Abstract
Site-specific genome engineering technologies are increasingly important tools in the post-genomic era, where biotechnological objectives often require organisms with precisely modified genomes. Rare-cutting endonucleases, through their capacity to create a targeted DNA strand break, are one of the most promising of these technologies. However, realizing the full potential of nuclease-induced genome engineering requires a detailed understanding of the variables that influence resolution of nuclease-induced DNA breaks. Here we present a genome engineering reporter system, designated Traffic Light, that supports rapid flow cytometric analysis of repair pathway choice at individual DNA breaks, quantitative tracking of nuclease expression and donor template delivery, and high throughput screens for factors that bias the engineering outcome. We applied the Traffic Light system to evaluate the efficiency and outcome of nuclease-induced genome engineering in human cell lines and identified strategies to facilitate isolation of cells in which a desired engineering outcome has occurred.
Introduction
The explosive accumulation of genomic sequence data is driving demand for technologies to site-specifically engineer genomes1. One promising approach for genome engineering is the use of rare-cutting endonucleases to exploit endogenous DNA repair pathways2-4. However, nuclease-induced DNA breaks may engage any one of several DNA repair pathways which can produce distinct genetic outcomes5,6. Therefore, an important technological goal is to understand how experimental variables influence DNA repair pathway choice, and to develop methods to bias break resolution towards a desired outcome. While several factors influencing repair pathway utilization following a DNA break are known, including cell cycle status7, DNA repair protein expression and post-translational modification8, availability of donor templates6,9, and use of single vs. double strand breaks10, their application to bias outcome in a genome engineering context have not been systematically explored. An important limitation in developing such applications has been the lack of a method to rapidly assess different types of repair outcomes occurring at an individual DNA breakpoint. Although a variety of nuclease-induced double-strand break repair reporters have been developed9,11-17, none afford the ability to directly measure the efficiency and competition between DNA repair pathways that resolve a DNA break.
We constructed a reporter, Traffic Light, that generates a flow cytometric readout of homology-directed repair (HDR) mediated gene targeting and mutagenic non-homologous end joining (mutNHEJ) mediated gene disruption occurring at an individual DNA breakpoint. The reporter was integrated into a system that provides for quantitative single cell tracking of nuclease and donor template delivery, and supports efficient siRNA-mediated manipulation of endogenous DNA repair pathways. We use this system to demonstrate that high donor template concentration can concurrently promote gene targeting rates while suppressing mutNHEJ, that single strand breaks can induce gene targeting without eliciting mutNHEJ, and that limiting the classical NHEJ pathway through DNA-PKcs silencing can increase the efficiency of gene targeting.
Results
Fluorescent Reporter for HDR and mutNHEJ
We designed a construct in which a double strand break is produced at an embedded nuclease cleavage site (in this case, an I-SceI site), and repair of the break generates distinct fluorescent signals upon resolution either through HDR with an exogenous donor template or through mutNHEJ (Fig. 1a,b): in the former case, a functional green fluorescent protein (eGFP) open reading frame is restored by the exogenously provided donor template to signal gene targeting9; in the latter case a frameshift places an mCherry coding sequence in frame to signal gene disruption. By design, the GFP coding sequence contains an alternative reading frame in the +3 translation (Supplementary Fig. 1), and the T2A “dis-linker” enables the downstream mCherry to escape degradation of the out of frame, mis-folded GFP+3 (Supplementary Fig. 2) We have designated the construct as the Traffic Light Reporter (TLR).
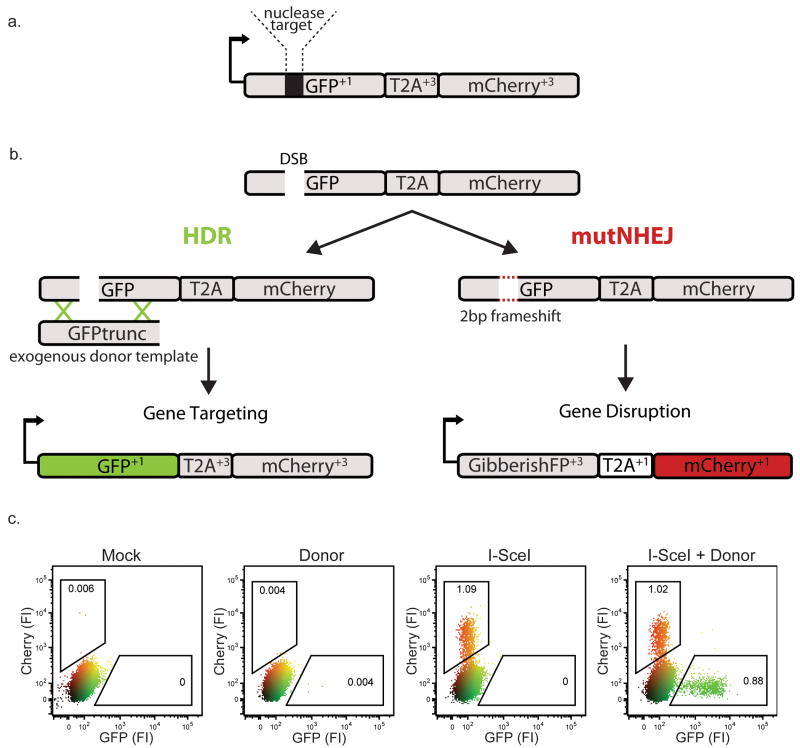
Figure 1. The Traffic light reporter.
(a) Diagram of the TLR. Arrow represents promoter and initial GFP start codon. Reading frames relative to the initial GFP start codon are indicated in superscript. (b) Schematic depicting different engineering outcomes following the induction of a site specific double stranded break (DSB). If the break is resolved through the HDR pathway the full GFP sequence will be reconstituted and cells will fluoresce green; if the break undergoes mutagenic non-homologous end joining (mutNHEJ), GFP will be translated out of frame (GibberishFP+3) and the T2A and mCherry sequences are rendered in frame to produce red fluorescent cells. (c) Flow cytometric analysis of HEK293 TLRsce cells 3 days post-transduction with the indicated lentiviral constructs. “FI” = relative fluorescence intensity.
We derived a polyclonal population of human HEK293 cells containing a single copy of the TLR integrated into the genome (HEK TLRsce), and expressed I-SceI with or without donor template. We analyzed the HEK TLRsce cells by flow cytometry (Fig. 1c). As expected, cells transduced with I-SceI alone produced only mCherry+ cells, indicative of mutNHEJ at the reporter locus. Cells co-transduced with I-SceI and donor template yielded either mCherry+ or eGFP+ positive cells. Sequence analysis confirmed that mCherry+ cells were generated by a variety of mutagenic events at the I-SceI target site that resulted in +3 frameshifts, and that +3 frameshifts represent about 1/3 of all the mutagenic events (Supplementary Fig. 3). We performed similar experiments in several single cell clones (Supplementary Fig. 4), and observed that all clones were capable of break resolution through either HDR or mutNHEJ, although some exhibited relative preferences for a specific pathway, suggesting a potential influence of genomic location or local chromatin environment on double strand break repair6.
Effect of Break Frequency and Donor Availability on TLR Readout
We evaluated the effect of an increasing dose of I-SceI and donor template, both delivered with a single integrating lentivirus (Fig. 2a). At low viral dose, we observed relatively few fluorescent cells, and nearly all were mCherry+, indicating that the repair events occurred via mutNHEJ pathways. As the amount of virus is increased, we observed the expected dose-dependent increase in the total number of repair events, with an increasing fraction of events accounted for by the HDR pathway (Fig. 2b,c).
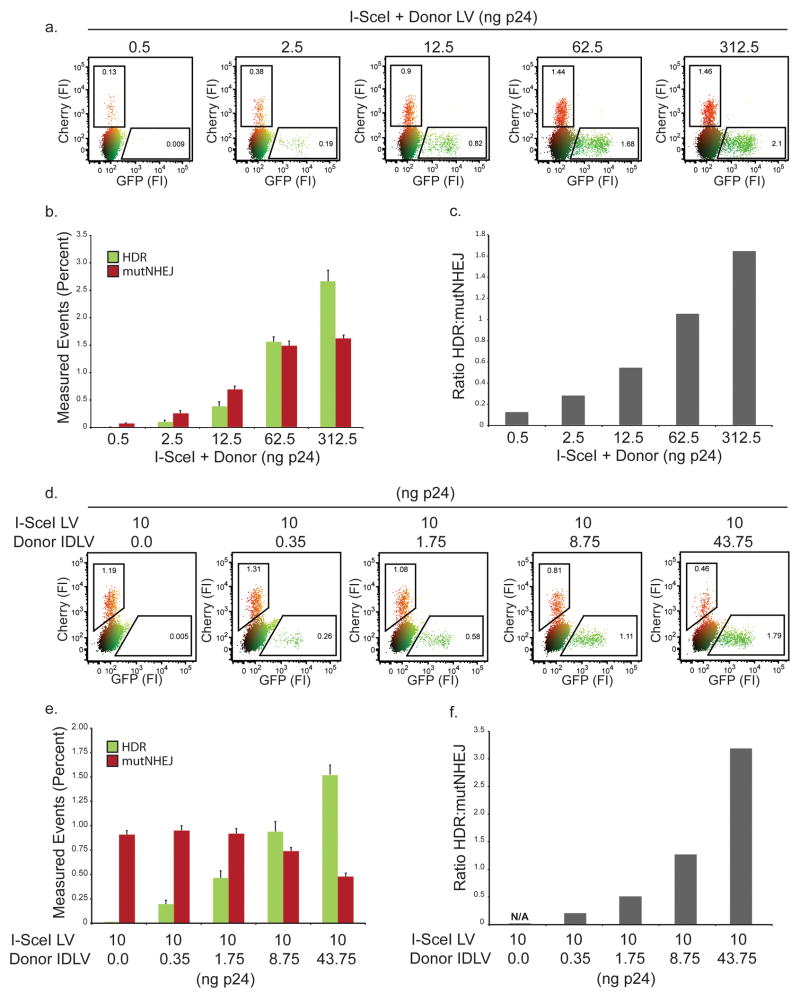
Figure 2. Titration of nuclease and donor template.
(a) Representative flow plot following transduction of HEK293 TLRsce cells with increasing amounts of I-SceI + Donor LV (b) Quantification of data from panel a. Bar graphs represent a minimum of 3 independent experiments performed in duplicate, with standard error shown. (c) Ratio of HDR to mutNHEJ based on data in panel b. (d) Representative flow plot following titration of Donor LV in 5 fold increments while holding I-SceI LV dose constant on HEK293 TLRsce cells. (e) Quantification of data from panel d. Bars represent a minimum of 3 independent experiments performed in duplicate, with standard error shown. (f) Ratio of HDR to mutNHEJ in panel e.
We reasoned that the bias towards HDR at high viral doses could be explained by increasing copies of donor template, as this has been previously shown to increase gene targeting rates at the population level9. To test this hypothesis, we used an integration-deficient lentiviral vector (IDLV18) to provide increasing amounts of episomal donor template, while holding the dose of I-SceI encoding lentivirus constant. As expected, gene targeting events increased in correlation with the amount of template transduced, however, we also observed a concomitant decrease in mutNHEJ events (Fig. 2d-f). The loss of mutNHEJ events with increasing donor template is an important observation, as it suggests that inadequate delivery of a suitable donor can lead to failed homology searches that default to mutNHEJ repair. This is consistent with recent work suggesting that mutagenic alternative NHEJ pathways may share a resection step with HDR19. Therefore, if HDR based genome modification is desired, high-level donor delivery is a key variable for concurrently promoting gene targeting and suppressing undesirable mutNHEJ.
Tracking Endonuclease and Donor Template Delivery
As genome engineering efficiency and outcome are both dependent on nuclease and donor template delivery, we tagged the nuclease and donor template with unique fluorescent markers, an infrared fluorescent protein, IFP1.4, and mTagBFP respectively to allow their simultaneous readout with the TLR (Supplementary Fig. 5). We used viral transduction to deliver I-SceI-T2A-IFP and Donor-T2A-BFP into HEK293 TLRsce cells and evaluated the four fluorescent parameters (eGFP, mCherry, IFP, and BFP) by flow cytometry (Fig. 3). Mock transduced cells were non-fluorescent, whereas cells transduced with the Donor-T2A-BFP IDLV were blue, with no detectable repair events (Fig. 3a). Cells transduced with integrating I-SceI-T2A-IFP lentivirus fluoresced in the IFP channel, and resulted in mCherry positive mutNHEJ events. Upon co-transduction of Sce-T2A-IFP and Donor-T2A-BFP, we observed a spectrum of cells with a variety of mean fluorescent intensities (MFI) that indicated both mutNHEJ and gene targeting repair events. The aggregate mutNHEJ and gene targeting repair event frequency correlated well with experiments using untagged expression and donor template vectors, indicating that the inclusion of the fluorescent markers was not adversely influencing the TLR readout.
Figure 3. Four-color system to track nuclease and donor template delivery simultaneously with the TLR.
(a) Representative flow plot 3 days post transduction of HEK293 TLRsce cells with I-SceI-T2A-IFP LV and Donor-T2A-BFP IDLV. (b) Control gating analysis of HEK293 TLRsce cells co-transduced with I-SceI-T2A-IFP and Donor-T2A-BFP donor template. Inset flow plots show nuclease vs. donor template expression levels. Large plots show readout from the TLR as a function of the gate shown in the inset. (c) Quantification of TLR readout when applying a nuclease titration gating analysis in cells co-transduced with I-SceI-T2A-IFP and Donor-T2A-BFP. Bars represent the amount of gene targeting and mutNHEJ present in the indicated inset gates (lowest to highest MFI) normalized to the HDR and mutNHEJ values for the total ungated population (see methods). Average data of 3 independent experiments are shown with standard error. (d) Quantification of TLR readout when applying donor template titration gating analysis as indicated above.
Since MFI has been shown to correlate with both protein levels and vector copy number20, we hypothesized that cells with unique MFI's for nuclease and donor template following co-transduction would have unique repair profiles. We examined the TLR readout in a population of co-transduced cells as a function of nuclease and donor template levels by gating on cells with unique MFI for each parameter. Control gates agreed with previous experiments, where cells expressing I-SceI alone (IFP+, BFP-) predominately generated mutNHEJ events, and cells with high amounts of both I-SceI and donor template (IFP+, BFP+) exhibited both mutNHEJ and gene targeting signals (Fig. 3b). In a series of gates where donor template delivery holds constant and nuclease expression increases, the absolute number of engineering events is observed to increase, while the ratio of HDR to mutNHEJ remains nearly constant (Fig. 3c). Conversely, in a series of gates where nuclease expression holds constant and donor template delivery increases, the ratio of gene targeting to mutNHEJ changes drastically, with cells harboring high donor template increasingly trending towards gene targeting (Fig.3d). We also observed a shift from mutNHEJ to HDR as a function of donor delivery for a zinc finger nuclease-mediated DNA break (Supplementary Fig. 6).
Together, these results confirm that the efficiency and outcome of genome engineering vary substantially at the population level, with increased nuclease delivery a strong predictor of the likelihood of an engineering “event”, and increased donor delivery a strong predictor of break resolution via gene targeting. They also illustrate how quantitative tracking of nuclease and donor delivery at the single cell level may be applied for isolation of cell populations with high frequencies of desired engineering outcomes.
Single vs. Double Strand Break-Induced Genome Engineering
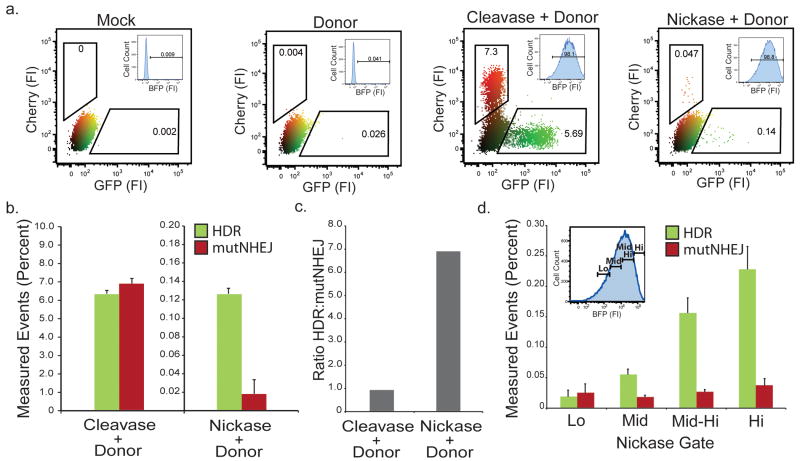
As single-strand breaks are a potential alternative approach to catalyzing site-specific HDR while minimizing mutagenic outcomes10,21,22, “nickases” may be useful for genome engineering when safety or fidelity is necessary. We compared single-strand and double-strand break-induced genome engineering using the TLR system (Fig. 4). We derived a TLR cell line harboring an I-AniI nuclease target site and initiated readout with either a double-strand break inducing nuclease, I-AniI Y223 (cleavase), or a variant that creates a single-strand break at the identical target site, I-AniI Y2 K227M21 (nickase) (Fig. 4a). To control for nuclease delivery, we added a T2A-linked BFP tracking fluorophore to both enzymes. We observed a 100-fold reduction in TLR event activity for the nickase at the population level (Fig. 4b). Importantly, of the repair events detected, the nickase exhibited a strong bias towards gene targeting over mutNHEJ (Fig. 4c). Of note, we also consistently detected a small portion of red fluorescent cells presumed to be mutNHEJ events following nickase expression, suggesting that a small fraction of events are processed through a DSB intermediate. Using the tracking BFP, we conducted a gating analysis to determine the effect of nickase expression levels on TLR readout (Fig. 4d). This revealed a dose dependent increase in gene targeting events, approaching 0.25% in the highest expressing cells, without an associated increase in mutNHEJ.
Figure 4. Effect of single vs. double-strand DNA breaks on engineering outcome.
(a) Representative flow plots showing TLR readout of HEK293 TLRani cells transduced with either the I-AniY2-T2A-BFP LV (cleavase) or I-AniIK227M-T2A-BFP (nickase). Inset plots show gating for nuclease expression to control for transduction levels. (b) Quantification of panel a from 3 independent experiments in duplicate. Percent measured events have had the background rates from cells transduced with donor alone subtracted out to control for the low numbers. (c) Comparison of the ratio of HDR to mutNHEJ between cleavase and nickase induced engineering. (d) Gating analysis showing TLR readout across nickase expression levels. Inset flow plot shows BFP histogram gated for relative nickase expression.
These results strongly support the concept that substituting a nickase in place of a cleavase is an effective means to induce HDR while minimizing mutNHEJ events. They also highlight a key utility of the TLR system: by providing concurrent quantitation of gene targeting and mutNHEJ events, we were able to rapidly analyze rare repair events occurring at a nuclease induced DNA single strand break.
siRNA Kinome Screen for Genome Engineering Enhancers
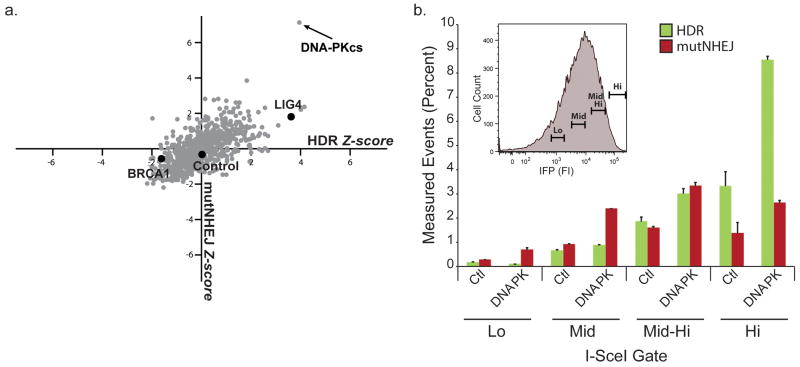
In many contexts, efficiency of genomic modification is likely to be the highest priority of a genome engineer. Therefore, we evaluated the TLR system as a high-throughput screening platform for DNA repair modulators that might improve engineering efficiency. As the DNA damage response is highly regulated by phosphorylation24, we evaluated a human kinome siRNA library as an initial screen. As seen in the Z score scatter plot (Fig. 5a, Supplementary Fig. 7), we observed a variety of HDR and mutNHEJ phenotypes. Several kinases known to play a role in DNA repair appeared at or near the top of ranked lists (Supplementary Table 1). These included DNA-PKcs, TLK-1, and several kinases in the higher order inositol phosphate pathway (IHPK3, IMPK, and PIK3C2B), which have recently been shown to play a role in classical NHEJ25,26. In addition, several kinases not previously implicated in DNA repair also scored positively.
Figure 5. High-throughput siRNA kinome screen to identify modifiers of engineering outcome.
(a) Scatter plot of gene targeting and mutNHEJ Z scores obtained from the siRNA screen. Library in gray, Control siRNAs in black. Control siRNA values are average of at least 3 independent transfections. (b) Gating analysis comparing TLR readout from control and DNA-PKcs siRNA treatment as a function of nuclease expression 72hrs after transduction of HEK293 TLRsce I-SceI-T2A-IFP LV. Inset histogram shows nuclease expression gates. Data are derived from 3 independent experiments performed in duplicate.
As we had observed suppression of mutNHEJ in cells with abundant donor template (Fig. 2e, Fig. 3c), we further hypothesized that coupling knockdown of a repair modulator with gating for cells with high enzyme and donor levels would allow for high rates of gene targeting while minimizing the associated mutNHEJ. To test this, we conducted a gating analysis across increasing transduction levels of HEK293 TLRsce cells pre-treated with DNA-PKcs siRNAs and co-transduced with I-SceI-T2A-IFP and donor virus (Fig. 5b). As transduction levels increased, we observed a 10-fold increase in gene targeting events with only a modest associated increase in mutNHEJ events, achieving absolute gene targeting rates of nearly 10% in highly transduced, DNA-PKcs knockdown cells, compared to < 1% in control siRNA treated cells.
While we consistently observed increases in gene targeting following knockdown of DNA-PKcs in multiple TLR clones (data not shown) and several different reporter systems using the I-SceI nuclease, the magnitude of the effect varied with the enzyme used to initiate the break (Supplementary Fig. 8).
Discussion
Modifying genomes through the induction of a site-specific DNA strand break is complicated by the existence of competing DNA repair pathways that resolve breaks to yield disparate engineering outcomes. In order to identify experimental variables that affect the processing of nuclease-induced breaks, we developed the Traffic Light reporter system. This system reports repair outcomes from nuclease-induced breaks at a single target in single cells and provides quantitative single cell tracking of nuclease and donor template delivery.
The TLR system possesses two key advantages over previously described systems (e.g.9,11-17): 1) It provides a rapid positive signal readout of both mutNHEJ and gene targeting occurring at a single break site, allowing direct assessment of the competition between repair mechanisms, and 2) It is combined with the capacity to track and control for both nuclease and donor template delivery. Previously described reporters have all focused on either NHEJ or HDR. Assessing repair profiles in cell lines with multiple reporters is complicated by epigenetic effects at different reporter loci, a requirement for multiple breakpoints, and a lack of direct competition for resolution27. Although it would be possible to obtain information on competition between repair processes at a single DNA break site by using one of the previously described HDR reporters, doing so would require high throughput DNA sequencing of PCR amplified loci to obtain statistically meaningful information, an approach incompatible with even medium throughput screening for genome engineering outcome modulators.
Our siRNA kinome screen for modulators of genome engineering outcome identified several genes whose silencing resulted in enhanced rates of both mutNHEJ and gene targeting. Prominent among these was DNA-PKcs, a component of the classical NHEJ complex that has previously been shown to improve homologous recombination rates28,29. This observation is consistent with a growing literature indicating that nuclease-induced breaks are often subject to precise rejoining of ends19,15, preventing them from engaging “engineering productive” pathways. While the effect of DNA-PKcs knockdown was consistently observed in several DNA repair contexts, it varied among a panel of nucleases with varying affinities and activities toward the same target site, suggesting that the biochemical and biophysical properties of the nuclease may influence processing of the break by the host DNA repair machinery. This is an observation with important implications for nuclease induced genome engineering, as certain enzymes may have a propensity to engage particular DNA repair pathways, or be more or less responsive to repair pathway manipulations. It also raises important considerations for investigators studying DNA repair using nuclease induced breaks, who have largely focused on breaks induced by a single enzyme, I-SceI.
In conclusion, the TLR is a flexible and powerful tool for analyzing nuclease-induced genome engineering. It should be applicable in diverse organisms for identifying new proteins involved in DNA repair or pathway choice, evaluating new approaches to inducing targeted breaks, screening for small molecule modulators of specific repair pathways, and rapidly and comprehensively vetting “third party” manipulations aimed at increasing endonuclease-induced engineering efficiency.
Methods
Construct Assembly
All constructs were cloned into the “RRL”(addgene #12252) or “CVL” (unpublished) lentiviral backbones using standard molecular biology techniques. A vector key is supplied (Supplementary Fig. 9). Plasmids and maps have been deposited in Addgene (www.addgene.org). The mCherry sequence was mutated by standard site directed mutagenesis to remove internal start codons that contributed to background fluorescence M9S, M16L) (primers in Supplementary Table 2).
Lentivirus Generation
Lentivirus was produced by transient co-transfection of HEK293T cells in 10cm dishes in 10mls media using PEI transfection reagent (Polysciences, Inc.) with 6 ug RRL or CVL backbone plamids, 1.5 ug pMD2G envelope plasmid (VSV-G), and 3 ug psPAX2 for integrating lentivirus (LV) and psPAX2 D64V for integration deficient lentivirus (IDLV), per plate. Viral supernatants were concentrated 100× by overnight centrifugation at 8,000 g, followed by aspiration of supernatant, and resuspension in 1/100th the original volume. 100× stocks were aliquoted and stored at -80 C. Virus was titered using Lenti-x p24 rapid titer ELISA kit (Clontech) according to manufacturer's protocol. MOI was calculated for fluorescent tagged virus experiments by dividing the amount of infectious units added by the number of cells plated. Infectious units were calculated by transducing 0.2 × 10ˆ6 HEK293 cells with increasing amounts of viral stocks and analyzing them on a flow cytometer 72 hrs post transduction and applying the following formula for volumes of virus that yielded between 5-20% fluorescent positive cells ((0.2×10ˆ6 × percent fluorescent positive cells)/100)/ volume viral stock added).
Cell Line Generation
Cell lines harboring the traffic light reporter were generated by transducing 0.2 × 10ˆ6 HEK293 cells with 1uL of un-concentrated reporter lentivirus, typically yielding ∼5% transduction based on fluorescent lentivirus prepared in parallel. 3 days post-transduction, cells with integrated reporters were selected for in 1 ug/ml puromycin for 5 days. Puro resistant cells were then sorted to remove the ∼0.1% of cells exhibiting mCherry fluorescence, thought to be due to integration errors.
Transduction
0.1 × 10ˆ6 HEK293 cells were seeded in a 24 well plate 24 hrs prior to transduction. Cells were transduced with indicated amounts of lentivirus and 4ug polybrene. 24 hrs post transduction, media was changed, cells were split, and analyzed 72 hrs post transduction. For Figures 3-4, we used 25 ng of p24 (HIV core protein) for all transductions – for integrating viral vector preparation this corresponded to an MOI of 8 for the four color experiments in Figure 3, and an MOI of 13, for the experiments in Figures 4 and 5.
Flow Cytometry
Cells were harvested 72 hrs post transduction and analyzed on a BD LSRII or BD Facs ARIA. GFP was measured using a 488 nm laser for excitation and a 530/30 filter. mCherry was excited using a 561 nm laser and acquired with a 610/20 filter. mTagBFP was excited on a 405 nm laser with a 450/50 filter, and IFP1.4 was measured with a 640 nm laser and acquired with a 710/50 filter. Biliverdin was not used for detecting IFP1.4, as it was determined unnecessary for detection at high transduction levels (data not shown). Data was analyzed using FloJo software. For sorting experiments, cells were sorted on BD Facs ARIA.
High-throughput siRNA Kinome Screen
RNA inhibition was carried out using a library against the human kinome (Sigma) containing 3 pooled siRNA's per gene in 384 well format containing positional constant controls on every plate. Using predetermined optimized conditions (over 80% transfection efficiency), 6 replicate transfections per gene were performed. 0.32 ul 100× lentivirus containing both I-SceI and donor template was aliquoted to each well 24hours post siRNA tranfection. 72 hrs post infection the cells were lifted via the addition of 10 mM EDTA and the 6 replicates were pooled into a single well for flow cytometry analysis. All automation was performed using a CyBio Vario and a Thermo Fisher Wellmate. Cytometry data was analyzed on FloJo software. Wells with less than 2,000 total cell counts were excluded from the analysis. HDR and mutNHEJ gates were initially applied to all samples universally, then inspected manually and shifted as necessary. Annotated Data from flow cytometry analysis underwent Z score analysis for each metric obtained and was used to generate ranks. Screening hits beyond these thresholds were followed up on via the determinants of novel biological interest and validation of previously reported trends. Hit validation was initially performed with 3 pooled siRNAs for each target from Qiagen in triplicate.
siRNA Knockdown
For knockdown experiments, 0.05 × 10ˆ6 HEK293 TLR cells were plated 24 hrs prior to transfection in a 24 well plate with 0.5 mls of media. A total of 10nM of siRNA pool was used per well, and transfected using RNAi-max reagent (Invitrogen) according to the manufacturers protocol. 24 hrs post transfection, media was changed, and cells were transduced as indicated above and analyzed 72 hrs post transduction. A minimum of two experimentally validated siRNA's (shown by Qiagen to knockdown greater than 70% mRNA) per indicated target were obtained from Qiagen and pooled. Control siRNA was a “universal” control from Qiagen.
Statistical Analysis
Error bars on bar graphs represent the standard error, as calculated by dividing the standard deviation of replicates by the square root of the number of total replicates. For normalized data, the percent measured value for HDR and mutNHEJ following flow cytometric analysis of a given treatment was divided by the percent measured value of HDR and mutNHEJ for the indicated parent population (control siRNA or ungated population). Z score was calculated by subtracting the average of a metric (HDR or mutNHEJ) from the raw score of a given siRNA, and dividing by the standard deviation of the same metric. Metric averages and standard deviations were calculated from the library values where more than 2,000 cells were acquired during the screen.
Supplementary Material
Acknowledgments
M.T.C. was supported in part by Public Health Service, National Research Service Award, T32 GM07270, from the National Institute of General Medical Sciences. Additional funding from NIH RL1CA133832, UL1DE019582, R01-HL075453, PL1-HL092557, RL1-HL092553, and Seattle Children's Center for Immunity and Immunotherapies. We would like to thank Cherie Ramirez and Keith Jyoung for Zinc Finger Nucleases (Harvard University, Massachusetts General Hospital) and all members of the Northwest Genome Engineering Consortium (NGEC) (http://ngec-seattle.org) for their many insightful discussions.
Footnotes
Competing Financial Interests: Authors declare no competing financial interests.
Contributions: M.T.C. designed and performed experiments, analyzed data, and wrote the paper; B.Y.R., J.E.A. and M.G. performed experiments; J.V.J. and D.J.R. designed experiments; and A.M.S. designed experiments and wrote the paper.
References
- 1.Carr PA, Church GM. Genome engineering. Nat Biotechnol. 2009;27:1151–1162. doi: 10.1038/nbt.1590. [DOI] [PubMed] [Google Scholar]
- 2.Pâques F, Duchateau P. Meganucleases and DNA double-strand break-induced recombination: perspectives for gene therapy. Curr Gene Ther. 2007;7:49–66. doi: 10.2174/156652307779940216. [DOI] [PubMed] [Google Scholar]
- 3.Durai S, et al. Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucl Acids Res. 2005;33:5978–5990. doi: 10.1093/nar/gki912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Porteus MH, Carroll D. Gene targeting using zinc finger nucleases. Nat Biotech. 2005;23:967–973. doi: 10.1038/nbt1125. [DOI] [PubMed] [Google Scholar]
- 5.Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet. 2008;9:619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- 6.Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- 7.Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nat Rev Mol Cell Biol. 2008;9:297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- 8.Cann KL, Hicks GG. Regulation of the cellular DNA double-strand break response. Biochem Cell Biol. 2007;85:663–674. doi: 10.1139/O07-135. [DOI] [PubMed] [Google Scholar]
- 9.Porteus MH, Baltimore D. Chimeric Nucleases Stimulate Gene Targeting in Human Cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- 10.Metzger MJ, McConnell-Smith A, Stoddard BL, Miller AD. Single-strand nicks induce homologous recombination with less toxicity than double-strand breaks using an AAV vector template. Nucleic Acids Research. 2011;39:926–935. doi: 10.1093/nar/gkq826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4:e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stark JM, Pierce AJ, Oh J, Pastink A, Jasin M. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol Cell Biol. 2004;24:9305–9316. doi: 10.1128/MCB.24.21.9305-9316.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagaraju G, Hartlerode A, Kwok A, Chandramouly G, Scully R. XRCC2 and XRCC3 regulate the balance between short- and long-tract gene conversions between sister chromatids. Mol Cell Biol. 2009;29:4283–4294. doi: 10.1128/MCB.01406-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brenneman MA, Wagener BM, Miller CA, Allen C, Nickoloff JA. XRCC3 controls the fidelity of homologous recombination: roles for XRCC3 in late stages of recombination. Mol Cell. 2002;10:387–395. doi: 10.1016/s1097-2765(02)00595-6. [DOI] [PubMed] [Google Scholar]
- 15.Guirouilh-Barbat J, Rass E, Plo I, Bertrand P, Lopez BS. Defects in XRCC4 and KU80 differentially affect the joining of distal nonhomologous ends. Proc Natl Acad Sci USA. 2007;104:20902–20907. doi: 10.1073/pnas.0708541104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aubert M, et al. Successful Targeting and Disruption of an Integrated Reporter Lentivirus Using the Engineered Homing Endonuclease Y2 I-AniI. PLoS ONE. 2011;6:e16825. doi: 10.1371/journal.pone.0016825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarkis C, Philippe S, Mallet J, Serguera C. Non-integrating lentiviral vectors. Curr Gene Ther. 2008;8:430–437. doi: 10.2174/156652308786848012. [DOI] [PubMed] [Google Scholar]
- 19.Bennardo N, Gunn A, Cheng A, Hasty P, Stark JM. Limiting the persistence of a chromosome break diminishes its mutagenic potential. PLoS Genet. 2009;5:e1000683. doi: 10.1371/journal.pgen.1000683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kustikova OS, et al. Dose finding with retroviral vectors: correlation of retroviral vector copy numbers in single cells with gene transfer efficiency in a cell population. Blood. 2003;102:3934–3937. doi: 10.1182/blood-2003-05-1424. [DOI] [PubMed] [Google Scholar]
- 21.McConnell Smith A, et al. Generation of a nicking enzyme that stimulates site-specific gene conversion from the I-AniI LAGLIDADG homing endonuclease. Proceedings of the National Academy of Sciences. 2009;106:5099–5104. doi: 10.1073/pnas.0810588106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee GS, Neiditch MB, Salus SS, Roth DB. RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell. 2004;117:171–184. doi: 10.1016/s0092-8674(04)00301-0. [DOI] [PubMed] [Google Scholar]
- 23.Takeuchi R, Certo M, Caprara MG, Scharenberg AM, Stoddard BL. Optimization of in vivo activity of a bifunctional homing endonuclease and maturase reverses evolutionary degradation. Nucleic Acids Res. 2009;37:877–890. doi: 10.1093/nar/gkn1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuoka S, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 25.Ma Y, Lieber MR. Binding of Inositol Hexakisphosphate (IP6) to Ku but Not to DNA-PKcs. Journal of Biological Chemistry. 2002;277:10756–10759. doi: 10.1074/jbc.C200030200. [DOI] [PubMed] [Google Scholar]
- 26.Kumar A, Fernandez-Capetillo O, Carrera AC. Nuclear phosphoinositide 3-kinase β controls double-strand break DNA repair. Proceedings of the National Academy of Sciences. 2010;107:7491–7496. doi: 10.1073/pnas.0914242107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guirouilh-Barbat J, et al. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol Cell. 2004;14:611–623. doi: 10.1016/j.molcel.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 28.Słabicki M, et al. A Genome-Scale DNA Repair RNAi Screen Identifies SPG48 as a Novel Gene Associated with Hereditary Spastic Paraplegia. PLoS Biol. 2010;8:e1000408. doi: 10.1371/journal.pbio.1000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pierce AJ, Hu P, Han M, Ellis N, Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 2001;15:3237–3242. doi: 10.1101/gad.946401. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.