Abstract
The Pim protein kinases play important roles in cancer development and progression, including prostate tumors and hematologic malignancies. To investigate the potential role of these enzymes as anticancer drug targets, we have synthesized novel benzylidene-thiazolidine-2,4-diones that function as potent Pim protein kinase inhibitors. With IC50 values in the nanomolar range, these compounds block the ability of Pim to phosphorylate peptides and proteins in vitro and, when added to DU145 prostate cancer cells overex-pressing Pim, inhibit the ability of this enzyme to phosphorylate a known substrate, the BH3 protein BAD. When added to prostate cancer cell lines, including PC3, DU145, and CWR22Rv1, and human leukemic cells, MV4;11, K562, and U937 cells, these compounds induce G1-S cell cycle arrest and block the antiapoptotic effect of the Pim protein kinase. The cell cycle arrest induced by these compounds is associated with an inhibition of cyclin-dependent kinase 2 and activity and translocation of the Pim-1 substrate p27Kip1, a cyclin-dependent kinase 2 inhibitory protein, to the nucleus. Furthermore, when added to leukemic cells, these compounds synergize with the mammalian target of rapamycin inhibitor rapamycin to decrease the phosphorylation level of the translational repressor 4E-BP1 at sites phosphorylated by mammalian target of rapamycin. Combinations of rapamycin and the benzylidene-thiazolidine-2,4-diones synergistically block the growth of leukemic cells. Thus, these agents represent novel Pim inhibitors and point to an important role for the Pim protein kinases in cell cycle control in multiple types of cancer cells.
Introduction
The Pim-1 and Pim-2 serine/threonine protein kinases are implicated as potential causative enzymes in the growth and progression of multiple cancer types. In human tumor samples, Pim-1 overexpression is reported in diffuse B-cell lymphoma, chronic lymphocytic leukemia, acute myelogenous leukemia, head and neck cancer, and prostate cancer (1, 2). Expression of Pim-1 is absent to low in benign prostatic hyperplasia, moderate to strong in high-grade prostatic intraepithelial neoplasia, and further increased in prostate adenocarcinoma (3, 4). This increase in Pim levels is correlated with higher Gleason scores and progression to a more aggressive disease (3).
The Pim protein kinases were first cloned as proviral insertions in murine T-cell lymphomas induced by the c-Myc oncogene (5, 6). In additional murine and cell culture models, there appears to be a close biological interaction between the c-Myc protein and the Pim kinases. Overexpression of either Pim-1 or Pim-2 protein kinases in transgenic mice containing elevated levels of c-Myc results in a high incidence of lymphoma (7, 8). As well, transgenic expression of c-Myc in the prostate elevates the level of Pim protein in this organ (4). Furthermore, human prostate cancer PC3 cells overexpressing Pim showed statistically significant higher levels of c-Myc mRNA compared with control PC3 cells (9). Recently, c-Myc has been shown to recruit Pim-1 to the E-boxes of c-Myc target genes and to phosphorylate histone H3 to facilitate Myc-dependent transcription (10). Additionally, Pim-1 has been shown to stabilize c-Myc and increase the levels of this protein by phosphorylating Ser62 resulting in enhanced transcriptional activity of the c-Myc protein (11).
The Pim protein kinases appear to play a key role in cell cycle progression and apoptosis in multiple cell types. The Pim kinases phosphorylate the proapoptotic protein Bad at Ser112 causing its inactivation, leading to enhancement of Bcl-2 activity, thus promoting cell survival (12–14). Thus, murine interleukin (IL)-3-dependent FDCP1 cells expressing Pim-1 are more resistant to apoptosis when starved of growth factors (15). Alternatively, Pim has been shown to regulate nuclear factor-κB activity and so doing has the potential to regulate additional downstream proteins involved in apoptosis, that is, Bax (16). Pim protein kinase has been shown to phosphorylate substrates involved in cell cycle progression including Cdc25A, p21, p27Kip1, NuMA, C-TAK1, and Cdc25C, the phosphorylation of which results in G1-S and/or G2-M progression (1, 17–19). Also, Pim-2 has been shown to regulate the phosphorylation of 4E-BP1 causing it to dissociate from eIF-4E, suggesting a potential indirect control mechanism of cell growth. In tissue culture, serum-starved PC3 cells showed cell cycle arrest in G1, whereas PC3-Pim cells showed much lower extent of arrest (9). When these cells were grown as s.c. tumors in mice, PC3 prostate cancer cells overexpressing Pim-1 grew significantly faster than cells expressing vector control, again pointing to a role of Pim in enhancing cell growth rate (9).
To explore the possibility that the Pim protein kinases would be an excellent target for small-molecule cancer chemotherapy and to better discern the biologic activity of this enzyme in tumor cells, we have screened a 50,000 compound library for inhibitors and identified and synthesized novel benzylidene-thiazolidine-2,4-diones as nanomolar inhibitors of these enzymes (20). In this report, we show that these compounds inhibit Pim-mediated phosphorylation in intact cancer cells and block cell growth of prostate cancer and leukemic cells in the G1 phase of the cell cycle. This cell cycle block is associated with decreased cyclin-dependent kinase 2 (Cdk2) activity and translocation of the known Pim substrate, p27Kip1, to the nucleus. Additionally, in leukemic cells, we find a synergistic interaction of the benzyli-dene-thiazolidine-2,4-diones and rapamycin in the inhibition of both cell growth and phosphorylation of 4E-BP1, a known mammalian target of rapamycin (mTOR) substrate. Thus, these agents point to Pim protein kinase as a cell cycle control protein and show that they have the potential to act as antitumor agents.
Materials and Methods
Reagents
The benzylidene-thiazolidine-2,4-dione Pim inhibitors were synthesized and characterized as described elsewhere (20). Recombinant Pim-1-GST and the Pim-1 peptide substrate (RSRHSSYPAGT; corresponding to amino acids 107–117 of Bad) were purchased from Millipore. Recombinant 4E-BP1 was purchased from Calbiochem, p27Kip1 was obtained from Novus Biologicals, and rapamycin was supplied by LC Laboratories. The following antibodies were purchased from Cell Signaling Technology: anti-phospho-Bad (Ser112; 5284), anti-Bad (9292), anti-c-Myc (9402), anti-p27Kip1 (2552), anti-β-tubulin (2146), anti-phospho-4E-BP1 (Thr37/Thr46; 9459), and anti-4E-BP1 (9452). The following were from Santa Cruz Biotechnology: anti-Pim-1 (SC-13513), anti-actin (SC-8432), anti-Cdk2 (SC-163), and anti-lamin B1 (SC-56144).
Treatment of Cell Lines with Pim Inhibitors
DU145 and CWR22Rv1 (22Rv1) human prostate cancer cells overexpressing Pim-1 cDNAs were produced through retroviral transduction as described previously (21). Briefly, the coding region of the human Pim-1 gene was cloned into the pLNCX retroviral vector (Clontech). To produce infectious virus, the GP-293 packaging cell line was cotransfected with retroviral plasmids (pLNCX or pLNCX/Pim-1) along with pVSV-G. After 48 h of incubation, the virus particles were concentrated by centrifugation from the medium. Prostate cells were plated at 1 × 105 per 60 mm plate 16 to 18 h before infection. Cells were infected with 5 × 104 viral particles/plate in the presence of 8 μg/mL polybrene. After 6 h of incubation, stable pools of G418-resistant cells were selected for 10 days and the expression of the Pim-1 protein was verified by Western blot analysis. Human prostate cancer cell lines PC3, DU145, DU145-vector, DU145-Pim, 22Rv1-vector, 22Rv1-Pim, and LNCaP and human leukemia cell lines MV4;11, K562, and U937 were maintained in RPMI 1640 with 10% FCS and 1% penicillin-streptomycin at 37°C in 5% CO2. The IL-3-dependent murine cell line FDCP1-Pim described previously (15) was grown in RPMI 1640 with 10% FCS, 1% penicillin-streptomycin, and IL-3 (2 ng/mL) at 37°C in 5% CO2.
To measure growth inhibition by Pim inhibitors, prostate cancer cell lines or leukemic cells were seeded in a 96-well plate (1 × 104 per 0.1 mL) and then treated with inhibitors or DMSO for 72 h. Leukemic cells were treated immediately, whereas prostate cancer cells were first allowed to attach overnight. Cell growth inhibition was measured using the CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay (Promega) according to the manufacturer’s instructions.
For the calculation of combination index values, MV4;11 growth inhibition was determined as described above at multiple concentrations of benzylidene-thiazolidine-2,4-diones, in combination with varied amounts of rapamycin, and the resulting data were analyzed according to the method described by Chou (22).
For cell cycle analysis, cells were harvested, washed once in PBS, fixed in cold 70% ethanol, stained with propidium iodide, and analyzed by flow cytometry (Medical University of South Carolina Flow Cytometry & Cell Sorting Core Facility).
Pim-1 Kinase Activity Assay
IC50 values for Pim inhibitors were measured by a coupled kinase assay as described previously (23) with the following changes: assays were carried out in 20 mmol/L MOPS (pH 7) containing 100 mmol/L NaCl, 10 mmol/L MgCl2, 2.5 mmol/L phosphoenolpyruvate, 0.2 mmol/L NADH, 30 μg/mL pyruvate kinase, 10 μg/mL lactate dehydrogenase, 2 mmol/L DTT, and 25 nmol/L Pim-1 with 100 μmol/L peptide (RSRHSSYPAGT). Activity was measured by monitoring NADH oxidation as the decrease at 340 nm in a VersaMax microplate reader (Molecular Devices) at 25°C. Reactions were initiated by the addition of ATP (100 μmol/L), and inhibitors (final 1% DMSO) were added just before the addition of ATP. IC50 values were determined using nonlinear regression with the program GraphPad Prism. This assay determines kinase activity by measuring the amount of ADP produced, which is coupled to NADH oxidation by lactate dehydrogenase and pyruvate kinase. The ability of Pim-1 kinase to phosphorylate full-length protein substrates 4E-BP1 and p27Kip1 was determined as follows: purified Pim-1 (1 ng) was added along with 4E-BP1 (2 μg) or p27Kip1 (2 μg) and Pim inhibitor in assay buffer [20 mmol/L MOPS (pH 7) containing 100 mmol/L NaCl, 10 mmol/L MgCl2, and 2 mmol/L DTT]. Assays were initiated by the addition of ATP (100 μmol/L) and [γ-32P]ATP (10 μCi). Reactions were allowed to proceed for 15 min at 30°C and then separated by SDS-PAGE. 32P-phosphorylated substrates were visualized by autoradiography and quantified by densitometry.
Western Blotting
Cells were harvested, washed with PBS, and resuspended in lysis buffer [20 mmol/L Tris-HCl (pH 7.5) containing 1% SDS, 50 mmol/L NaCl, 1 mmol/L EDTA, 1 mmol/L phe-nylmethysulfonyl fluoride, 10 mmol/L sodium fluoride, and 1 mmol/L sodium orthovanadate]. Samples were then incubated on ice for 30 min followed by 15 min centrifugation. Supernatants were separated by SDS-PAGE and transferred to nitrocellulose membranes. Membranes were blocked in 5% nonfat milk in TBST [20 mmol/L Tris-HCl (pH 7.5) containing 150 mmol/L NaCl and 0.1% Tween 20] for 1 h with agitation and washed, and primary antibodies were added (1:1,000 dilution in 5% bovine serum albumin in TBST) and membranes were incubated overnight at 4°C with agitation. Membranes were washed and incubated with horseradish peroxidase-conjugated secondary antibodies (1:5,000 dilution in 5% nonfat milk in TBST) for 2 h at room temperature with agitation. Proteins were detected using the ECL Western Blotting Detection Reagent (GE Healthcare).
p27Kip1 Location and Cdk2 Kinase Activity Assays
To examine p27Kip1 location, K562, U937, or MV4;11 cells (1 × 105/mL) were incubated for 72 h in complete medium with DMSO or benzylidene-thiazolidine-2,4-diones, 4a or 16a. Cells were harvested and washed in PBS and cytoplasmic and nuclear fractions were prepared using the NE-PER Nuclear and Cytoplasmic Extraction kit (Pierce Biotechnology) according to the manufacturer’s instructions, followed by SDS-PAGE and Western blotting with anti-p27Kip1 antibody, as described above. To measure Cdk2 activity, this protein was immunoprecipitated from K562, U937, or MV4;11 cells treated for 72 h with Pim inhibitors and lysed in buffer [50 mmol/L Tris-HCl (pH 8.0) containing 5 mmol/L EDTA, 150 mmol/L NaCl, 1% NP-40, and 1 mmol/L phenylmethylsulfonyl fluoride] followed by the addition of Cdk2 antibody (2 μg). Samples were then rotated overnight at 4°C, and Cdk2 was immunoprecipitated by the addition of protein G beads (Pierce Biotechnology) with rotation at room temperature for 1 h. Beads were washed three times with PBS and resuspended in assay buffer [10 mmol/L MOPS (pH 7.2) containing 1 mmol/L EDTA, 15 mmol/L MgCl2, 10 mmol/L sodium fluoride, and 1 mmol/L sodium orthovanadate] containing histone H1 (3 μg; Millipore) as a Cdk2 substrate, ATP (100 μmol/L), and [γ-32P]ATP (10 μCi). Reactions were allowed to proceed for 15 min at 37°C and then analyzed by SDS-PAGE. 32P-phosphorylated histone H1 was visualized by autoradiography, and Cdk2 protein levels were detected by Western blotting as described above.
To examine p27Kip1 location by fluorescence microscopy, DU145-vector and DU145-Pim cells were transfected with plasmids pEYFP-C1, pEYFP-p27Kip1, pEYFP-p27Kip1 (T157A), or pEYFP-p27Kip1 (T198A; a generous gift from Dr. Joyce Slingerland, University of Miami Miller School of Medicine; 1 μg DNA/well in a 6-well dish) using Lipofectamine 2000 (Invitrogen). Forty-eight hours after transfection, cells were treated with 4a or 16a (5 μmol/L) in DMEM containing 1% FCS for 24 h. The expression of enhanced yellow fluorescent protein (EYFP)-p27Kip1 in live cells was visualized on a Leica TCS SP2 laser scanning confocal microscope (Leica Microsystems).
The recombinant HA-tagged p27, wild-type and mutants, were generated by PCR, sequenced, and cloned into pcDNA3.1 between HindIII and EcoRV restriction sites. The plasmids were transfected into K562 cells with Lipofectamine 2000, harvested after 48 h of incubation, and subjected to cytosolic and nuclear fractionation.
Results
Identification of Benzylidene-Thiazolidine-2,4-Dione Inhibitors of Pim-1
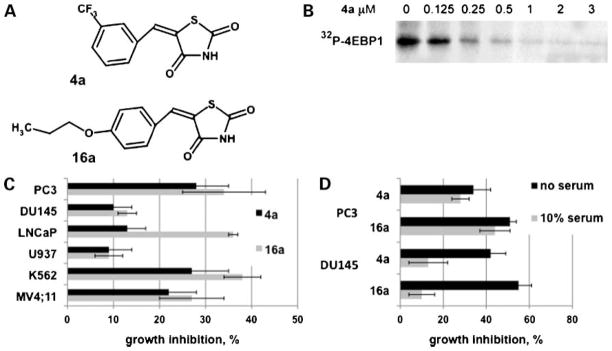
To identify small-molecule Pim protein kinase inhibitors, the DIVERSet collection from the ChemBridge was screened using the S6 kinase/RSK2 peptide as a substrate. Of the 50,000 compounds screened, 10 compounds with an IC50 of ≤20 μmol/L were identified (20). Using a cell-based assay, which examined the ability of these compounds to inhibit the autophosphorylation of Pim-1 protein kinase transfected in HEK 293 cells, the benzylidene-thiazolidine-2,4-dione chemotype (Supplementary Table S1)8 was identified as a cell-permeable Pim inhibitor (20). Subsequently, a series of compounds in which substitutions at the meta- and para-positions on the benzene ring were synthesized as described elsewhere (20). To determine which compound(s) warranted further investigation in cell-based assays, the percent growth inhibition of each compound was determined using the prostate cancer cell line PC3 at a single dose of 5 μmol/L after 24 h. This dose was previously determined to be well above the IC50 value for Pim-1 (20). The IC50 values for compounds that showed the highest percent PC3 growth inhibition along with the original screening hit, 4a, was determined in a coupled kinase assay using a peptide corresponding to amino acids 107 to 117 of the proapoptotic protein Bad (RSRHSSYPAGT) a known in vivo substrate of Pim kinase (14). Compounds 4a and 16a (Fig. 1A) were found to be the most potent Pim-1 inhibitors based on IC50 value (17 ± 7 nmol/L for 4a and 63 ± 11 nmol/L for 16a). Both of these compounds were competitive with respect to ATP (20), suggesting that they bind within the ATP-binding pocket. Additionally, 4a inhibited the in vitro phosphorylation by Pim-1 of the known substrate, the translational repressor 4E-BP1 (Fig. 1B; ref. 9). The ability of 4a and 16a to inhibit the growth of various cancer cell lines was evaluated after treatment for 72 h in culture. Prostate cancer and leukemic cell lines were chosen because Pim-1 has been shown to play an integral role in the development of prostate carcinogenesis and hematologic malignancies (2–4, 24–26). A dose of 5 μmol/L was chosen for cell studies because this concentration of compound had been used in the in vitro screening of potential protein kinase substrates (20). As shown in Fig. 1C, 4a and 16a caused similar growth inhibition of each cell line, except the prostate cancer cell line LNCaP in which 16a was considerably more inhibitory than 4a. U937 leukemia and DU145 prostate cells were less sensitive to either Pim inhibitor. Both 4a and 16a were shown to be serum-bound (data not shown); therefore, growth inhibition in serum versus no serum was determined using PC3 and DU145 cells. The sensitivity to 4a and 16a was not affected by withdrawal of serum from PC3 cells; however, DU145 cells became considerably more sensitive under serum-free conditions (Fig. 1D). This suggests that the response to the Pim inhibitors depends on both the specific cell line tested and the presence or absence of serum.
Figure 1.
Benzylidene-thiazolidine-2,4-diones inhibit Pim kinase activity and prostate cancer and leukemic cell growth. A, structures of 4a and 16a. B, dose-dependent inhibition of Pim-1 kinase activity by 4a. Recombinant Pim-1 activity was measured as described in Materials and Methods using the full-length substrate 4E-BP1 in the presence of increasing concentrations of 4a and a fixed concentration of [γ32P]ATP. C, 4a and 16a inhibit prostate cancer cell (PC3, DU145, and LNCaP) and leukemic cell (U937, K562, and MV4;11) growth in the presence of 10% serum. Cells were incubated for 72 h in complete medium in the presence of 5 μmol/L 4a, 16a, or DMSO (control) followed by a MTS assay (see Materials and Methods). Data are represented as the percent growth inhibition relative to DMSO. Average (SD) of 4 independent assays. D, comparison of the inhibitory effect of 4a and 16a in 10% serum-containing medium versus serum-free medium. PC3 or DU145 cells were plated in 10% serum-containing medium and allowed to attach for 24 h. Medium was then replaced with serum-free medium and 5 μmol/L 4a, 16a, or DMSO (control) was added and cells were incubated for an additional 72 h. Data are represented as in C.
Figure 4.
Pim inhibitors 4a and 16a cause translocation of p27Kip1 to the nucleus and reduce Cdk2 activity. A, 4a and 16a reduce the Pim-1-mediated phosphorylation of p27Kip1. The ability of Pim-1 to phosphorylate p27Kip1 in the presence of DMSO (control), 4a (5 μmol/L), or 16a (5 μmol/L) was assayed in the presence of [γ32P]ATP as described in Materials and Methods. Autoradiography of a SDS-PAGE gel is shown. B, 4a or 16a treatment causes the translocation of p27Kip1 to the nucleus. K562, U937, and MV4;11 cells were treated with DMSO (control), 4a (5 μmol/L), or 16a (5 μmol/L) for 72 h in RPMI containing 10% FCS. Cytoplasmic (C) and nuclear (N) fractions were prepared as described in Materials and Methods, and the level of p27Kip1 in the cytoplasmic and nuclear fraction was determined by SDS-PAGE followed by Western blotting. β-Tubulin and lamin B1 serve as cytoplasmic and nuclear markers, respectively. C, Cdk2 activity in K562 cells treated with DMSO, 4a (5 μmol/L), or 16a (5 μmol/L) for 72 h in RPMI containing 10% FCS. Cdk2 was immunoprecipitated from cell lysates and its activity was measured using histone H1 as a substrate in the presence [γ32P]ATP as described in Materials and Methods. Scanning densitometry of the 32P-labeled histone H1 band was used to quantitate the degree of inhibition of activity induced by 4a and 16a addition to cells. Representative of 3 experiments showing similar results.
We showed previously that 4a and 16a were highly specific for inhibition of Pim-1 (20). Of the 60 kinases profiled, 16a, but not 4a, showed inhibition of only one other kinase, the dual-specificity tyrosine phosphorylation-regulated protein kinase 1a (DYRK1a). To examine overlapping biochemical activities between DYRK1a and Pim, we determined the activity of the DYRK1a enzyme activity using the Bad peptide as the substrate. Compared with the preferred DYRK1a peptide substrate Woodtide (27), we found no detectable DYRK1a activity using the Bad peptide (Supplementary Fig. S1A).8 Conversely, Pim-1 showed no activity using the peptide Woodtide as a substrate. We did confirm in our coupled kinase assay the inhibitory activity of 16a, but not 4a, toward recombinant DYRK1a activity (Supplementary Fig. S1A).8
Pim-1 Inhibitors Reduce Phosphorylation of the Pim Target Bad in Prostate and Hematopoietic Cells
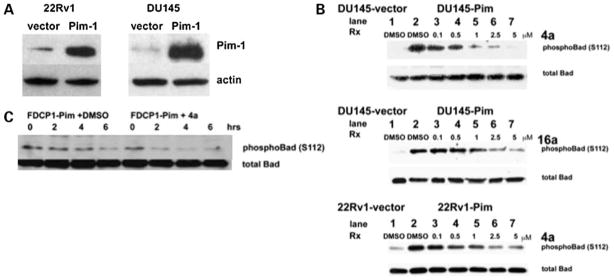
To confirm that 4a and 16a inhibit Pim-1 activity in cells, the phosphorylation level of the Pim target Bad was determined by Western blotting using prostate cancer and hematopoietic cells stably transfected with Pim-1 (Fig. 2). The 22Rv1-vector cells show more endogenous Pim-1 protein compared with DU145-vector cells (Fig. 2A) and therefore more endogenous phospho-Bad protein (Fig. 2B). The level of phospho-Bad decreased in a dose-dependent manner in both 22Rv1-Pim and DU145-Pim cells treated with 4a or 16a for 1 h under serum-free conditions, whereas the level of total Bad protein remained constant. A significant (>80%) reduction of phospho-Bad was observed at a concentration of 5 μmol/L of 4a or 16a; therefore, this concentration was chosen for subsequent cellular experiments. Under IL-3-starved conditions, the FDCP1-Pim cell line has been shown to survive longer with fewer apoptotic cells compared with the FDCP1-vector cell line, which may be attributed to the retention of Bad phosphorylation (15). Therefore, the level of phospho-Bad was examined over a time course in the he-matopoietic cell line FDCP1 stably transfected with Pim-1 in the absence (DMSO) or presence of 4a (5 μmol/L) in serum-and IL-3-free conditions. A significant reduction in phos-pho-Bad levels was observed in Pim inhibitor-treated FDCP1-Pim cells by 2 h compared with DMSO-treated cells (Fig. 2C). Other kinases known to phosphorylate Bad at Ser112 are RSK1 and protein kinase A; however, the kinase selectivity profile showed that neither of these are targets for inhibition by 4a or 16a (20).
Figure 2.
Benzylidene-thiazolidine-2,4-diones inhibit Pim kinase activity in prostate cancer cells. A, Western blot showing the level of Pim-1 in endogenous (vector) and Pim overexpressor cell lines. B, dose-dependent reduction of Bad phosphorylation by 4a and 16a. Prostate cancer cells, DU145 or 22Rv1, were starved in serum-free medium overnight and then treated with the indicated amount of 4a or 16a or DMSO for 1 h. The level of phospho-Bad (Ser112) and total Bad were determined by SDS-PAGE followed by Western blotting. The level of endogenous phospho-Bad protein is significantly lower in vector-transfected cells compared with Pim-1-expressing cells (compare lanes 1 and 2). C, time course-dependent reduction of phospho-Bad (Ser112) in 4a- or DMSO-treated FDCP1-Pim cells. FDCP1-Pim cells were washed in PBS and placed in IL-3- and serum-free medium and 4a (5 μmol/L) or DMSO was added as a control. Lysates were prepared at the indicated time points and subjected to SDS-PAGE followed by Western blotting to measure phospho-Bad (Ser112) and total Bad.
Pim Inhibitors Cause Cell Cycle Arrest and Reverse the Antiapoptotic Activity of Pim-1
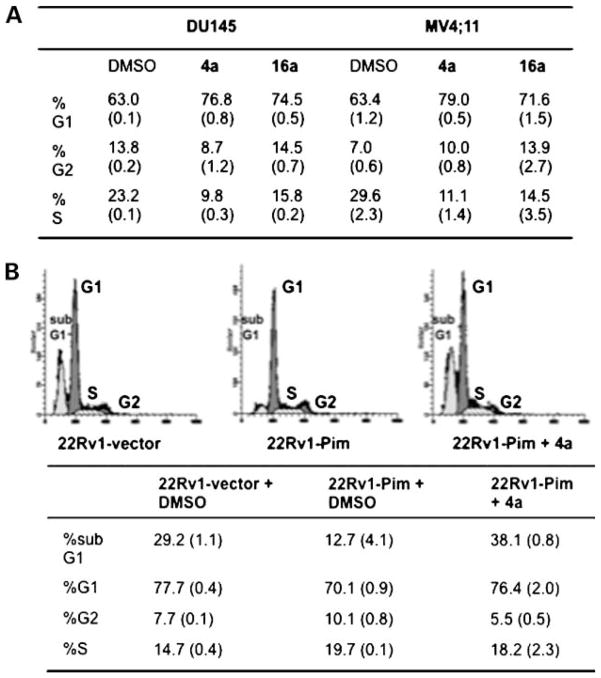
Many Pim-1 substrates play a role in cell cycle progression including Cdc25A, p21, p27Kip1, NuMA, C-TAK1, and Cdc25C, which when phosphorylated result in G1-S and/or G2-M progression (1, 17–19). Therefore, the ability of 4a and 16a to affect the cell cycle distribution of both prostate cancer and hematopoietic cells was determined. DU145 growing in 2% serum and MV4;11 cells plated in 10% serum were treated with 4a or 16a at 5 μmol/L for 72 h followed by fluorescence-activated cell sorting analysis. As shown in Fig. 3A, both Pim inhibitors caused a significant G1 cell cycle arrest compared with the DMSO control. No significant sub-G1 population (apoptotic cells) was observed in either cell line. However, the apoptotic effect of 4a was clearly shown using the 22Rv1-vector and 22Rv1-Pim cell lines. In Fig. 3B, cells were treated with DMSO or 4a (5 μmol/L) for 72 h under serum-free conditions. Serum starvation of 22Rv1-vector (+DMSO) clearly resulted in a considerable amount of apoptosis (sub-G1 29.2%); however, expression of Pim-1 decreased the percent of apoptotic cells (12.7%) consistent with its prosurvival role as determined previously in myeloid cells (15). Treatment of 22Rv1-Pim cells with 4a reversed the antiapoptotic effect of Pim-1, as the sub-G1 population increased to 38.1% (compared with 12.7% for DMSO-treated cells). Additionally, the cell cycle analysis shows that overexpression of Pim-1 decreases the percent-age of cells in G1 and increases the number in S and G2. This Pim-1 effect is reversed by treatment with 4a or 16a, showing their ability to induce a G1 block.
Figure 3.
Pim inhibitors 4a and 16a cause cell cycle arrest and apoptosis. A, DU145 (2% FCS) and MV4;11 (10% FCS) cells were treated with 4a (5 μmol/L), 16a (5 μmol/L), or DMSO (control) for 72 h followed by staining with propidium iodide and cell cycle analysis by flow cytometry. Data are represented as the percent of cells in each phase. Average (SD) of 3 independent experiments. B, 4a reverses the antiapoptotic activity (% sub-G1) of Pim-1 in 22Rv1 cells. 22Rv1-vector or 22Rv1-Pim cells were treated in serum-free medium with 4a (5 μmol/L) or DMSO for 72 h followed by staining with propidium iodide and cell cycle analysis by flow cytometry (top). Data shown in the table are the percent of cells in each phase of the cell cycle. Average (SD) of 3 independent assays.
Pim Inhibitors Increase the Amount of p27Kip1 in the Nucleus
Recently, Pim-1 has been shown to phosphorylate the cyclin-dependent kinase inhibitor p27Kip1 resulting in its nuclear export and degradation (19). Because 4a and 16a induced cell cycle arrest (Fig. 3A and B), the localization of p27Kip1 in Pim inhibitor-treated cells was investigated as a potential mechanism explaining the biological activity of these agents. First, the ability of Pim-1 to phosphorylate p27Kip1 and the ability of 4a and 16a (5 μmol/L) to reduce phosphorylation of this substrate was shown in vitro (Fig. 4A). Next, the leukemic cell lines K562, U937, and MV4;11 were treated with 4a or 16a for 72 h in medium containing 10% FCS followed by detection of p27Kip1 levels in cytoplasmic and nuclear fractions (Fig. 4B). Both Pim inhibitors caused an increase in the amount of p27Kip1 in nuclear fractions in all three cell lines. Given these results and the previous observation that overexpression of Pim-1 in K562 cells promoted cell cycle progression by up-regulating Cdk2 activity (19), the affect of Pim-1 inhibition by 4a and 16a on Cdk2 activity was determined. K562 cells were treated under the same conditions in Fig. 4B, Cdk2 was immunoprecipitated, and its kinase activity was determined using histone H1 as the substrate (Fig. 4C). Cdk2 immunoprecipitated from 4a- or 16a-treated cells showed ~50% and 60%, respectively, decreased activity. Kinase selectivity profiling showed that 4a and 16a do not inhibit Cdk2 activity (20). Thus, the results are consistent with inhibition of endogenous Pim-1 by these agents causing increased nuclear p27Kip1 levels and inhibiting Cdk2 activity.
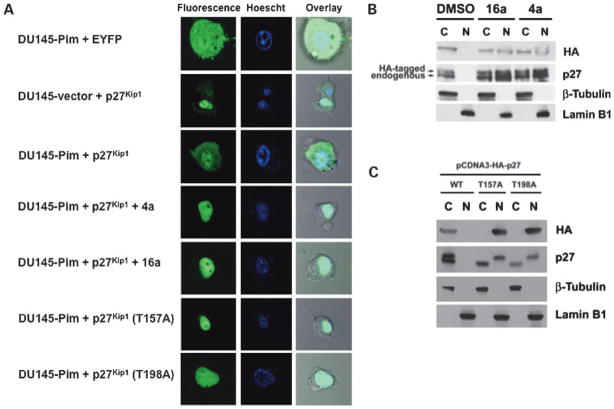
To determine the effect of Pim-1 overexpression on p27Kip1 localization, DU145-vector and DU145-Pim cells were transfected with a plasmid expressing p27Kip1 fused to EYFP and p27Kip1 was then visualized by fluorescence microscopy. As shown in Fig. 5A, the control vector expressing EYFP alone is distributed throughout the nucleus and cytosol, whereas the fusion with p27Kip1 localizes the fluorescence in the nucleus as shown by overlay with Hoechst dye, which stains nuclei. Overexpression of Pim-1 in the DU145 cells increased the amount of p27Kip1 located in the cytosol. Treatment of these cells with the Pim inhibitors 4a or 16a reversed this Pim-mediated effect as shown by the decreased cytosolic p27Kip1 after treatment. Mutation of T157A or T198A, two hypothesized Pim phosphorylation sites, targeted this protein to the nucleus. Using cell fractionation and Western blotting, similar results were obtained in K562 leukemia cells transfected with HA-tagged p27Kip1 (Fig. 5B). Pim-1 has been shown to phosphorylate p27Kip1 at T157 and T198, which is postulated to promote p27Kip1 nuclear export (19). Accordingly, mutation of either T157 or T198 to alanine resulted in a mutant p27Kip1 that localized exclusively to the nucleus in K562 cells, showing similar results to the Pim-1-overexpressing DU145 cells (Fig. 5A and C). These results are consistent with the inhibition of Cdk2 phosphorylation by 4a or 16a causing nuclear retention of p27Kip1 (Fig. 5).
Figure 5.
Pim-1 overexpression increases the amount of cytoplasmic p27Kip1 and 4a and 16a reduce this Pim-mediated effect. A, DU145-vector and DU145-Pim cells were transfected with the control EYFP, EYFP-p27Kip1, EYFP-p27Kip1 (T157A), or EYFP-p27Kip1 (T198A) plasmids. Transfected DU145-Pim cells were then treated with 4a (5 μmol/L) or 16a (5 μmol/L) for 24 h. p27Kip1 was visualized in live cells using a fluorescence microscope as described in Materials and Methods. The overlay lane places the fluorescent image on the top of the phase-contrast microscopic image in identical cells. B, K562 cells were transfected with HA-tagged p27Kip1 and treated with 4a (5 μmol/L) or 16a (5 μmol/L) for 24 h. Cytoplasmic and nuclear fractions were prepared as described in Materials and Methods, and the level of p27Kip1 in the cytoplasmic and nuclear fraction was determined by SDS-PAGE followed by Western blotting. β-Tubulin and lamin B1 serve as cytoplasmic and nuclear markers, respectively. C, K562 cells were transfected with HA-tagged wild-type p27Kip1 (WT), T157A-p27Kip1, or T198A-p27Kip1. Cytoplasmic and nuclear fractions were prepared as described in Materials and Methods, and the level of wild-type and mutant p27Kip1 protein in the cytoplasmic and nuclear fractions were determined by SDS-PAGE followed by Western blotting. The two bands of different size observed in the blot probed with the antibody to p27 in both B and C correspond to HA-tagged (top) and endogenous (bottom) p27Kip1.
Pim Inhibitors Synergize with Rapamycin to Cause Significant Growth Inhibition of Leukemic Cells
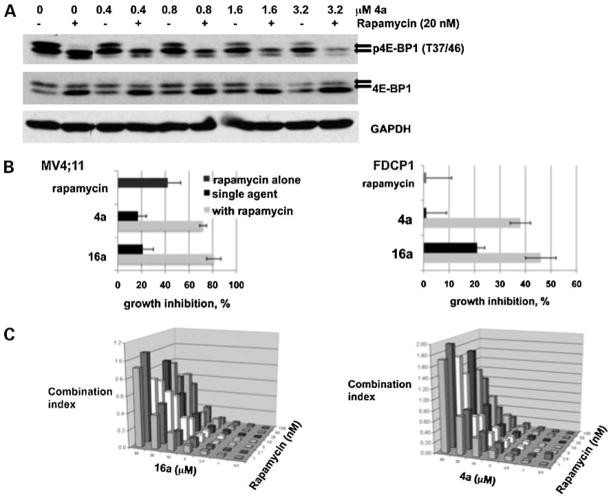
On addition of serum or growth factors, the translational repressor 4E-BP1 is inactivated by hyperphosphorylation, in part through the activity of mTOR on Thr37 and Thr46 of 4E-BP1, allowing for increased protein synthesis (28). Phosphorylation of these sites is sensitive to treatment with the mTOR inhibitor rapamycin (9). 4E-BP1 is a known in vitro target of the Pim kinases (9, 26), although the mechanism by which Pim affects this protein in vivo is not clearly defined. We have shown that 4a and 16a (Fig. 1B; data not shown) inhibit the in vitro Pim-mediated phosphorylation of 4E-BP1. Interestingly, Pim-2 has been shown to confer rapamycin resistance in hematopoietic cells (26). To evaluate the role of combined treatment of rapamycin and benzyli-dene-thiazolidine-2,4-dione inhibitors, we have used the FDCP1 cell line that is IL-3 dependent. To evaluate the effects of 4a and rapamycin, these cells were starved of serum and IL-3 for 1 h during which rapamycin (20 nmol/L) or 4a at various concentrations or in combination were added. At the end of this incubation, IL-3 was added to stimulate 4E-BP1 phosphorylation. The cells were centrifuged and extracts were subjected to SDS-PAGE and Western blotting. Using an antibody to the Thr37/Thr46 phosphorylation site of 4E-BP1, increasing 4a concentrations reduced the level of the most highly phosphorylated form of 4E-BP1 (Fig. 6A, top arrow) and when combined with rapamycin also decreased the less phosphorylated forms of 4E-BP1 (Fig. 6A, bottom arrow). This combined effect is seen in the 4E-BP1 blot as an increase in the lower band. Similar regulation of 4E-BP1 phosphorylation was seen with MV4;11 cells (data not shown). Furthermore, the combined treatment of rapamycin with 4a or 16a for 72 h caused significant growth inhibition of MV4;11 and FDCP1 cells, with 16a showing slightly more combined inhibitory effect than 4a (Fig. 6B). To examine the potential synergistic growth-inhibitory effect between 4a or 16a and rapamycin in MV4;11 cells, a combination index analysis was carried out (Fig. 6C). These results show that, at low doses of benzylidene-thiazolidine-2,4-diones and rapamycin, the combined effect of these agents is highly synergistic, whereas, at higher concentrations of 4a and 16a, this synergism is lost. Considering the inhibition of DYRK1a by 16a (20), we determined if inhibition of DYRK1a by 16a contributed to the synergism of rapamycin and 16a using the DYRK inhibitor harmine (29). MV4;11 cells treated with harmine and rapamycin showed essentially identical growth inhibition compared with treatment with harmine alone (Supplementary Fig. S1B).8 This suggests that the synergism observed between 4a or 16a and rapamycin is secondary to inhibition of the Pim protein kinases. In addition, because the mTOR pathway regulates the levels of c-Myc protein, we evaluated the effect of treatment with these Pim protein kinase inhibitors on c-Myc levels. We determined that treatment of the BCR/ABL-positive human leukemia cell line K562 with 4a or 16a for 4 h reduced the levels of c-Myc protein (Supplementary Fig. S3).8 This finding is consistent with the fact that inhibition of Pim-1 signaling has been shown to attenuate cell proliferation in this cell line (19).
Figure 6.
Pim inhibitors 4a and 16a act synergistically with rapamycin to inhibit cell growth. A, 4a and 16a combined with rapamycin to effectively reduce the level of phospho-4E-BP1 (Thr37/Thr46). FDCP1 cells were starved of IL-3 and serum for 1 h during which cells were treated with rapamycin or 4a or a combination of the two agents. After 1 h of treatment, IL-3 (2 ng/mL) was added for 5 min to stimulate 4E-BP1 phosphorylation. Cells were pelleted and the level of phospho-4E-BP1 (Thr37/Thr46), 4E-BP1, and GAPDH was determined by SDS-PAGE followed by Western blotting. Arrows, phospho-4E-BP1 protein. B, combination of 4a or 16a with rapamycin effectively inhibits the growth of MV4;11 (left) and FDCP1 (right) cells. Cells were incubated for 72 h in RPMI + 10% FCS (IL-3 included in FDCP1 cells) with rapamycin (5 nmol/L), 4a (5 μmol/L), 16a (5 μmol/L), or the combination. Data are represented as the percent growth inhibition relative to DMSO. Average (SD) of 4 independent experiments. C, combination index values show synergism between rapamycin and 4a or 16a in MV4;11 cells. Combination index analyses were carried out as described in Materials and Methods.
Discussion
We have identified a new chemotype of Pim protein kinases inhibitors, the benzylidene-thiazolidine-2,4-diones. In vitro, these small molecules are capable of blocking the ability of Pim kinase to phosphorylate peptides with IC50 values in the nanomolar range and inhibit the Pim protein kinase directed phosphorylation of two known substrates, 4E-BP1 and p27Kip1. Members of this chemotype can be Pim-1, or Pim-2, specific or dual inhibitors blocking the activity of both of these enzymes (20). As suggested by a screen of 60 additional serine, threonine, and tyrosine protein kinases (20), benzylidene-thiazolidine-2,4-diones are highly specific inhibitors of the Pim protein kinase. Our compounds differ in structure from previously described small-molecule inhibitors of this enzyme. These inhibitors include ruthenium half-sandwich complexes, which mimic staurosporine (30, 31), imidazo[1,2-b]pyridazines (23, 32), substituted pyridones (33), and specific flavinoids (34, 35). It is well known that staurosporine analogues and flavinoids inhibit several other kinases, leaving the usefulness of such compounds as Pim-1 inhibitors unclear.
The addition of 4a or 16a to two different prostate cancer cell lines inhibited the ability of Pim kinase to phosphorylate the proapoptotic Bad protein on Ser112. This is consistent with the previous observation that either Pim-1 or Pim-2 is capable of phosphorylating this residue and thus inhibiting apoptotic cell death of growth factor-starved FDCP1 cells (12). Phospho-Bad protein is sequestered by 14-3-3 proteins, which blocks its ability to cause apoptotic cell death. Possibly through this mechanism, Pim promotes survival of chemotherapy-treated prostate cancer (21) and regulates cardiomyocyte survival and T-cell survival (36, 37). We find that this inhibitor chemotype is capable of reversing the prosurvival phenotype induced by Pim overexpression, thus suggesting that these compounds could be useful as chemotherapeutic agent in tumors with enhanced survival secondary to overexpression of this enzyme.
Our data show that addition of Pim inhibitors to several prostate and leukemic cell lines induces a G1 cell cycle block, although, in other cell lines, especially in low-serum conditions, these inhibitors are capable of inducing apoptosis (data not shown). The cell cycle block induced by these agents is consistent with our previous observation that PC3 cells overexpressing Pim express fewer cells in the G1 phase of the cell cycle (9). Recently, Morishita et al. also showed that overexpression of Pim in K562 cells decreased the number of cells in G1 and increased the cells found in S phase. Further, they showed that Pim is capable of phosphorylating p27Kip1, decreasing the cellular levels of this protein and translocating this protein to the cytosol, thus stimulating cell growth (19). In leukemic cells, the benzylidene-thiazoli-dine-2,4-diones inhibit the activity of Cdk2, and in both leukemic and prostate cancer cells, the Pim inhibitors reverse the effects of Pim by transferring at least a portion of the cytoplasmic enzyme to the nucleus.
Both 4a and 16a were shown to be highly selective for Pim-1 based on profiling a total of 60 kinases; however, in this in vitro assay, 16a, but not 4a, showed significant inhibition of the DYRK1a (20). Other kinase inhibitors have shown potent inhibition of all members of the Pim kinase family (Pim-1, Pim-2, and Pim-3) and members of the DYRK family (DYRK1a, DYRK2, and DYRK3; ref. 38). Based on these results, it is expected that the Pim and DYRK kinases would share some structural similarity within their ATP-binding domains because inhibitors common to both Pim and DYRK kinases were shown to be ATP competitive (38). An amino acid alignment of Pim-1, Pim-2, and DYRK1a shows a considerable number of conserved residues, including residues known to facilitate inhibitor interactions in the ATP-binding domain of Pim-1 (Supplementary Fig. S2; ref. 39).8 Interestingly, both 4a and the recently identified Pim-1 inhibitor K00135 share a trifluoromethyl substituent, which was suggested to interact with Arg122 (23). This Arg corresponds to Met or Leu in the DYRKs and may contribute to the observed lack of inhibitory activity of 4a toward DYRK1a.
The phosphatidylinositol/Akt/mTOR pathway is often activated in leukemia and lymphoma downstream of a variety of oncogenes including receptor tyrosine kinases (40). Therefore, several clinical trials have investigated using the mTOR inhibitor rapamycin and second-generation rapamycin analogues for the treatment of hematologic malignancies including acute myelogenous leukemia, chronic myelogenous leukemia, B-cell lymphoma, mantle cell lym-phoma, and multiple myeloma (41). The results of one clinical trial showed that (a) mTOR is activated in 23 acute myelogenous leukemia patient samples and the downstream mTOR target 4E-BP1 is phosphorylated, (b) rapamycin treatment of these samples reduced 4E-BP1 phosphorylation, and (c) rapamycin showed significant clinical response in 4 of 9 patients with acute myelogenous leukemia (42). We have shown that the Pim inhibitors 4a and 16a act synergistically with the mTOR inhibitor rapamycin to reduce the level of phospho-4E-BP1 and inhibit leukemic cell growth in FDCP1 cells and the MV4;11 cell line containing the activating internal tandem duplication mutation of the tyrosine kinase FLT3. Additionally, we have shown that 4a and 16a decrease the level of c-Myc, the transcription and translation of which are known to be rapamycin sensitive. Based on the observations that the Pim kinases have been shown to promote rapa-mycin resistance in hematopoietic cells (26, 37) and are frequently up-regulated along with c-Myc in leukemia and lymphoma (1), the combination of Pim kinase and mTOR inhibitors represents a potential novel treatment option for hematologic malignancies and other tumor types that show reduced sensitivity to rapamycin.
Supplementary Material
Acknowledgments
Grant support: Supported by DOD W8IXWH-08 and 1P30-CA138313.
We thank the Hollings Cancer Center flow cytometry shared resource facility for carrying out fluorescence-activated cell sorting analysis, Dr. Margaret Kelly (Hollings Cancer Center Cell and Molecular Imaging Core) and Drs. Elizabeth Garrett-Mayer and Georgiana Oniescu (Hollings Cancer Center) for help in determining combination index values, Dr. Suiquan Wang for carrying out initial studies, Dr. Danyelle Townsend (Drug Metabolism and Clinical Pharmacology Facility of the Hollings Cancer Center) for helping analyze benzylidene-thiazolidine-2,4-dione serum binding, and Dr. Joyce Slingerland (University of Miami Miller School of Medicine) for the plasmids pEYFP-C1 and pEYFP-p27Kip1.
Footnotes
Supplementary material for this article is available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Amaravadi R, Thompson CB. The survival kinases Akt and Pim as potential pharmacological targets. J Clin Invest. 2005;115:2618–24. doi: 10.1172/JCI26273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cibull TL, Jones TD, Li L, et al. Overexpression of Pim-1 during progression of prostatic adenocarcinoma. J Clin Pathol. 2006;59:285–8. doi: 10.1136/jcp.2005.027672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dhanasekaran SM, Barrette TR, Ghosh D, et al. Delineation of prognostic biomarkers in prostate cancer. Nature. 2001;412:822–6. doi: 10.1038/35090585. [DOI] [PubMed] [Google Scholar]
- 4.Ellwood-Yen K, Graeber TG, Wongvipat J, et al. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell. 2003;4:223–38. doi: 10.1016/s1535-6108(03)00197-1. [DOI] [PubMed] [Google Scholar]
- 5.Hoover D, Friedmann M, Reeves R, Magnuson NS. Recombinant human pim-1 protein exhibits serine/threonine kinase activity. J Biol Chem. 1991;266:14018–23. [PubMed] [Google Scholar]
- 6.Cuypers HT, Selten G, Quint W, et al. Murine leukemia virus-induced T-cell lymphomagenesis: integration of proviruses in a distinct chromosomal region. Cell. 1984;37:141–50. doi: 10.1016/0092-8674(84)90309-x. [DOI] [PubMed] [Google Scholar]
- 7.van Lohuizen M, Verbeek S, Krimpenfort P, et al. Predisposition to lymphomagenesis in pim-1 transgenic mice: cooperation with c-myc and N-myc in murine leukemia virus-induced tumors. Cell. 1989;56:673–82. doi: 10.1016/0092-8674(89)90589-8. [DOI] [PubMed] [Google Scholar]
- 8.Breuer ML, Cuypers HT, Berns A. Evidence for the involvement of pim-2, a new common proviral insertion site, in progression of lymphomas. EMBO J. 1989;8:743–8. doi: 10.1002/j.1460-2075.1989.tb03434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen WW, Chan DC, Donald C, Lilly MB, Kraft AS. Pim family kinases enhance tumor growth of prostate cancer cells. Mol Cancer Res. 2005;3:443–51. doi: 10.1158/1541-7786.MCR-05-0007. [DOI] [PubMed] [Google Scholar]
- 10.Zippo A, De Robertis A, Serafini R, Oliviero S. PIM1-dependent phosphorylation of histone H3 at serine 10 is required for MYC-dependent transcriptional activation and oncogenic transformation. Nat Cell Biol. 2007;9:932–44. doi: 10.1038/ncb1618. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, Wang Z, Li X, Magnuson NS. Pim kinase-dependent inhibition of c-Myc degradation. Oncogene. 2008;27:4809–19. doi: 10.1038/onc.2008.123. [DOI] [PubMed] [Google Scholar]
- 12.Yan B, Zemskova M, Holder S, et al. The PIM-2 kinase phosphorylates BAD on serine 112 and reverses BAD-induced cell death. J Biol Chem. 2003;278:45358–67. doi: 10.1074/jbc.M307933200. [DOI] [PubMed] [Google Scholar]
- 13.Macdonald A, Campbell DG, Toth R, McLauchlan H, Hastie CJ, Arthur JS. Pim kinases phosphorylate multiple sites on Bad and promote 14-3-3 binding and dissociation from Bcl-XL. BMC Cell Biol. 2006;7:1. doi: 10.1186/1471-2121-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aho TL, Sandholm J, Peltola KJ, Mankonen HP, Lilly M, Koskinen PJ. Pim-1 kinase promotes inactivation of the pro-apoptotic Bad protein by phosphorylating it on the Ser112 gatekeeper site. FEBS Lett. 2004;571:43–9. doi: 10.1016/j.febslet.2004.06.050. [DOI] [PubMed] [Google Scholar]
- 15.Lilly M, Kraft A. Enforced expression of the Mr 33,000 Pim-1 kinase enhances factor-independent survival and inhibits apoptosis in murine myeloid cells. Cancer Res. 1997;57:5348–55. [PubMed] [Google Scholar]
- 16.Hammerman PS, Fox CJ, Cinalli RM, et al. Lymphocyte transformation by Pim-2 is dependent on nuclear factor-κB activation. Cancer Res. 2004;64:8341–8. doi: 10.1158/0008-5472.CAN-04-2284. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, Wang Z, Magnuson NS. Pim-1 kinase-dependent phosphorylation of p21Cip1/WAF1 regulates its stability and cellular localization in H1299 cells. Mol Cancer Res. 2007;5:909–22. doi: 10.1158/1541-7786.MCR-06-0388. [DOI] [PubMed] [Google Scholar]
- 18.Bachmann M, Moroy T. The serine/threonine kinase Pim-1. Int J Biochem Cell Biol. 2005;37:726–30. doi: 10.1016/j.biocel.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 19.Morishita D, Katayama R, Sekimizu K, Tsuruo T, Fujita N. Pim kinases promote cell cycle progression by phosphorylating and down-regulating p27Kip1 at the transcriptional and posttranscriptional levels. Cancer Res. 2008;68:5076–85. doi: 10.1158/0008-5472.CAN-08-0634. [DOI] [PubMed] [Google Scholar]
- 20.Xia Z, Knaak C, Ma J, et al. Synthesis and evaluation of novel inhibitors of Pim-1 and Pim-2 protein kinases. J Med Chem. 2009;52:74–86. doi: 10.1021/jm800937p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zemskova M, Sahakian E, Bashkirova S, Lilly M. The PIM1 kinase is a critical component of a survival pathway activated by docetaxel and promotes survival of docetaxel-treated prostate cancer cells. J Biol Chem. 2008;283:20635–44. doi: 10.1074/jbc.M709479200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006;58:621–81. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 23.Pogacic V, Bullock AN, Fedorov O, et al. Structural analysis identifies imidazo[1,2-b]pyridazines as PIM kinase inhibitors with in vitro antileukemic activity. Cancer Res. 2007;67:6916–24. doi: 10.1158/0008-5472.CAN-07-0320. [DOI] [PubMed] [Google Scholar]
- 24.Kim KT, Baird K, Davis S, et al. Constitutive Fms-like tyrosine kinase 3 activation results in specific changes in gene expression in myeloid leukaemic cells. Br J Haematol. 2007;138:603–15. doi: 10.1111/j.1365-2141.2007.06696.x. [DOI] [PubMed] [Google Scholar]
- 25.Adam M, Pogacic V, Bendit M, et al. Targeting PIM kinases impairs survival of hematopoietic cells transformed by kinase inhibitor-sensitive and kinase inhibitor-resistant forms of Fms-like tyrosine kinase 3 and BCR/ABL. Cancer Res. 2006;66:3828–35. doi: 10.1158/0008-5472.CAN-05-2309. [DOI] [PubMed] [Google Scholar]
- 26.Hammerman PS, Fox CJ, Birnbaum MJ, Thompson CB. Pim and Akt oncogenes are independent regulators of hematopoietic cell growth and survival. Blood. 2005;105:4477–83. doi: 10.1182/blood-2004-09-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woods YL, Rena G, Morrice N, et al. The kinase DYRK1A phosphorylates the transcription factor FKHR at Ser329 in vitro, a novel in vivo phosphorylation site. Biochem J. 2001;355:597–607. doi: 10.1042/bj3550597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hara K, Yonezawa K, Kozlowski MT, et al. Regulation of eIF-4E BP1 phosphorylation by mTOR. J Biol Chem. 1997;272:26457–63. doi: 10.1074/jbc.272.42.26457. [DOI] [PubMed] [Google Scholar]
- 29.Bain J, Plater L, Elliott M, et al. The selectivity of protein kinase inhibitors: a further update. Biochem J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bregman H, Meggers E. Ruthenium half-sandwich complexes as protein kinase inhibitors: an N-succinimidyl ester for rapid derivatizations of the cyclopentadienyl moiety. Org Lett. 2006;8:5465–8. doi: 10.1021/ol0620646. [DOI] [PubMed] [Google Scholar]
- 31.Debreczeni JE, Bullock AN, Atilla GE, et al. Ruthenium half-sandwich complexes bound to protein kinase Pim-1. Angew Chem Int Ed Engl. 2006;45:1580–5. doi: 10.1002/anie.200503468. [DOI] [PubMed] [Google Scholar]
- 32.Bullock AN, Debreczeni JE, Fedorov OY, Nelson A, Marsden BD, Knapp S. Structural basis of inhibitor specificity of the human protooncogene proviral insertion site in Moloney murine leukemia virus (PIM-1) kinase. J Med Chem. 2005;48:7604–14. doi: 10.1021/jm0504858. [DOI] [PubMed] [Google Scholar]
- 33.Cheney IW, Yan S, Appleby T, et al. Identification and structure-activity relationships of substituted pyridones as inhibitors of Pim-1 kinase. Bioorg Med Chem Lett. 2007;17:1679–83. doi: 10.1016/j.bmcl.2006.12.086. [DOI] [PubMed] [Google Scholar]
- 34.Holder S, Lilly M, Brown ML. Comparative molecular field analysis of flavonoid inhibitors of the PIM-1 kinase. Bioorg Med Chem. 2007;15:6463–73. doi: 10.1016/j.bmc.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 35.Holder S, Zemskova M, Zhang C, et al. Characterization of a potent and selective small-molecule inhibitor of the PIM1 kinase. Mol Cancer Ther. 2007;6:163–72. doi: 10.1158/1535-7163.MCT-06-0397. [DOI] [PubMed] [Google Scholar]
- 36.Muraski JA, Rota M, Misao Y, et al. Pim-1 regulates cardiomyocyte survival downstream of Akt. Nat Med. 2007;13:1467–75. doi: 10.1038/nm1671. [DOI] [PubMed] [Google Scholar]
- 37.Fox CJ, Hammerman PS, Thompson CB. The Pim kinases control rapamycin-resistant T cell survival and activation. J Exp Med. 2005;201:259–66. doi: 10.1084/jem.20042020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pagano MA, Bain J, Kazimierczuk Z, et al. The selectivity of inhibitors of protein kinase CK2. An update Biochem J. 2008;415:353–65. doi: 10.1042/BJ20080309. [DOI] [PubMed] [Google Scholar]
- 39.Kumar A, Mandiyan V, Suzuki Y, et al. Crystal structures of proto-oncogene kinase Pim1: a target of aberrant somatic hypermutations in diffuse large cell lymphoma. J Mol Biol. 2005;348:183–93. doi: 10.1016/j.jmb.2005.02.039. [DOI] [PubMed] [Google Scholar]
- 40.Matsumura I, Mizuki M, Kanakura Y. Roles for deregulated receptor tyrosine kinases and their downstream signaling molecules in hematologic malignancies. Cancer Sci. 2008;99:479–85. doi: 10.1111/j.1349-7006.2007.00717.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altman JK, Platanias LC. Exploiting the mammalian target of rapamycin pathway in hematologic malignancies. Curr Opin Hematol. 2008;15:88–94. doi: 10.1097/MOH.0b013e3282f3deaa. [DOI] [PubMed] [Google Scholar]
- 42.Recher C, Beyne-Rauzy O, Demur C, et al. Antileukemic activity of rapamycin in acute myeloid leukemia. Blood. 2005;105:2527–34. doi: 10.1182/blood-2004-06-2494. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.