy human subjects have CD4+ T cells directed against H5N1 influenza virus. J Immunol. 2008;180:1758–1768. doi: 10.4049/jimmunol.180.3.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gioia C, et al. Cross-subtype immunity against avian influenza in persons recently vaccinated for influenza. Emerg Infect Dis. 2008;14:121–128. doi: 10.3201/eid1401.061283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kashyap AK, et al. Combinatorial antibody libraries from survivors of the Turkish H5N1 avian influenza outbreak reveal virus neutralization strategies. Proc Natl Acad Sci U S A. 2008;105:5986–5991. doi: 10.1073/pnas.0801367105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Throsby M, et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One. 2008;3:e3942. doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okuno Y, Isegawa Y, Sasao F, Ueda S. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J Virol. 1993;67:2552–2558. doi: 10.1128/jvi.67.5.2552-2558.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smirnov YA, et al. An epitope shared by the hemagglutinins of H1, H2, H5, and H6 subtypes of influenza A virus. Acta Virol. 1999;43:237–244. [PubMed] [Google Scholar]