Abstract
Background
It has been hypothesised that seizure induced neuronal loss and axonal damage in medial temporal lobe epilepsy (MTLE) may lead to the development of aberrant connections between limbic structures and eventually result in the reorganisation of the limbic network. In this study, limbic structural connectivity in patients with MTLE was investigated, using diffusion tensor MRI, probabilistic tractography and graph theory based network analysis.
Methods
12 patients with unilateral MTLE and hippocampal sclerosis (five left and seven right MTLE) and 26 healthy controls were studied. The connectivity of 10 bilateral limbic regions of interest was mapped with probabilistic tractography, and the probabilistic fibre density between each pair of regions was used as the measure of their weighted structural connectivity. Binary connectivity matrices were then obtained from the weighted connectivity matrix using a range of fixed density thresholds. Graph theory based properties of nodes (degree, local efficiency, clustering coefficient and betweenness centrality) and the network (global efficiency and average clustering coefficient) were calculated from the weight and binary connectivity matrices of each subject and compared between patients and controls.
Results
MTLE was associated with a regional reduction in fibre density compared with controls. Paradoxically, patients exhibited (1) increased limbic network clustering and (2) increased nodal efficiency, degree and clustering coefficient in the ipsilateral insula, superior temporal region and thalamus. There was also a significant reduction in clustering coefficient and efficiency of the ipsilateral hippocampus, accompanied by increased nodal degree.
Conclusions
These results suggest that MTLE is associated with reorganisation of the limbic system. These results corroborate the concept of MTLE as a network disease, and may contribute to the understanding of network excitability dynamics in epilepsy and MTLE.
Introduction
Hippocampal sclerosis is the most common pathological finding in patients with medial temporal lobe epilepsy (MTLE).1 Nonetheless, in vivo imaging studies suggest that hippocampal damage does not represent an isolated injury in MTLE. Studies employing quantitative measures obtained from MRI have consistently demonstrated extra-hippocampal and extra-temporal limbic atrophy in patients with medication refractory MTLE.2–5
While routine clinical MRI can be used to visually identify signs that are associated with hippocampal sclerosis,6 the use of quantitative techniques can improve the sensitivity of MRI in detecting subtle abnormalities.7 The initial studies employing quantitative measures of structural integrity focused on hippocampal and medial temporal lobe abnormalities2 6 8 but improvements in methodology later enabled objective evaluation of the whole brain. In particular, the advent of voxel based morphometry9 and cortical thickness measurements10 facilitated a more comprehensive assessment of MTLE related brain damage. Whole brain MRI studies utilising voxel based morphometry or cortical thickness measurements repeatedly demonstrated that extra-hippocampal structures and extra-temporal limbic structures are significantly atrophied in patients with MTLE.2–5 11 12
The discovery of damage extending beyond the hippocampus in patients with MTLE also highlighted that regional brain damage in MTLE is more likely to involve structures within the limbic system, particularly regions that are anatomically and functionally related to the hippocampus.3 13 This observation led to the hypothesis that MTLE is a disease affecting not only the hippocampus but also involving a network of interrelated structures, such as the entorhinal and perirhinal cortices,2 8 14 thalamus,15 16 anterior cingulate11 and cortical association areas.5
The concept of MTLE as a ‘network disease’ has been further supported by studies employing diffusion tensor MRI to study white matter pathways. These studies have consistently demonstrated that limbic networks are significantly disrupted in MTLE. Patients with MTLE display traditional findings of neuronal fibre loss—for example, increased mean diffusivity and reduced fractional anisotropy—in limbic white matter pathways.13 17–19 Furthermore, reduced connectivity demonstrated by tractography is associated with reduced regional grey matter volumes.13 20
While these findings suggest that structural damage can occur beyond the hippocampus in patients with MTLE, they do not confirm whether MTLE is inherently associated with an abnormal configuration of limbic networks. It has been hypothesised that seizure induced neuronal loss and axonal damage may lead to the development of aberrant connections between limbic structures and eventually result in the reorganisation of the limbic network.14 Specifically, this progressive restructuring could lead to a higher tendency for clustering of connections between adjacent structures, also self-reinforcing excitation, thereby facilitating epileptogenicity and recurrent seizures. Consequently, the ability to non-invasively identify network reorganisation patterns could lead to better disease quantification, prognosis assessment and therapy monitoring.
In this study, we aimed to evaluate whether and how limbic networks are configured differently in patients with MTLE compared with healthy controls. Specifically, we aimed to test whether white matter pathways in MTLE are associated with structural configurations that indicate a tendency towards higher clustering. We tested this hypothesis utilising graph theoretical analysis of structural network configuration, as defined by diffusion tensor imaging (DTI).
Methods
Subjects
We studied 12 consecutive patients who were diagnosed with MTLE according to the parameters defined by the International League Against Epilepsy.21 All patients underwent a careful neurological assessment, including neurological examination and video electroencephalography monitoring. All patients had unilateral hippocampal atrophy based on diagnostic high resolution MRI, and the side of hippocampal atrophy was concordant with the side of seizure onset. Seven patients had right MTLE and five had left MTLE. The mean age of patients was 37.5±9.8 years, and eight patients were women.
We also studied a control group of 26 healthy individuals without any significant past medical, neurological or psychiatric history. The mean age of controls was 34.3±8.8 years. Sixteen controls were women and the control group was similar to the patient group in age (t(20)=0.96, p=0.34) and gender distribution (Yates' χ2=0.003, p=0.76).
The institutional review board committee of the Medical University of South Carolina approved this study. All subjects signed an informed consent to participate in this study.
Imaging
DTI scans were acquired on a 3 T Philips scanner equipped with an eight channel head coil. The scans were obtained with the routine clinical sequence and consisted of a b=0 image and 15 diffusion weighted images with a b value of 1000 s/mm2. Other scans parameters were TR=8933 ms, TE=82 ms, FOV=224×224 mm2 and voxel size=2×2×3 mm3.
DTI preprocessing
DICOM images were converted into the NIfTI format and diffusion gradient directions were extracted with the software dcm2nii (http://www.mccauslandcenter.sc.edu/mricro/mricron/dcm2nii.html). We then employed FMRIB Software Library (FSL)'s Diffusion Toolkit (FDT) (http://www.fmrib.ox.ac.uk/fsl) for the preprocessing of diffusion images and to estimate the diffusion tensor. The diffusion weighted images underwent eddy current correction through affine transformation of each diffusion weighted image to the base b=0 T2 weighted image using FDT's eddy current correction. Variations in acquisition geometry were corrected and gradients were updated using the software dcm2nii. FDT's dtifit was then used to perform voxel-wise calculation of the diffusion tensor within the brain, where the brain mask was obtained with FSL's Brain Extraction Tool (BET). Voxel-wise calculation of the magnitude (MD) and the directionality (FA) of molecular displacement was calculated with FSL's (FMRIB's Software Library, http://www.fmrib.ox.ac.uk/fsl) FDT.
Probabilistic tractography
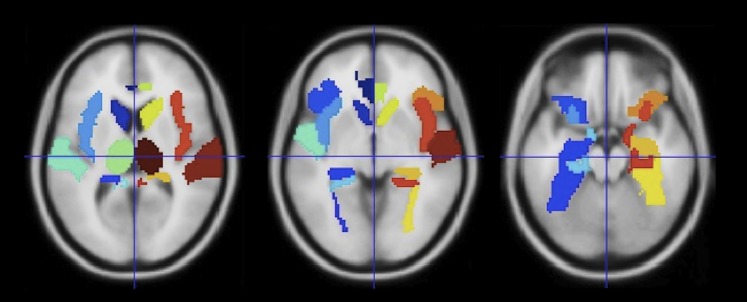
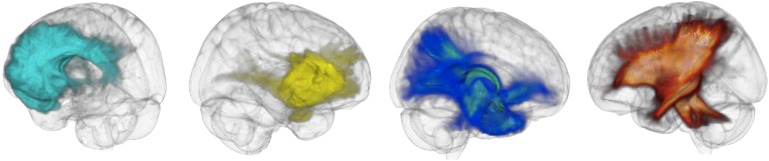
Probabilistic tractography was performed on DTI data after voxel-wise calculation of the diffusion tensor. In this study, we opted to employ probabilistic tractography to define the strength of connections between regions as probabilistic tractography is capable of, to some extent, resolving intra-voxel fibre crossings,22 23 and one of the limitations of the current study is the limited number of direction in the DTI sequence (ie, 15 directions). FDT's BEDPOST was used to build up distributions of diffusion parameters at each voxel. Probabilistic tractography was obtained using FDT's Probtrackx. Seed regions for tractography were obtained from the Anatomical Automatic Labelling template24 (http://www.cyceron.fr/freeware/), corresponding to 10 limbic and perilimbic structures in each hemisphere (figure 1, table 1) for a total of 20 regions. These seed regions were chosen because they represent the locations where structural changes have been consistently observed in patients with MTLE.11 The seed masks in standard space were transformed into each subject's native space using an affine transformation obtained with FSL's FLIRT. Figure 2 and supplementary figure 1 (supplementary figure 1 is available online only) demonstrate the probabilistic tractography pathways from these structures.
Figure 1.
Locations of the regions of interest employed in this study overlaid onto a T1 weighted template in standard space.
Table 1.
Names, coordinates of centre of mass and volumes of the regions of interest employed in this study. The numbering of the regions of interest is consistent with the numbering used in figure 3
| Side | No | Structure | X | Y | Z | Volume |
| Ipsilateral | 1 | Anterior cingulate | −4.8 | 38.4 | −1.53 | 2832 |
| 2 | Caudate | −11.5 | 11.9 | 8.78 | 7392 | |
| 3 | Fusiform | −32.2 | −41 | −17.5 | 13 704 | |
| 4 | Hippocampus | −25.3 | −20.8 | −10 | 7584 | |
| 5 | Inferior orbitofrontal | −33.2 | 25.9 | −12 | 8192 | |
| 6 | Insula | −35.5 | 7.12 | 3.09 | 14 456 | |
| 7 | Parahippocampal | −21.5 | −16.6 | −20.1 | 7536 | |
| 8 | Posterior cingulate | −6.61 | −42.5 | 17.5 | 1600 | |
| 9 | Superior temporal | −53.4 | −19.9 | 6.78 | 17792 | |
| 10 | Thalamus | −11.2 | −17.4 | 8.3 | 8664 | |
| Contralateral | 11 | Anterior cingulate | 7.06 | 37.4 | 0.766 | 2192 |
| 12 | Caudate | 14.4 | 12.7 | 9.11 | 7736 | |
| 13 | Fusiform | 33.9 | −39.5 | −17.2 | 14 136 | |
| 14 | Hippocampus | 29.1 | −19.7 | −10.2 | 7640 | |
| 15 | Inferior orbitofrontal | 37.8 | 26.6 | −12.7 | 7136 | |
| 16 | Insula | 38.8 | 6.49 | 2.33 | 14 272 | |
| 17 | Parahippocampal | 25.1 | −15.3 | −20.2 | 8904 | |
| 18 | Posterior cingulate | 7.22 | −40.2 | 16.5 | 1504 | |
| 19 | Superior temporal | 57 | −22.2 | 7.19 | 23 568 | |
| 20 | Thalamus | 12.7 | −17.3 | 8.48 | 8384 |
Figure 2.
Results from probabilistic tractography obtained from different regions of interest (ROIs) averaged over all subjects. Each three-dimensional brain rendering illustrates the voxel-wise probabilistic map of connectivity from one ROI (from left to right: left anterior cingulate, left inferior orbital region, left hippocampus and left thalamus).
Connectivity matrix
The next step involved the definition of the weighted connectivity between all pairs of the 20 seed regions. The raw connectivity between regions A and B was determined as follows. First, the density of fibres originating from either region A or B and passing through or terminating in the other region (B or A) was obtained. For each seed region, Probtrackx generates a volume containing the fibre count for all voxels in the brain. For each of the remaining 19 regions, the fibre density was obtained as the mean fibre count taken over all voxels in that region. This was accomplished using the software MRIcron (http://www.mccauslandcenter.sc.edu/mricro/mricron/). The connectivity between regions A and B was then defined as the average of the two estimates of fibre density (A to B and B to A).
Binary connectivity matrices were then obtained by applying a series of thresholds to the 20×20 weighted connectivity matrix of each subject. The thresholds were set to 95th, 90th, 85th, …, and 45th percentile of the weights in the matrix, resulting in 11 binary matrices with densities of 5%, 10%, …, and 55%. The density of a binary connection matrix is defined as the number of connections in the matrix divided by the number of possible connections. This ‘thresholding’ system ensured that networks in both groups have the same number of connections and that the between group differences reflected alterations in network organisation rather than differences in absolute connectivity. This thresholding system also ensures that at high thresholds the networks retain a known degree of connectivity.25
For consistency, the matrices obtained from patients were organised so that the first 10 structures represented the ipsilateral side of seizure origin (left for left MTLE and right for right MTLE). A two sample one tailed t test was conducted to determine differences in MD, FA and fibre counts between MTLE and controls. Statistical significance was set at p<0.05. Results were evaluated without and with correction for multiple comparisons utilising false discovery rate (FDR),26 as described in the results section.
Graph theoretic measures
Structural network properties were characterised using graph theory based measures. We employed the brain connectivity toolbox27 (https://sites.google.com/a/brain-connectivity-toolbox.net/bct/) to obtain graph measures of interest, focusing the analysis on basic graph theoretical properties directly relevant to MTLE. A large number of graphical measures can be calculated from connectivity matrices; however, their relevance in MTLE is not defined. Therefore, we opted to investigate measures with a tangible theoretical relationship with the network abnormalities that could be speculated to be intrinsic to MTLE. We investigated measures related to (1) node properties, where each node corresponds to one of the 20 anatomical ROIs; and (2) global network properties. Specifically, we focused the whole network analyses on average clustering coefficient (which can be understood as the fraction of connected loops or triangles around a node), average efficiency (which represents the average inverse shortest path length in the network, and can be understood as a measure of effectiveness of information flow) and average betweenness centrality (which represents the number of shortest paths from all nodes to all others that pass through that node, and can be understood as the ‘importance’ of that node in the network). Regarding node properties, we focused on regional clustering coefficient, degree (which represents the number of connections to the node), local efficiency and betweenness centrality. The measures employed in this study were chosen as they represent basic graph properties and are directly relevant to MTLE. They are summarised in table 2. Graph theoretic properties of controls and patients were compared using the Wilcoxon signed rank test in MATLAB. Statistical significance was set at p<0.05.
Table 2.
Graph theoretical properties that were investigated in this study
| Graph theoretical property | Explanation |
| Network measurements | |
| Global efficiency | Average of inverse nodal shortest path length |
| Average clustering coefficient | Average of nodal clustering coefficients |
| Average betweenness centrality | Average of nodal betweenness centrality |
| Node measures | |
| Nodal degree | No of links connected to the node |
| Local efficiency | Average of the inverse shortest path length in the neighbourhood of the node |
| Clustering coefficient | Fraction of a node's neighbours that are neighbours of each other |
| Betweenness centrality | No of shortest paths from and to all nodes that pass this node |
Results
Strength of connections between nodes
Compared with controls, patients exhibited a significant decrease in FA within perilimbic regions, notably involving the parahippocampal and temporal subcortical white matter, as well as the anterior and posterior cingulate regions. The differences were more intense in the side ipsilateral to the side of seizure onset. There were no areas of increased FA in patients compared with controls. These results were corrected by multiple comparisons with FDR and are summarised in supplementary table 1 and supplementary figure 2 (supplementary table 1 and supplementary figure 2 are available online only). There were no significant differences in MD between groups.
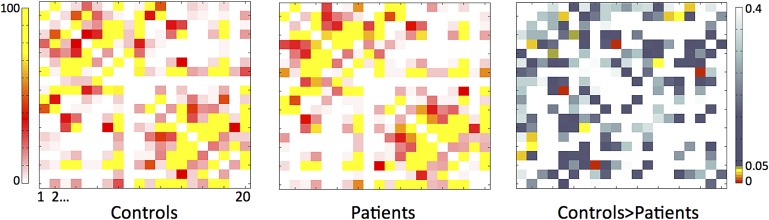
Patient also exhibited a reduction in the number of probabilistic connections between temporal and extra-temporal regions. Specifically, there was a significant reduction in the absolute connectivity between: (1) the ipsilateral anterior cingulate and the contralateral parahippocampal region; (2) the ipsilateral caudate and the bilateral inferior orbitofrontal regions; (3) the ipsilateral caudate and the contralateral insula; (4) the ipsilateral inferior orbitofrontal region and the bilateral thalami; (5) the ipsilateral insula and the contralateral fusiform area; (7) the contralateral hippocampus and the ipsilateral parahippocampal region; and the (8) ipsilateral and contralateral posterior cingulate regions. There was also an increase in connectivity between the contralateral hippocampus and the insula. These results are summarised in figure 3. Importantly, these results were not corrected for multiple comparisons but are shown here to provide an overview of differences between groups. When the FDR26 correction was applied, the connection between the ipsilateral and contralateral posterior cingulate regions remained significant, with reduction of connections in patients.
Figure 3.
Regional brain connectivity, as depicted by weighted connectivity matrices for controls and patients, and the ensuing matrix of statistical differences. The connectivity matrices for patients and controls illustrate the weight of connection between regions, whereby each cell entry represents a corrected weight of connection between the region depicted on the y and x axes. The scale (0–100) represents the average voxel-wise number of probabilistic defined fibre counts between regions. To facilitate visualisation, regions are number from 1 to 20, representing the numbering adopted in table 1 (with left representing ipsilateral and right contralateral). The difference matrix illustrates the p values obtained from a two sample t test performed between connectivity matrices, showing areas with higher connectivity in controls compared with patients with medial temporal lobe epilepsy.
Whole network graphical properties
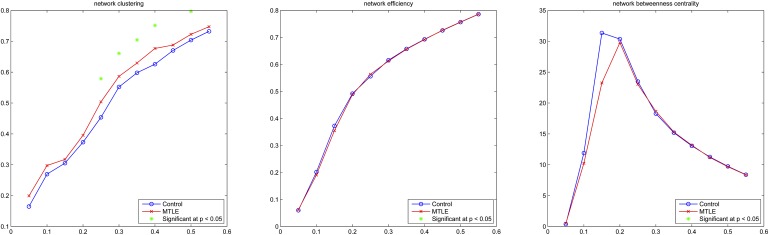
There were significant differences in the network properties between patients and controls. Patients with MTLE exhibited an increase in the mean network clustering coefficient for a consecutive range of binary thresholds, compared with controls. In contrast, global network efficiency and betweenness centrality were not significantly different across thresholds between the two groups. These results are summarised in table 3 and displayed in detail in figure 4.
Table 3.
Summary of the findings from network and node based graph theoretical analyses
| Region | Findings | |
| Network measurements | ||
| Average clustering coefficient | Increased in MTLE | |
| Network efficiency | No significant differences | |
| Node measurements | ||
| Nodal degree | Hippocampus | Increased in MTLE |
| Insula | No significant differences | |
| Superior temporal | Increased in MTLE | |
| Thalamus | No significant differences | |
| Local efficiency | Hippocampus | Decreased in MTLE |
| Insula | Increased in MTLE | |
| Superior temporal | Increased in MTLE | |
| Thalamus | Increased in MTLE | |
| Clustering coefficient | Hippocampus | Decreased in MTLE |
| Insula | Increased in MTLE | |
| Superior temporal | Increased in MTLE | |
| Thalamus | Increased in MTLE | |
| Betweenness centrality | Hippocampus | Increased in MTLE |
| Insula | Decreased in MTLE | |
| Superior temporal | No significant differences | |
| Thalamus | No significant differences | |
MTLE, medial temporal lobe epilepsy.
Figure 4.
Results from analyses related to the whole network. The y axis represents the value for the network measure and the x axis represents the fixed density threshold utilised to define a binary matrix. MTLE, medial temporal lobe epilepsy.
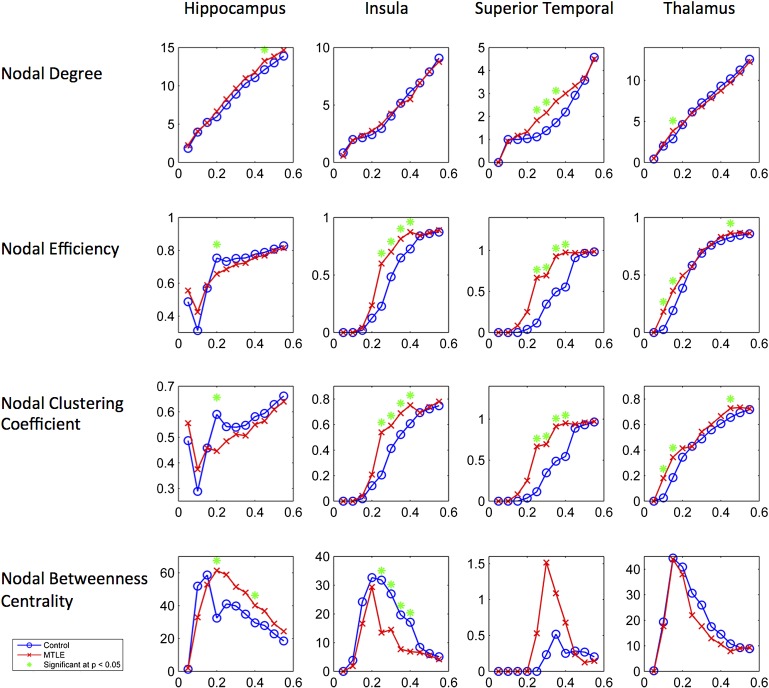
Node graphical properties
Nodal abnormalities were also observed. There was an increase in nodal degree in the ipsilateral hippocampus, superior temporal region and thalamus in patients. The nodal efficiency from patients was increased in the ipsilateral insula, superior temporal region and thalamus but it was reduced in the ipsilateral hippocampus in patients.
In patients, nodal clustering coefficient was increased in the ipsilateral insula, superior temporal region and thalamus but it was also reduced in the ipsilateral hippocampus. Finally, nodal betweenness centrality in patients was increased in the ipsilateral hippocampus but decreased in the ipsilateral insula. These results are summarised in table 3 and figure 5.
Figure 5.
Results from the node properties graphical analyses. For each graph, the y axis represents the network measure. The x axis represents the fixed density threshold utilised to define a binary matrix. Each column demonstrates the results for a particular anatomical structure. MTLE, medial temporal lobe epilepsy.
No significant nodal changes were observed in this dataset regarding the nodes contralateral to the side of seizure onset.
In summary, these results demonstrate a regional decrease in the absolute connectivity among limbic regions in MTLE, accompanied by a paradoxical increase in the average limbic network clustering coefficient. MTLE is also associated with an increase in nodal efficiency, degree and clustering of the ipsilateral superior temporal region and the thalamus, while the hippocampus exhibits a higher degree but reduced clustering coefficient and efficiency.
Discussion
In this study, we observed that patients with MTLE exhibited an overall reorganisation of limbic circuitry. Even though MTLE is associated with regional loss of connections (demonstrated by a reduction in absolute connectivity between regions), it is also associated with higher limbic clustering coefficient and a higher influence of specific nodes over the network, as demonstrated by the higher efficiency, clustering and degree of the thalamus, insula and superior temporal regions. Interestingly, these changes were observed at a ‘regional’ level. The overall efficiency of the network was not changed, even though a global increase in whole network clustering coefficient was observed. The global increase in clustering coefficient, without changes in efficiency, may represent a higher per cent of re-entrant connections that do not overall influence the proficiency of the whole network to transfer information. The core of network alterations in MTLE is likely more restricted to the medial temporal subnetworks of the limbic network.
The results from this study corroborate previous findings suggesting that MTLE is not restricted to the hippocampus but may encompass a network of limbic structures. These results also confirm that limbic connectivity in MTLE is configured differently compared with controls. In particular, previous studies have consistently demonstrated that MTLE is associated with a significant reduction of fibres, and also laterality of connectivity. Importantly, these results can be related to clinical performance. For example, language networks can be partially lateralised to the non-dominant hemisphere in patients with left MTLE.28 Also, memory performance is related to the integrity of medial temporal fibres,29 30 and the progressive loss of limbic connectivity plays a role in the cognitive decline observed in chronic localisation related epilepsy.31
Previous studies have focused on the quantification of indirect measurements of axonal fibres but questions about specific graphical topological network reorganisation depend on graph theory analysis. Therefore, the results from our study suggest that the structural organisation of the limbic network in MTLE can lead to aberrant connections, which may play a role in the pathophysiology of seizure generation and maintenance. In particular, the findings from this study suggest that perihippocampal structures such as the insula, thalamus and superior temporal region are sites of altered connectivity in the limbic network of patients with MTLE. This observation may suggest that periepileptogenic regions have a significant impact on limbic networks.
These are preliminary findings and the relationship between structural network changes and clinical manifestations of epilepsy were not tested in this manuscript. Furthermore, we did not investigate whether structural changes in MTLE networks occur as a consequence of seizures, leading to abnormal plasticity, or whether these changes are the cause of seizures. Other limitations of this study include the use of relatively low number directions in DTI images, possibly underestimating connections, and the otherwise limitations of DTI images, which may not overcome complex fibre crossing, or non-gaussian distribution of diffusion data in a rich microstructural environment.32
Another important limitation of this study is related to the lack of application of correction for multiple comparisons during the analyses of node parameters. This is a challenging issue regarding graph theoretical analyses of neuroimaging data as the large number of ROIs, connections, graphical measures and binary thresholds may lead to an overly prohibitive corrected threshold with a very high likelihood of type II statistical errors. One way of overcoming this issue was the strategy applied in this study, which involved: first, evaluating whether the main results were compatible with the a priori hypothesis—namely, changes in medial temporal connectivity—and second, observing whether differences across groups were consistently observed across multiple binary fixed density thresholds (ie, the curves are consecutively distanced apart across multiple thresholds). If type II errors were responsible for most of the significant changes, one would expect significant changes randomly distributed across regions and thresholds. Therefore, the reproducibility of the results, which also match the a priori hypothesis, likely indicates its true biological meaning. In particular, the use of binary thresholds provides an indirect measurement of the effect size of the differences observed, and this may be more meaningful in studies employing graph theory than the standard FDR and Bonferroni comparisons used with voxel-base imaging studies. Further methodological studies could certainly be targeted to address this issue.
The observation that MTLE is associated with structural network reorganisation may have a profound impact in future disease quantification and tracking of disease progression. In this study, we observed a significant pattern of reduced connectivity between regions; however, the reduced connectivity was not as widespread as in previous studies employing conventional DTI measurements such as FA and MD.13 19 This is likely due to the fact that the present analyses were restricted to a limbic and paralimbic structures and not applied to the whole brain. In addition, the present sample size was more limited compared with other studies. The analyses performed in this study were driven by the hypothesis that network reorganisation may be related to the local influence of the medial temporal lobe; hence a more liberal threshold was employed, with an arbitrary lower statistical threshold. This approach is similar to previous studies evaluating the influence of suspected abnormal regions in MTLE when the testable hypothesis is driven by a biological premise.30
The observation that components of the limbic network in epilepsy may exhibit a higher level of clustering, efficiency, degree and betweenness centrality may lead to the conjecture that these aberrant connections may represent hyperexcitable circuits that initiate and maintain seizures. Computational modelling of epilepsy suggests that hyperconnected hubs may play a considerable role in the pathophysiology of epilepsy.33 Information from DTI alone may not enable the conclusive evidence towards excitability as excitatory and inhibitory fibres are indistinguishable. Nonetheless, modelling based on known properties of anatomical location and the combination of these findings from experimental data may enable in the future a direct quantification of the epileptic potential of subnetworks. Furthermore, the evaluation of ‘hub’ components of the network through their susceptibility to attack, or between centrality of weighted connections, may aid the evaluation of the epileptogenic potential of the network.
In conclusion, the analysis of graphical reorganisation of limbic networks in MTLE may be used as a biomarker for assessment of treatment response and disease severity. Network changes may become a promising additional form of disease quantification of MTLE and other forms of epilepsy.
Supplementary Material
Footnotes
Contributors: All authors have contributed to the production of this study and are in agreement with its content. As per the International Committee of Medical Journal Editors, authorship credit should be based on: (1) substantial contributions to conception and design, acquisition of data, or analysis and interpretation of the data; (2) drafting the article or revising it critically for important intellectual content; and (3) final approval of the version to be published. The authors of the this paper met the following conditions: LB: (1), (2), (3); TN: (2); GUM: (1), (2); JEJ: (1), (2), (3); MVS: (2); JCE: (2); AT: (1), (2), (3).
Funding: This work was supported in part by the National Science Foundation/EPSCoR under grant No EPS-0919440. This publication was supported by the South Carolina Clinical and Translational Research (SCTR) Institute, with an academic home at the Medical University of South Carolina, through NIH grant Nos UL1 RR029882 and UL1 TR000062.
Competing interests: None.
Ethics approval: The study was approved by the institutional review board of the Medical University of South Carolina.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Babb TL, Brown WJ. Pathological findings in epilepsy. In: Engel J, Jr, ed. Surgical treatment of the epilepsies. New York: Raven Press, 1987:511–40 [Google Scholar]
- 2.Bonilha L, Kobayashi E, Rorden C, et al. Medial temporal lobe atrophy in patients with refractory temporal lobe epilepsy. J Neurol Neurosurg Psychiatry 2003;74:1627–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernasconi N, Duchesne S, Janke A, et al. Whole-brain voxel-based statistical analysis of gray matter and white matter in temporal lobe epilepsy. Neuroimage 2004;23:717–23 [DOI] [PubMed] [Google Scholar]
- 4.Keller SS, Mackay CE, Barrick TR, et al. Voxel-based morphometric comparison of hippocampal and extrahippocampal abnormalities in patients with left and right hippocampal atrophy. Neuroimage 2002;16:23–31 [DOI] [PubMed] [Google Scholar]
- 5.McDonald CR, Hagler DJ, Jr, Ahmadi ME, et al. Regional neocortical thinning in mesial temporal lobe epilepsy. Epilepsia 2008;49:794–803 [DOI] [PubMed] [Google Scholar]
- 6.Jack CR, Jr, Sharbrough FW, Marsh WR. Use of MR imaging for quantitative evaluation of resection for temporal lobe epilepsy. Radiology 1988;169:463–8 [DOI] [PubMed] [Google Scholar]
- 7.Cendes F, Andermann F, Gloor P, et al. MRI volumetric measurement of amygdala and hippocampus in temporal lobe epilepsy. Neurology 1993;43:719–25 [DOI] [PubMed] [Google Scholar]
- 8.Bernasconi N, Bernasconi A, Caramanos Z, et al. Mesial temporal damage in temporal lobe epilepsy: a volumetric MRI study of the hippocampus, amygdala and parahippocampal region. Brain 2003;126:462–9 [DOI] [PubMed] [Google Scholar]
- 9.Ashburner J, Friston KJ. Why voxel-based morphometry should be used. Neuroimage 2001;14:1238–43 [DOI] [PubMed] [Google Scholar]
- 10.Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci U S A 2000;97:11050–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keller SS, Roberts N. Voxel-based morphometry of temporal lobe epilepsy: an introduction and review of the literature. Epilepsia 2008;49:741–57 [DOI] [PubMed] [Google Scholar]
- 12.Bonilha L, Rorden C, Castellano G, et al. Voxel-based morphometry reveals gray matter network atrophy in refractory medial temporal lobe epilepsy. Arch Neurol 2004;61:1379–84 [DOI] [PubMed] [Google Scholar]
- 13.Bonilha L, Edwards JC, Kinsman SL, et al. Extrahippocampal gray matter loss and hippocampal deafferentation in patients with temporal lobe epilepsy. Epilepsia 2010;51:519–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spencer SS. Neural networks in human epilepsy: evidence of and implications for treatment. Epilepsia 2002;43:219–27 [DOI] [PubMed] [Google Scholar]
- 15.Bonilha L, Rorden C, Castellano G, et al. Voxel-based morphometry of the thalamus in patients with refractory medial temporal lobe epilepsy. Neuroimage 2005;25:1016–21 [DOI] [PubMed] [Google Scholar]
- 16.Mueller SG, Laxer KD, Cashdollar N, et al. Voxel-based optimized morphometry (VBM) of gray and white matter in temporal lobe epilepsy (TLE) with and without mesial temporal sclerosis. Epilepsia 2006;47:900–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin JJ, Riley JD, Juranek J, et al. Vulnerability of the frontal-temporal connections in temporal lobe epilepsy. Epilepsy Res 2008;82:162–70 [DOI] [PubMed] [Google Scholar]
- 18.Riley JD, Franklin DL, Choi V, et al. Altered white matter integrity in temporal lobe epilepsy: association with cognitive and clinical profiles. Epilepsia 2010;51:536–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Focke NK, Yogarajah M, Bonelli SB, et al. Voxel-based diffusion tensor imaging in patients with mesial temporal lobe epilepsy and hippocampal sclerosis. Neuroimage 2008;40:728–37 [DOI] [PubMed] [Google Scholar]
- 20.Ellmore TM, Pieters TA, Tandon N. Dissociation between diffusion MR tractography density and strength in epilepsy patients with hippocampal sclerosis. Epilepsy Res 2011;93:197–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Proposal for revised classification of epilepsies and epileptic syndromes Commission on Classification and Terminology of the International League Against Epilepsy. Epilepsia 1989;30:389–99 [DOI] [PubMed] [Google Scholar]
- 22.Nucifora PG, Verma R, Lee SK, et al. Diffusion-tensor MR imaging and tractography: exploring brain microstructure and connectivity. Radiology 2007;245:367–84 [DOI] [PubMed] [Google Scholar]
- 23.Behrens TE, Berg HJ, Jbabdi S, et al. Probabilistic diffusion tractography with multiple fibre orientations: what can we gain? Neuroimage 2007;34:144–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tzourio-Mazoyer N, Landeau B, Papathanassiou D, et al. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. NeuroImage 2002;15:273–89 [DOI] [PubMed] [Google Scholar]
- 25.van Wijk BC, Stam CJ, Daffertshofer A. Comparing brain networks of different size and connectivity density using graph theory. PloS One 2010;5:e13701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Genovese CR, Lazar NA, Nichols T. Thresholding of statistical maps in functional neuroimaging using the false discovery rate. Neuroimage 2002;15:870–8 [DOI] [PubMed] [Google Scholar]
- 27.Rubinov M, Sporns O. Complex network measures of brain connectivity: uses and interpretations. NeuroImage 2010;52:1059–69 [DOI] [PubMed] [Google Scholar]
- 28.Powell HW, Parker GJ, Alexander DC, et al. Abnormalities of language networks in temporal lobe epilepsy. Neuroimage 2007;36:209–21 [DOI] [PubMed] [Google Scholar]
- 29.Yogarajah M, Powell HW, Parker GJ, et al. Tractography of the parahippocampal gyrus and material specific memory impairment in unilateral temporal lobe epilepsy. Neuroimage 2008;40:1755–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDonald CR, Ahmadi ME, Hagler DJ, et al. Diffusion tensor imaging correlates of memory and language impairments in temporal lobe epilepsy. Neurology 2008;71:1869–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaessen MJ, Jansen JF, Vlooswijk MC, et al. White matter network abnormalities are associated with cognitive decline in chronic epilepsy. Cereb Cortex. Published Online First: 29 October 2011. doi:10.1093/cercor/bhr298 [DOI] [PubMed] [Google Scholar]
- 32.Tabesh A, Jensen JH, Ardekani BA, et al. Estimation of tensors and tensor-derived measures in diffusional kurtosis imaging. Magn Reson Med 2011;65:823–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Case M, Soltesz I. Computational modeling of epilepsy. Epilepsia 2011;52(Suppl 8):12–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.