Abstract
Model legumes such as Lotus japonicus have contributed significantly to the understanding of symbiotic nitrogen fixation. This insight is mainly a result of forward genetic screens followed by map-based cloning to identify causal alleles. The L. japonicus ecotype ‘Gifu’ was used as a common parent for inter-accession crosses to produce F2 mapping populations either with other L. japonicus ecotypes, MG-20 and Funakura, or with the related species L. filicaulis. These populations have all been used for genetic studies but segregation distortion, suppression of recombination, low polymorphism levels, and poor viability have also been observed. More recently, the diploid species L. burttii has been identified as a fertile crossing partner of L. japonicus. To assess its qualities in genetic linkage analysis and to enable quantitative trait locus (QTL) mapping for a wider range of traits in Lotus species, we have generated and genotyped a set of 163 Gifu × L. burttii recombinant inbred lines (RILs). By direct comparisons of RIL and F2 population data, we show that L. burttii is a valid alternative to MG-20 as a Gifu mapping partner. In addition, we demonstrate the utility of the Gifu × L. burttii RILs in QTL mapping by identifying an Nfr1-linked QTL for Sinorhizobium fredii nodulation.
Keywords: Lotus japonicus, Lotus burttii, RIL, QTL, Sinorhizobium fredii
1. Introduction
Symbiotic associations between plants and microorganisms have a large impact on ecosystems and plant production worldwide, and two prominent symbionts are phosphorus delivering fungi of the Glomeromycota,1 forming arbuscular mycorrhiza, and nitrogen fixing soil bacteria, rhizobia. Since the most widely used model plant Arabidopsis thaliana is unable to host these symbionts, two model legumes, Lotus japonicus and Medicago truncatula, were introduced to allow the genetic dissection of symbiotic processes.
These model legumes are diploid, inbreeding species with relatively small sequenced genomes,2,3 and they have allowed the isolation of a number of genes needed for mycorrhizal and rhizobial symbiosis.4 Most of these genes were isolated through map-based cloning, but transposon and retrotransposon tagging have also been used.5,6
Selection of mapping parents is important in order to establish F2 populations that allow successful causal gene identification by map-based cloning. Ideally, the number of inter-parental polymorphisms should be high, while the F2 populations should be characterized by low levels of segregation distortion and suppression of recombination. For Lotus, the first genetic maps were produced using amplified fragment length polymorphism and microsatellite markers to genotype F2 offspring. The mapping populations were established using L. japonicus Gifu as the common parent either in crosses with MG-20 and Funakura or in an interspecific cross with L. filicaulis.7–9 The level of polymorphism between L. filicaulis and Gifu is high but the F2 populations suffered from low viability and from severe segregation distortion in large genomic regions.9,10 Whereas Gifu and Funakura were genetically very similar, a relatively high level of polymorphism was found between Gifu and MG-20, and the F2 populations showed good viability combined with little segregation distortion.7 A dense linkage map with several hundred microsatellite markers was therefore developed, and most map-based cloning projects in Lotus have been based on crosses between Gifu and MG-20.2 The major disadvantage of these populations is the large translocation between the top of Gifu chromosome 1 and the bottom of MG-20 chromosome 2,7 which corresponds to a genomic region with complete suppression of recombination. In parallel with these early genetic studies, Gifu-based recombinant inbred lines (RILs) were developed from crosses with MG-20, Funakura and L. filicaulis.8,10,11
To allow map-based cloning in the intervals displaying the suppression of recombination in Gifu × MG-20 populations, L. burttii originating from West Pakistan was introduced as a fourth crossing partner.12,13 Compared with MG-20 and L. filicaulis, L. burttii showed an intermediate level of polymorphism with respect to Gifu. This high marker density combined with good viability in offspring from crosses with Gifu and with the lack of suppression of recombination at the top of chromosome 1 made L. burttii a promising source of genetic material.13 Despite these favourable characteristics, Gifu × L. burttii mapping populations have been sparsely used, probably because they have not been extensively characterized. This issue can be addressed by generating and analysing RILs, since they capture and stabilize a large number of recombination events, which can be exploited for the characterization of genome structure and recombination landscapes, as well as for quantitative trait locus (QTL) mapping.
Here, we describe the development and genotyping of RILs derived from a Gifu × L. burttii F2 population. By analysing RIL data, we show that L. burttii is a valid alternative to MG-20 as a crossing partner to Gifu in mapping schemes, and we demonstrate that the Gifu × L. burttii RILs can be used to guide the selection of mapping parents. To illustrate the use of the RILs in QTL mapping, we investigated the nodulation of L. burttii by Sinorhizobium fredii HH103, which is a fast-growing rhizobial strain that nodulates soybean and a number of other legumes.14,15 We found that L. burttii differs from Gifu and MG-20 in forming nitrogen-fixing nodules with S. fredii HH103 and demonstrated the utility of the RILs by identifying a major Nfr1-linked QTL controlling S. fredii compatibility.
2. Materials and methods
2.1. RIL propagation and genotyping
Inbred lines of L. japonicus Gifu B-12916,17 and L. burttii B-30313 were used for crossing. Gifu ×L. burttii RILs were developed from an F2 population by selfing until F8 and F9 using single-seed descent. A total of 163 lines have reached F8.
Microsatellite markers developed to detect polymorphism between Gifu and MG-202,18 were tested for polymorphism between Gifu and L. burttii (http://www.kazusa.or.jp/lotus/markerdb_index.html). Additional markers were developed for sequenced transformation competent artificial chromosome and bacterial artificial chromosome clones by searching for microsatellite sequences with the help of WebSat.19 Newly developed markers are listed in Supplementary Table S7. The markers were either analysed on 2–3% agarose gels or using an ABI3730 48 capillary sequencer (Applied Biosystems) in conjunction with the proprietary GeneMapper software (version 3.7) using fluorescently labelled primers. One hundred and forty-six lines were scored with 97 microsatellite markers distributed across all six chromosomes (Supplementary Table S1).
2.2. Analysis of RIL data and QTL mapping
The RIL genotyping data presented in Supplementary Tables S1 and S2 were imported into R/qtl20 version 1.21–2 (read.cross), converted to RIL format (convert2riself), and recombination fraction and logarithm of odds (LOD) scores plotted (plot.rf; Fig. 1A and B). For the QTL analysis, the genetic map was estimated (est.map and replace.map) and missing genotype values extrapolated (mqmaugment). QTL scans were then conducted using 1000 permutations to determine significance thresholds (mqmpermutation) and plotted (mqmplot.permutations).
Figure 1.
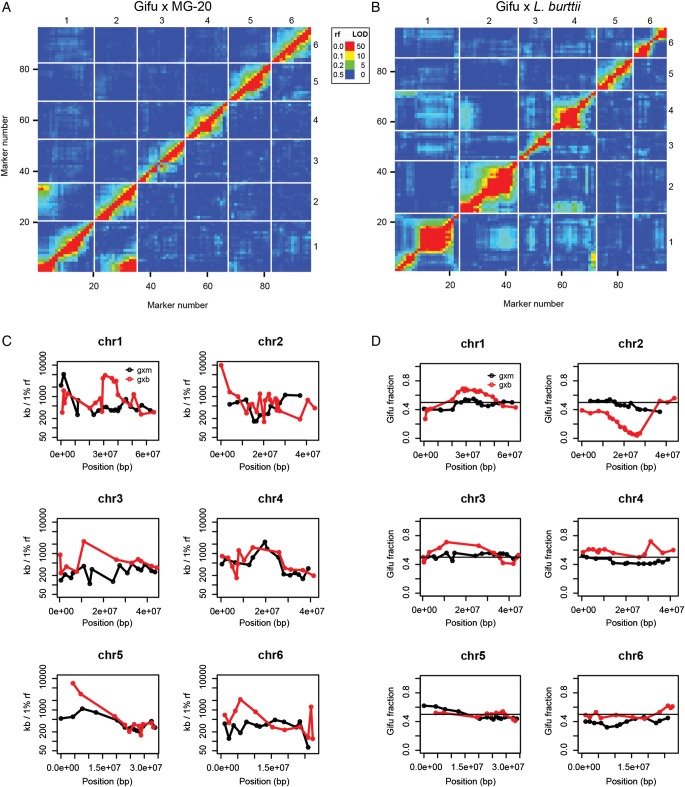
Marker linkage and recombination landscapes for Gifu × MG-20 and Gifu × L. burttii RIL populations. (A and B) Recombination fractions (rf) and LOD scores for all marker pairs. Recombination fractions are in the upper left triangle and the LOD scores are in the lower right triangle. The LOD scores are for a test of r = 0.5. Red corresponds to a large LOD or a small recombination fraction, indicating a strong marker linkage, while blue is the reverse. Missing values appear in light grey. All plotted values are listed in Supplementary Tables S3 and S4. (A) Gifu × MG-20 RILs. (B) Gifu × L. burttii RILs. (C) The physical distance (in kb) corresponding to a recombination fraction of 1% is plotted at each marker position. A high value indicates a low rate of recombination. (D) The fraction of L. japonicus Gifu genotypes at each marker position is plotted. A fraction of 0.5 (black line) is expected in the absence of segregation distortion.
To allow the comparison of physical distances and recombination frequencies, genotyping markers were anchored to the L. japonicus MG-20 genome version 2.5 using BLAST.21 Recombination fractions were calculated using MapMaker322 with error detection on. Physical distances per recombination fraction were then calculated (Supplementary Tables S5 and S6). To avoid inflated values during plotting (Fig. 1C), a minimum recombination fraction of 0.004 was used.
2.3. Nodulation assays
Seeds were germinated and plants grown on 1/4 B&D medium as described.23 Sinorhizobium fredii HH103 was grown in YMB medium for 2 days and then diluted to an OD600 of 0.01–0.02 before inoculating with 400 µl per plant by pipetting the suspension directly onto the root. The nodulation phenotype was scored 6 weeks post-inoculation.
3. Results and discussion
3.1. Generation and genotyping of 163 Gifu × L. burttii RILs
Characterization of populations derived from crosses between Gifu and L. burtti is required to promote the exploitation of the high level of Gifu/L. burttii polymorphism for genetic mapping. For this purpose, and to provide the legume community with a stable genetic resource for QTL and recombination rate analysis, Gifu × L. burttii RILs were generated by selfing. Starting from 200 F2 plants, 163 lines have reached generations 8 or 9. One hundred and forty-six of these lines were genotyped by scoring 97 microsatellite markers (Supplementary Table S1),2 and a Gifu × L. burttii genetic map, syntenic to the Gifu × MG-20 map,2,7 was constructed (Supplementary Table S1). The RILs have been deposited at LegumeBase, Miyazaki, Japan, where they are available for the scientific community (http://www.legumebase.brc.miyazaki-u.ac.jp/). The genotypes of the Gifu × MG-20 RILs can be found at ftp://ftp.kazusa.or.jp/pub/lotus/RIline/ and in Supplementary Table S2.
3.2. Gifu × L. burttii and Gifu × MG-20 mapping crosses yield complementary information
To assess whether Gifu × L. burttii populations would offer genetic resolution in chromosomal regions where the suppression of recombination is prominent in populations derived from crosses between Gifu and MG-20, we compared recombination landscapes based on RIL genotyping data. Analysing the pairwise recombination fractions and LOD scores for all marker combinations in the Gifu × MG-20 RILs resulted in a clear marker linkage signal at the previously described Gifu/MG-20 translocation between chromosomes 1 and 2 (Fig. 1A and Supplementary Table S3).7 This signal was absent in the Gifu × L. burttii RIL data (Fig. 1B and Supplementary Table S4), confirming previous observations.13 However, markers at the top of chromosome 1 and the bottom of chromosome 4 appeared linked in the Gifu × L. burttii population, suggesting an altered L. burttii chromosomal structure in this region (Fig. 1B and Supplementary Table S4).
To investigate if these differences in the genome structure were reflected in differential recombination rates, the genotyping markers were anchored on the MG-20 release 2.5 genome assembly, and physical distance per recombination fraction was calculated and plotted (Fig. 1C and Supplementary Tables S5 and S6).
Both a relatively high inter-marker distance and an incomplete genome assembly contribute to the uncertainty of this analysis, and conclusions should be drawn with care. We did, however, observe a higher recombination rate in the Gifu × L. burttii population than in the Gifu × MG20 population (Fig. 1C) in the genomic interval located at the top of chromosome 1 in the MG-20 reference sequence, as noted previously.13 A higher recombination frequency for the Gifu × L. burttii population was also found in the sym10 region near the top of chromosome 4. On the other hand, this population appeared to suffer from suppression of recombination in the central part of chromosome 1, but not in the chromosome 1 and 4 regions displaying the marker linkage. Overall, it appears that the two populations show comparable levels of recombination and that the recombination frequency varies significantly with both genomic location and population. In addition, the Gifu × L. burttii cross showed a balanced segregation of genotypes, except for the central region of chromosome 2 (Fig. 1D). In conclusion, L. burttii appears to be a valid alternative to MG-20 as a Gifu crossing partner for generating Lotus mapping populations.
3.3. Mapping of the symbiotic mutant sym10 demonstrates the utility of RIL-guided selection of mapping parents
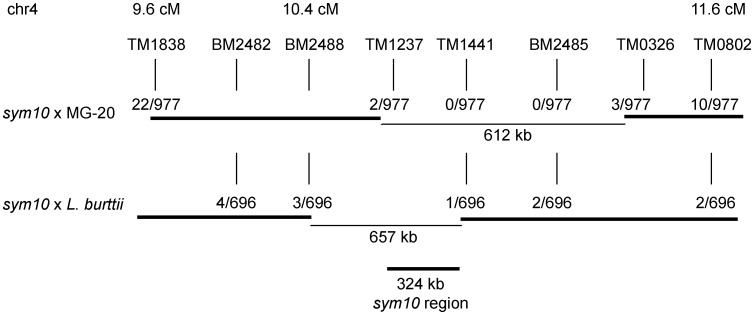
Fine-mapping of the sym10 locus located on chromosome 4 near 10 cM10 has proved challenging using a Gifu × MG-20 population, as no recombination events could be detected between the flanking markers TM1237 and TM0326 in a total of 977 individuals. This limited the mapping resolution to ∼600 kb. The Gifu × L. burttii RILs documented here represent a genetic resource, which can be used to test the applicability of a mapping cross in a given genomic region. For the sym10 region, the high local rate of recombination indicated by the RIL data suggested that a Gifu × L. burttii population could be used to increase genetic mapping resolution (Fig. 1C).
Motivated by this result, we generated and genotyped a sym10 × L. burttii mapping population. Although 977 individuals from the sym10 × MG-20 population allowed us to narrow down the candidate region to ∼600 kb, the 696 individuals from the sym10 × L. burttii population provided additional information, limiting the candidate region to ∼300 kb (Fig. 2).
Figure 2.
sym10 mapping resolution with crosses to L. japonicus MG-20 and L. burttii. The number of informative recombinants is shown in the cross(es) where the marker is polymorphic. The delimited size of the sym10 region is shown for the two crosses individually and with the combined mapping information.
In this case, the high recombination rate in the region of interest suggested by the RIL data was reflected in the F2 mapping population, leading to increased genetic resolution when combining information from the two populations. We propose that taking advantage of the RIL resources to probe recombination frequencies in genomic regions of interest prior to the selection of the mapping partner for F2 fine-mapping populations could allow faster mapping progress in a number of cases.
High-throughput sequencing of bulked mapping populations generated either by interspecies or by back-crosses has proved effective in identifying causal ethyl methanesulfonate (EMS) mutations.24,25 For the sequencing analysis of interspecies crosses, marker density and recombination frequencies remain important parameters, and selecting the optimal mapping population cross is still important. Deep sequencing back-crossing methods also require genetic mapping information, and the induced EMS mutations have been used as markers in place of interspecies polymorphisms.25 A number of interesting symbiotic mutants in Lotus are not generated by EMS mutagenesis but rather by unknown genomic changes induced by tissue culture regeneration,26 and accurate genetic mapping information remains critical for identifying these causal mutations.
3.4. A major QTL near Nfr1 controls S. fredii compatibility
In addition to facilitating the choice of mapping crosses for the identification of causal mutations, the genotyped Gifu × L. burttii RILs should also be well suited for exploiting Lotus natural variation to identify genetic determinants of a wide range of traits through QTL analysis.
Since we had observed a clear difference in nodulation with S. fredii HH103 between L. burttii and Gifu (Fig. 3), we chose this trait as an example for QTL analysis and proceeded to record the S. fredii nodulation phenotype of all RILs. Twenty plants were scored per line, and the trait was quantified as the ‘average number of pink nodules per plant’.
Figure 3.

S. fredii infection phenotypes for L. burttii and Gifu. Six-week-old plate-grown plants inoculated with S. fredii. (A) L. burttii. (B) L. japonicus Gifu. Arrow points to red root nodule.
Analysing the data using R/qtl initially revealed two significant peaks. One was present near the marker TM0002, which is closely linked to the Nod factor receptor Nfr1,27 and another was found at the bottom of chromosome 4 near marker TM0617 (Fig. 4A). Further, a minor peak was seen at the top of chromosome 5. Rerunning the QTL analysis including the markers corresponding to the three highest peaks as cofactors eliminated the peak on chromosome 4 (Fig. 4B), indicating that it was an artefact caused by marker linkage between the top of chromosome 1 and the bottom of chromosome 4 (Fig. 1B). The vast majority of the phenotypic variation, therefore, appeared to be explained by the QTL on chromosome 1 near Nfr1.
Figure 4.
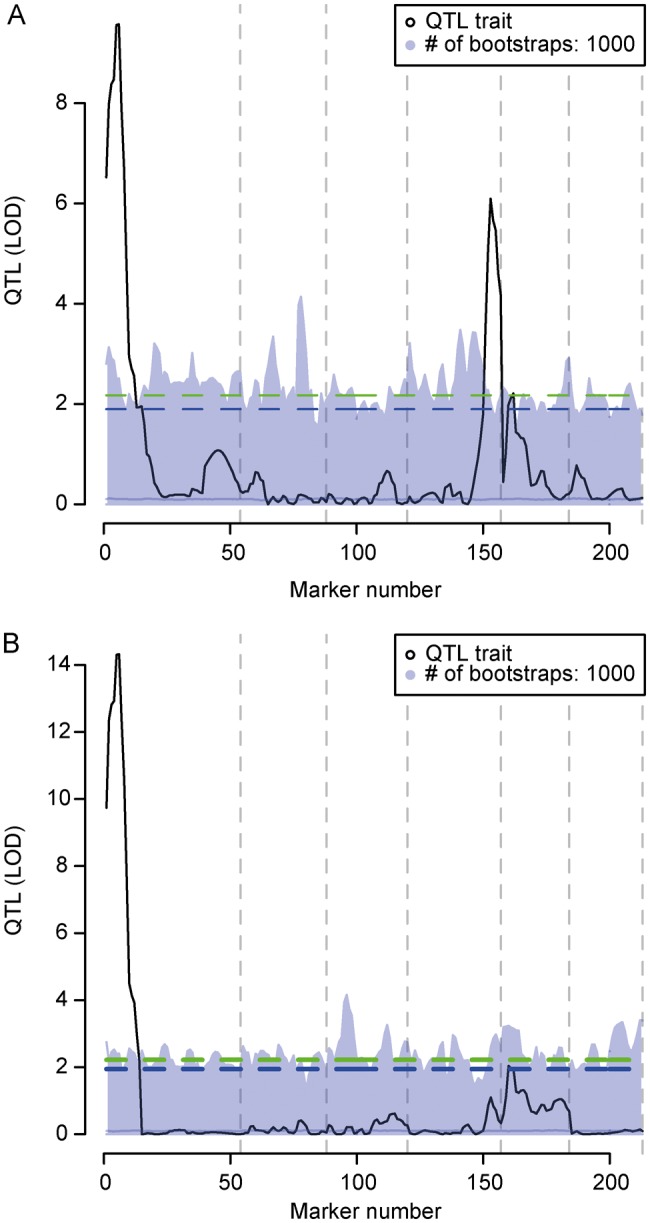
QTL analysis of S. fredii nodulation. The trait analysed is the average number of nodules on Gifu × L. burttii RILs inoculated with S. fredii. (A) QTL analysis without cofactors. (B) QTL analysis including markers TM0002, TM0617, and TM0494, corresponding to the three significant peaks in (A), as cofactors. Five and 10% false discovery rate levels are indicated by the upper and lower dashed lines, respectively. Light grey area delimits the results of 1000 QTL analyses on permuted data, which were used to determine the significance thresholds. Dashed vertical grey lines indicate chromosome ends.
In conclusion, the Gifu × L. burttii RILs have allowed us to identify an Nfr1-linked locus, which supports an unusual rhizobial–Lotus symbiotic interaction. In contrast to the Nfr5-dependent host-range extension shown for L. filicaulis previously,28 the Nfr5 locus does not seem to influence the interaction between L. burttii and S. fredii.
The present set of Gifu × L. burttii RILs expands both the genotypic and the phenotypic space available to Lotus researchers and promises to deliver new insight into the genetic mechanisms governing natural variation in rhizobial–legume interactions. In addition, the RIL population displays variation in flower colour, leaf shape, flowering time, and pod shattering, making it well suited for QTL mapping of these traits.
Supplementary data
Supplementary data are available online at www.dnaresearch.oxfordjournals.org.
Funding
This work was supported by Danish National Research Foundation, project number P07-CVI-02506 of the Andalusia Government and Kazusa DNA Research Institute Foundation.
Supplementary Material
Acknowledgements
We are grateful to F. Pedersen for plant care. Keisuke Yokota, Sabine Zitzenbacher, Katarzyna Szyrajew, Rafal Zgadzaj, and Pawel Mikulski are thanked for their contributions to the sym10 data and Hanne Busk is thanked for help with RIL microsatellite markers.
References
- 1.Kruger M., Kruger C., Walker C., Stockinger H., Schussler A. Phylogenetic reference data for systematics and phylotaxonomy of arbuscular mycorrhizal fungi from phylum to species level. New Phytol. 2012;193:970–84. doi: 10.1111/j.1469-8137.2011.03962.x. [DOI] [PubMed] [Google Scholar]
- 2.Sato S., Nakamura Y., Kaneko T., et al. Genome structure of the legume, Lotus japonicus. DNA Res. 2008;15:227–39. doi: 10.1093/dnares/dsn008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Young N.D., Debelle F., Oldroyd G.E., et al. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature. 2011;480:520–4. doi: 10.1038/nature10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kouchi H., Imaizumi-Anraku H., Hayashi M., et al. How many peas in a pod? Legume genes responsible for mutualistic symbioses underground. Plant Cell Phys. 2010;51:1381–97. doi: 10.1093/pcp/pcq107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schauser L., Roussis A., Stiller J., Stougaard J. A plant regulator controlling development of symbiotic root nodules. Nature. 1999;402:191–5. doi: 10.1038/46058. [DOI] [PubMed] [Google Scholar]
- 6.Yokota K., Fukai E., Madsen L.H., et al. Rearrangement of actin cytoskeleton mediates invasion of Lotus japonicus roots by Mesorhizobium loti. Plant Cell. 2009;21:267–84. doi: 10.1105/tpc.108.063693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayashi M., Miyahara A., Sato S., et al. Construction of a genetic linkage map of the model legume Lotus japonicus using an intraspecific F2 population. DNA Res. 2001;8:301–10. doi: 10.1093/dnares/8.6.301. [DOI] [PubMed] [Google Scholar]
- 8.Jiang Q., Gresshoff P.M. Classical and molecular genetics of the model legume Lotus japonicus. Mol. Plant Microbe Interact. 1997;10:59–68. doi: 10.1094/MPMI.1997.10.1.59. [DOI] [PubMed] [Google Scholar]
- 9.Sandal N., Krusell L., Radutoiu S., et al. A genetic linkage map of the model legume Lotus japonicus and strategies for fast mapping of new loci. Genetics. 2002;161:1673–83. doi: 10.1093/genetics/161.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sandal N., Petersen T.R., Murray J., et al. Genetics of symbiosis in Lotus japonicus: recombinant inbred lines, comparative genetic maps, and map position of 35 symbiotic loci. Mol. Plant Microbe Interact. 2006;19:80–91. doi: 10.1094/MPMI-19-0080. [DOI] [PubMed] [Google Scholar]
- 11.Gondo T., Sato S., Okumura K., Tabata S., Akashi R., Isobe S. Quantitative trait locus analysis of multiple agronomic traits in the model legume Lotus japonicus. Genome. 2007;50:627–37. doi: 10.1139/g07-040. [DOI] [PubMed] [Google Scholar]
- 12.Borsos O.S., Somaroo B.H., Grant W.F. New diploid species of Lotus (Leguminosae) in Pakistan. Can. J. Botany. 1972;50:1865–1870. [Google Scholar]
- 13.Kawaguchi M., Pedrosa-Harand A., Yano K., et al. Lotus burttii takes a position of the third corner in the Lotus molecular genetics triangle. DNA Res. 2005;12:69–77. doi: 10.1093/dnares/12.1.69. [DOI] [PubMed] [Google Scholar]
- 14.Margaret I., Becker A., Blom J., et al. Symbiotic properties and first analyses of the genomic sequence of the fast growing model strain Sinorhizobium fredii HH103 nodulating soybean. J. Biotechnol. 2011;155:11–9. doi: 10.1016/j.jbiotec.2011.03.016. [DOI] [PubMed] [Google Scholar]
- 15.Weidner S., Becker A., Bonilla I., et al. Genome sequence of the soybean symbiont Sinorhizobium fredii HH103. J Bacteriol. 2012;194:1617–18. doi: 10.1128/JB.06729-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Handberg K., Stougaard J. Lotus japonicus, an autogamous, diploid legume species for classical and molecular genetics. Plant J. 1992;2:487–96. [Google Scholar]
- 17.Stougaard J., Beuselinck P. Registration of GIFU B-129-S9 Lotus japonicus germplasm. Crop Sci. 1996;36:476. [Google Scholar]
- 18.Kawaguchi M. Lotus japonicus Miyakojima MG-20: an early-flowering accession suitable for indoor handling. J. Plant Res. 2000;113:507–9. [Google Scholar]
- 19.Martins W.S., Lucas D.C., Neves K.F., Bertioli D.J. WebSat—a web software for microsatellite marker development. Bioinformation. 2009;3:282–3. doi: 10.6026/97320630003282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arends D., Prins P., Jansen R.C., Broman K.W. R/qtl: high-throughput multiple QTL mapping. Bioinformatics. 2010;26:2990–2. doi: 10.1093/bioinformatics/btq565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 22.Lander E.S., Green P., Abrahamson J., et al. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–81. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- 23.Heckmann A.B., Sandal N., Bek A.S., et al. Cytokinin induction of root nodule primordia in Lotus japonicus is regulated by a mechanism operating in the root cortex. Mol. Plant Microbe Interact. 2011;24:1385–95. doi: 10.1094/MPMI-05-11-0142. [DOI] [PubMed] [Google Scholar]
- 24.Schneeberger K., Ossowski S., Lanz C., et al. SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat. Meth. 2009;6:550–1. doi: 10.1038/nmeth0809-550. [DOI] [PubMed] [Google Scholar]
- 25.Zuryn S., Le Gras S., Jamet K., Jarriault S. A strategy for direct mapping and identification of mutations by whole-genome sequencing. Genetics. 2010;186:427–30. doi: 10.1534/genetics.110.119230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schauser L., Handberg K., Sandal N., et al. Symbiotic mutants deficient in nodule establishment identified after T-DNA transformation of Lotus japonicus. Mol. Gen. Genet. 1998;259:414–23. doi: 10.1007/s004380050831. [DOI] [PubMed] [Google Scholar]
- 27.Radutoiu S., Madsen L.H., Madsen E.B., et al. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature. 2003;425:585–92. doi: 10.1038/nature02039. [DOI] [PubMed] [Google Scholar]
- 28.Radutoiu S., Madsen L.H., Madsen E.B., et al. LysM domains mediate lipochitin-oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J. 2007;26:3923–35. doi: 10.1038/sj.emboj.7601826. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.