Abstract
Abnormal bone marrow (BM) suppression is one of the hallmarks of dengue virus (DENV) infection in patients. Although the etiology remains unclear, direct viral targeting of the BM has been reasoned to be a contributing factor. The present studies were carried out in an effort to determine the potential effect of DENV infection on the cellularity of BM using a previously established nonhuman primate model of DENV-induced coagulopathy. BM aspirates were collected at various times from the infected nonhuman primate and cells were phenotypically defined and isolated using standard flow cytometry (fluorescence-activated cell sorting). These isolated cells were subjected to detection of DENV utilizing quantitative real-time reverse transcription polymerase chain reaction, electron microscopy, and immunostaining techniques. DENV RNA was detectable by quantitative real-time reverse transcription polymerase chain reaction in BM specimens and the presence of DENV-like particles within platelet was confirmed by electron microscopy. Enumeration of BM cells revealed a transient surge in cellularity at day 1, followed by a gradual decline from days 2 to 10 post infection. Detailed phenotypic studies showed similar kinetics in the frequencies of CD41+CD61+ cells, regardless of CD34 and CD45 expression. The CD61+ cells were not only the predominant cells that stained for DENV antigen but fluorescence-activated cell sorting–assisted isolation of CD61+ cells from the BM were shown to contain infectious DENV by coculture with Vero cells. These data support the view that intravenous infection of nonhuman primate with DENV leads to direct infection of the BM, which is likely to be a contributing factor for transient cell suppression in the peripheral blood characteristic of acute DENVinfection.
Dengue virus (DENV) infection has often been referred to as “breakbone fever” because of the intense pain within joints that are characteristics of DENV infection. The bone marrow (BM) has thus been reasoned to be either directly and/or indirectly involved in dengue pathogenesis. One early investigation on the cellularity of BM revealed that early BM suppression in dengue patients is a common phenomenon [1]. DENV has been isolated from autopsy BM from patients dying of dengue shock syndrome and from BM suspensions of several dengue hemorrhagic fever patients who survived the infection [2]. In addition, BM-associated aplasia in dengue patients, although infrequent, has also been documented [3–5]. Ex vivo experimental studies have revealed that DENV can efficiently infect hematopoietic cells [6,7] and is only capable of replication in leukocytes derived from the BM and not from other lymphatic tissues (e.g., spleen, thymus, and lymph node) [8]. These earlier findings in humans are supported by data derived in monkeys, in which the BM was identified as an early site of DENV replication [9,10]. However, since these earlier studies, the role of the BM as a site for DENV replication has not been substantiated because of the difficulty in obtaining BM biopsies from dengue patients, given the increased risk of bleeding associated with such collections.
Despite the fact that detailed hematological profiling of the peripheral blood of dengue patients has been well documented [11], and some of the key findings have been validated, for example, leukopenia and thrombocytopenia, atypical lymphocytes, and abnormal ratio of immune cells[12,13], the precise mechanisms leading to these hematological changes remain ill-defined. In addition, although BM suppression has been well documented in dengue patients as early as the 1960s, there is clearly a paucity in the reports that are available on the pathophysiological effects and on the fate of BM cells during the course of DENV infection. The studies that do exist consist mainly of experiments involving in vitro DENV infection of BM specimens from normal donors [6,8,14] and, to some extent, studies of BM from the murine severe combined immunodeficient humanized model [7,15]. Results of these studies indicate that DENV replicates predominantly in hematopoietic progenitor cells derived from the BM or cord blood [6–8,14,15]. However, the lack of a suitable animal model that fully recapitulates the cardinal features of DENV infection has prevented detailed studies of the potential role of the BM hematopoietic progenitor cells in the pathogenesis of DENV infection. Recently, our laboratory, documented for the first time the induction of readily visible signs of hemorrhage in rhesus macaques infected with a high dose of DENV administered intravenously [16]. We have extended these previous studies using this nonhuman primate animal model to address the potential mechanisms involved by which dengue induces hemorrhage by detailed studies of BM progenitor cells during the course of DENV infection.
Materials and methods
Sample collections and preparations
Six rhesus monkeys (Macaca mulatta) of Indian origin that were part of two separate experiments (three animals each, designated animal ID RM #1 to 3 and RM #4 to 6 for Experiments 1 and 2) and were infected intravenously with DENV serotype 2 (strain 16681) as described previously [16], were the source of the samples described here. At different time points post infection, BM was aspirated from the iliac crest and supplemented with heparin. All experimental protocols and procedures were conducted after approval by the Emory Institutional Animal Care and Use Committee, and all animals were housed at the Yerkes National Primate Research Center of Emory University and cared for in conformance to the guidelines of the Committee on the Care and Use of Laboratory Animals of the Institute of Laboratory Animal Resources, National Research Council, and the Health and Human Services [17]. BM cellularity analyses were performed by the Clinical Pathology Laboratory at Emory University. Platelets and plasma were isolated from BM aspirates using OptiPrep density gradient centrifugation as described previously, with minor modifications [18]. BM mononuclear cells were purified using Lymphoprep separation medium (Mediatech, Inc., Manassas, VA) according to the manufacturer’s instructions.
Real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) for the detection of viral RNA
RNA was extracted from 140 mL plasma or culture supernatant fluids and from platelets isolated from the BM using QIAmp Viral RNA mini kit and RNeasy mini kit (QIAGEN, Germantown, MD, USA), respectively. The resultant RNA was then subjected to qRT-PCR using the Taqman RT kit (Perkin-Elmer Applied Biosystem, Waltham, MA, USA) and a Bio-Rad iCycler system according to a previously described method [16]. RNA from a viral stock of DENV was used as a control and the data obtained are reported as viral RNA copy number/mL of sample. The detection limit of this assay was about 100 copies of viral RNA genome equivalents per milliliter.
Electron microscopy
Highly enriched preparations of platelets from the blood of rhesus macaques were fixed with 2.5% glutaraldehyde or 4% paraformaldehyde in 0.1 M phosphate buffer overnight and then processed for electron microscopy (EM) by the Robert P. Apkarian Integrated Electron microscopy Core Facility Service. Immuno-EM was performed according to a previously described method with minor modifications [19]. An immunogold-conjugated mouse monoclonal antibody with specificity for dengue virus E (clone 3H5; Chemicon, Billerica, MA, USA) was utilized for detection of DENV particles.
Immunodetection of DENV antigen in megakaryocytic cells
To perform immunofluorescent staining, smears of BM or isolated mononuclear cells on glass slides were fixed with methanol for 5 minutes and rinsed with phosphate-buffered saline (PBS). Slides were then incubated with 10% human AB serum in PBS at room temperature for 15 minutes, followed by either mouse anti-E monoclonal antibody (clone 4G2) or isotype-matched control (IgG2a) antibody for 1 hour. Slides were washed and then incubated withphycoerythrin-conjugated goat anti-mouse IgG antibody (eBioscience, San Diego, CA, USA) at a dilution of 1:1000 for 1 hour. Slides were washed in PBS and then incubated with 10% normal mouse serum in PBS for 30 minutes at room temperature to block the remaining binding sites of the anti-mouse IgG antibody, followed by the addition of fluorescein isothiocyanate–conjugated mouse anti-CD61 antibody (eBioscience) for 1 hour. Slides were washed three times with PBS and mounted with 4′,6-diamidino-2-phenylindole mount reagent (Invitrogen, Carlsbad, CA, USA), and images were captured using a Zeiss microscope equipped with an Axis 5 digital camera. To perform immunohistochemical staining for both DENV antigen and CD41a, megakaryocytes were collected from the BM onto a nucleopore polycarbonate membrane of 5-mm pore diameter (Fisher Scientific, Pittsburgh, PA, USA) using a method previously described by Wilde et al. [20]. The membrane was then subjected to the staining procedure as described in more detail in the Supplementary Materials and Methods (online only, available at www.exphem.org).
Flow cytometry and cell sorting
Aliquots of heparinized BM samples were stained with a panel of monoclonal antibodies with specificity for CD34, CD45, CD14, CD41, CD61, and CD71 conjugated to different fluorochromes and analyzed by polychromatic flow cytometry using a BD LSRII (BD Biosciences, San Jose, CA, USA) according to a previously described method [16]. The acquired data were analyzed using FlowJo software (TreeStar Inc., Ashland, OR, USA) and statistical analysis (two-way analysis of variance) was subsequently performed using GraphPad Prism 5 (GraphPad Software, Inc., La Jolla, CA, USA). To specifically analyze subpopulations of BM cells potentially involved in DENV infection, a panel of labeled monoclonal antibodies with specificity for CD3, CD14, CD61, and CCR5 was used to stain aliquots of mononuclear cells isolated from the BM. For select experiments, a FACSAria II (BD Biosciences) was utilized to isolate highly enriched populations of specific lineages of BM cells as outlined in the text. An aliquot of the sorted cells harvested at the enrichment step was analyzed for the purity of the sorted population.
Immunohistochemical staining of sorted cells
Smears made from aliquots of the FACSAria II enriched population of mononuclear cells were fixed with methanol for 5 minutes and then treated with 0.3% H2O2 solution containing 0.1% sodium azide in PBS for 1 hour to block endogenous peroxidase, followed by a 30-minute incubation with 10% human AB serum. After two washes with PBS, the samples were then incubated with mouse anti-E monoclonal antibody (clone 4G2) or its isotype-matched control antibody at 4°C overnight. Samples were washed three times with PBS and incubated with biotinylated horse anti-mouse immunoglobulins (Vector Laboratories, Inc., Burlingame, CA, USA) at room temperature for 30 minutes, followed by three washes with the same buffer. The samples were then incubated for 30 minutes each with Vectastain ABC reagent (Vector) and diaminobenzidine as an enzyme substrate for peroxidase followed by counterstaining with hematoxylin.
Recovery of infectious DENV by coculture
A monolayer of Vero cells was incubated for 2 hours with three different subpopulations of enriched subsets of mononuclear cells, which were comprised of the CD61+CCR5−, CD61−CCR5+ and CD61+CCR5+ subsets, respectively. Thereafter, the cocultured cells were washed with PBS, and resuspended in RPMI media supplemented with 10% fetal bovine serum and incubated at 37°C in a humidified atmosphere. At different time points, supernatant fluids were collected and assayed for viral RNA (copies/mL) by real-time qRT-PCR as described previously [16].
Results
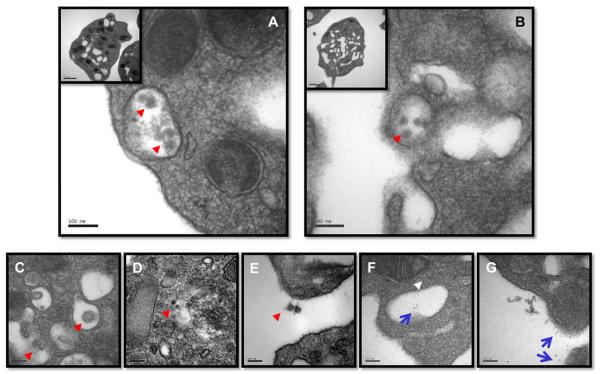
A number of studies have documented several aspects of platelet destruction in the peripheral blood during acute DENVinfection, which includes reports of a direct association of DENV with human platelets invivo [18,21,22]. In efforts to confirm this observation, platelets isolated from peripheral blood of DENV–infected rhesus monkeys (days 1 to 5 post infection) were subjected to EM and immuno-EM. As shown in Figure 1A and B, vesicles inside the platelets were seen to contain virus-like particles. Occasionally, events such as budding, maturation, and exocytosis of virus-like particles were noted (Fig.1C, D, and E). Immuno-EM confirmed that the platelets were positive for DENVenvelope antigen (Fig. 1F and G). In addition, immunohistochemical stainingusing the periodic acid-Schiff reagent revealed cells that are likely platelets, which contained readily detectable number of budding vesicles (Supplementary Figure E1A and E1B; online only, available at www.exphem.org). The morphology of dengue viral particles being shed from platelets to the exterior appeared to be distinct from that of classical dengue viral particle. Although the significance of the difference remains to be determined, ithas been reported that infectious dengueviral particles are present in supernatant fluid of of infected cells in a dynamic state ranging from a classical viral particle to viral particles, which are associated with lipids termed as subviral particles [23,24].
Figure 1.
Dengue viral particles in platelets. Platelets were purified from acutely infected rhesus monkeys. The purified platelets were subjected to procedures for morphological EM and immune-EM investigations as described in the Materials and Methods. (A) and (B) DENV-like particles (red arrowhead). (C, D) DENV-like particles observed in platelets (red arrowhead). (E) DENV-like particles shedding from platelets (red arrowhead). Note the circular substance left behind on the surface of the platelet. (F) Immuno-EM illustrating DENV-like particles (blue arrow) in a vesicular compartment and in the cytoplasm (white arrowhead). (G) Immuno-EM depicts dengue viral particles (blue arrow). The morphology is markedly different from the classical view shown in (A) and (B).
The megakaryocytic cell lineages are the direct progenitors of platelets that are derived through membrane demarcation of apoptotic cells and/or formation of proplatelets that are released into the blood [25,26]. Because BM suppression in dengue patients is a well-documented finding, it was reasoned that a study of the potential for megakaryocytes to serve as targets of DENV infection was indicated, because such virus targeting may be the basis for the abnormal cellular composition of the BM and peripheral blood mononuclear cells in the circulation of DENV-infected patients.
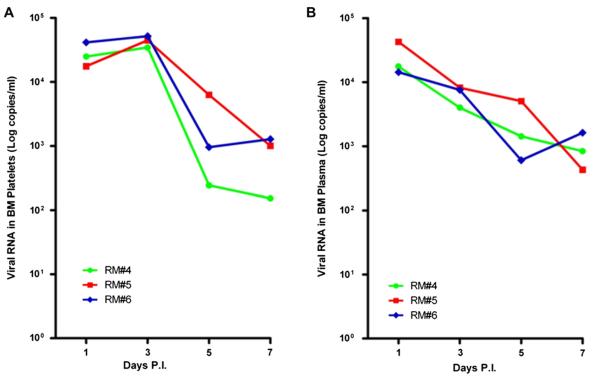
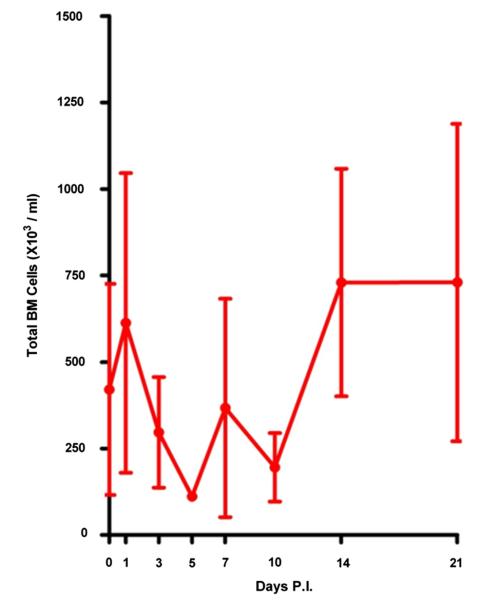
Two independent experiments comprising three DENV-infected rhesus macaque monkeys each were performed. Initially, real-time qRT-PCR was performed on aliquots of plasma and enriched population of platelets isolated from the BM of the DENV-infected rhesus monkeys. Results indicated that viral RNA could be readily detected in both the plasma and platelet specimens with the peak titer at early time points post infection (Supplementary Table E1; online only, available at www.exphem.org, Experiment 1, and Fig. 2, Experiment 2). Examination of the cellularity of the BM demonstrated a transient surge on day 1, followed by a gradual decrease during days 2 to 10 post infection, and a rebound to normal or hypercellularity beyond 10 days post infection (Fig. 3).
Figure 2.
Dengue viral load in platelets and plasma fraction isolated from BM. BM platelets and plasma were purified as previously described with minor modifications [18].Viral RNAs were quantified by qRT-PCR as described in the Materials and Methods. (A) Viral load in BM platelets. (B) Viral load in BM plasma. The RM# 4 to 6 indicates animal ID used in the study as previously described [16]. PI 5 post infection.
Figure 3.
BM cellularity in DENV-infected rhesus monkeys. Aspirated BM cellularity (RM#4 to RM#6) was estimated as described in the Materials and Methods. A transient drop of the cellularity was observed on days 2 to 10 after infection and rose to the normal level 2 weeks later. Error bar indicate mean 6 standard error of mean. PI 5 post infection.
Declines in BM mass accompanied the decline of viral load in the plasma and platelets from the BM (Figs. 2 and 3). In contrast, this decline appeared to be inversely correlated with the viral load detectable in the corresponding blood plasma fraction reported previously for the same DENV-infected monkeys [16]. These findings suggest that the dysregulation in the BM may be associated with increased DENV replication in vivo. A further study on the phenotypic changes of cells in the BM was performed by using polychromatic flow cytometry–based analysis of BM samples from these DENV-infected animals. A panel of antibodies with specificity for cell surface markers expressed uniquely by subsets of BM cells, which included CD34, CD45, CD41, and CD61 was used. Results revealed that regardless of CD34/CD45 expression, cells dually positive for CD41 and CD61 (markers for the megakaryocyte lineage) increased transiently on day 1 post infection (Table 1). Although, overall, CD34−CD45−CD41+CD61− cells appeared to significantly increase (p < 0.05) and stayed at an elevated level during the course of acute DENV infection, other subpopulations of BM cells tended to decrease or remained unchanged (Table 1).
Table 1.
Phenotypic analysis of cells in the BM by flow cytometry*
| Day post infection |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Cell phenotype | 0 | 1 | 3 | 5 | 7 | 10 | 14 | 21 | |
| CD34+CD45+ | CD41+CD61+ | 0.08 ± 0.04 | 0.23 ± 0.04 | 0.07 ± 0.04 | 0.18 ± 0.09 | 0.06 ± 0.02 | 0.03 ± 0.02 | 0.02 ± 0.00 | 0.04 ± 0.01 |
| CD41+CD61− | 2.95 ± 2.87 | 1.76 ± 1.55 | 0.23 ± 0.12 | 0.94 ± 0.92 | 0.66 ± 0.54 | 0.93 ± 0.89 | 0.58 ± 0.36 | 0.18 ± 0.06 | |
| CD41−CD61+ | 0.03 ± 0.02 | 0.06 ± 0.02 | 0.02 ± 0.01 | 0.05 ± 0.02 | 0.02 ± 0.00 | 0.01 ± 0.01 | 0.00 ± 0.00 | 0.01v0.00 | |
| CD41−CD61− | 0.35 ± 0.31 | 0.33 ± 0.28 | 0.04 ± 0.02 | 0.04 ± 0.03 | 0.04 ± 0.03 | 0.12 ± 0.11 | 0.10 ± 0.04 | 0.04 ± 0.02 | |
| CD34−CD45+ | CD41+CD61+ | 0.19 ± 0.02 | 0.26 ± 0.10 | 0.25 ± 0.13 | 0.06 ± 0.01 | 0.07 ± 0.01 | 0.13 ± 0.09 | 0.11 ± 0.03 | 0.04 ± 0.02 |
| CD41+CD61−† | 65.86 ± 11.28 | 40.66 ± 7.55 | 70.26 ± 2.46 | 69.62 ± 4.54 | 56.33 ± 11.04 | 50.87 ± 5.12 | 59.32 ± 11.77 | 56.01 ± 13.80 | |
| CD41−CD61+ | 0.03 ± 0.02 | 0.05 ± 0.03 | 0.01 ± 0.01 | 0.02 ± 0.01 | 0.01 ± 0.01 | 0.01 ± 0.00 | 0.01 ± 0.01 | 0.01 ± 0.00 | |
| CD41−CD61− | 9.86 ± 3.95 | 9.81 ± 4.89 | 3.61 ± 1.15 | 1.63 ± 0.08 | 1.93 ± 0.77 | 3.02 ± 0.87 | 4.91 ± 1.11 | 3.36 ± 0.59 | |
| CD34−CD45− | CD41+CD61+ | 0.27 ± 0.07 | 0.67 ± 0.35 | 0.36 ± 0.13 | 0.12 ± 0.06 | 0.33 ± 0.11 | 0.46 ± 0.18 | 0.47 ± 0.04 | 0.23 ± 0.10 |
| CD41+CD61−‡ | 11.23 ± 3.22 | 23.94 ± 3.14 | 17.71 ± 4.00 | 21.97 ± 4.53 | 27.04 ± 6.31 | 34.24 ± 3.17 | 22.41 ± 5.91 | 29.62 ± 7.12 | |
| CD41−CD61+ | 0.01 ± 0.00 | 0.04 ± 0.02 | 0.01 ± 0.01 | 0.01 ± 0.00 | 0.01 ± 0.01 | 0.00 ± 0.00 | 0.01 ± 0.01 | 0.01 ± 0.00 | |
| CD41−CD61− | 8.99 ± 5.20 | 22.05 ± 5.25 | 7.40 ± 1.94 | 5.29 ± 0.68 | 13.43 ± 4.96 | 10.00 ± 3.60 | 11.99 ± 5.22 | 10.41 ± 6.14 | |
Percentage of gated population (mean ± standard error of mean; n = 3, RM#4 to RM#6) with two-way analysis of variance.
Statistically predominant in comparison with other CD34−CD45+ cell populations (p < 0.001) at all time points tested.
Constantly elevated as compared to both CD34−CD45−CD41+CD61+ and CD34−CD45−CD41−CD61+ cells at days 1, 5, 7, 10, 14, and 21 (p < 0.001) and at day 3 (p < 0.01) post infection and to CD34−CD45−CD41−CD61− cells at day 5 (p < 0.01), day 7 (p < 0.05), days 10 and 21 (p < 0.001) post infection.
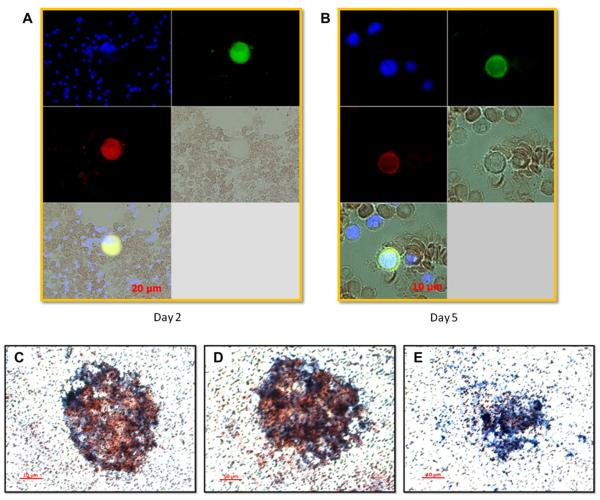
Attempts were made to identify the lineage of cells in the BM that were positive for dengue viral antigen utilizing a monoclonal antibody specific for dengue virus E (clone 3H5) and immunofluorescence techniques. Kinetic studies of smear slides prepared from BM samples were performed. Results as shown in Figure 4 demonstrated that the predominant cells positive for dengue viral antigen were CD61+ cells. Cells that were positive for dengue viral antigen appeared to have giant cell morphology that were observed starting on day 2 post infection (Fig. 4A), and those presenting a high nucleus to cytoplasm ratio became prominent on day 5 post infection (Fig. 4B). Additional experiments were conducted to recover intact giant cells (presumably megakaryocyte) from the BM by utilizing a previously established method [20]. Costaining with monoclonal antibodies for dengue virus E and CD41a indeed confirmed that megakaryocytes were positive for dengue viral antigen (note that the cell membrane appeared to be in the process of demarcation) (Fig. 4C, D). Altogether, data from these findings indicate that the DENV-antigen–positive cells belong to the megakaryocytic lineage as defined by the coexpression of CD41 and CD61. It has been documented previously that megakaryocytic cells, in addition to the expression of CD61 and CD41, also express the chemokine receptor CCR5, which becomes downregulated during the course of human immunodeficiency virus infection [27]. Therefore, it is possible that CCR5, which is the receptor for the chemokines macrophage inflammatory protein-1a, macrophage inflammatory protein-1b, and RANTES, may be an important molecule contributing to or preventing DENV disease in response to infection.
Figure 4.
Immunostaining of BM cells for megakaryocytic cell markers and dengue viral antigen. BM aspirates were smeared onto slides and stained with antibodies specific for cell markers and dengue antigen as described in the Materials and Methods. (A) A giant cell with megakaryocytic marker CD61 (green) was positive for dengue viral antigen (red) on day 2 after infection. (B) A cell with a high ratio of marker CD61 (green) on the cell surface was also positive for dengue viral antigen (red) on day 5 after infection. Large cells were collected from BM aspirate and subjected to double immunohistochemical staining with either dengue-specific antibody or isotype-matched control antibody and anti-CD41a antibody as described in the Materials and Methods. (C, D) Large cells with visible demarcation membrane were stained positive for both CD41a (blue), a megakaryocytic marker, and dengue antigen (red). (E) Cells stained with isotype control and CD41a-specific antibodies served as controls.
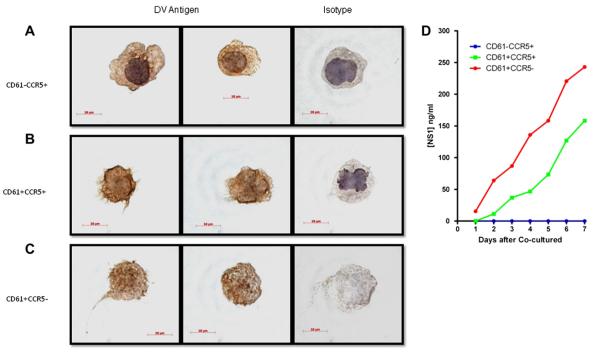
This view prompted us to carry out more detailed studies in efforts to determine the involvement of CD61 and CCR5 during DENV infection of the BM. Mononuclear cells isolated from the BM of a DENV-infected monkey were subjected to immunofluorescent staining and flow cytometry– assisted cell sorting to prepare highly enriched populations of CD61 and CCR5 coexpressing cells. The gating strategy utilized is illustrated in Supplementary Figure E2 (online only, available at www.exphem.org). As shown in Supplementary Figure E3 (online only, available at www. exphem.org), the grape-like appearing CD61+ cells from the post-enrichment fractions were positive for dengue viral antigen. In addition, highly enriched populations of cells expressing CD61+CCR5−, CD61+CCR5+, and CD61−CCR5+ were isolated (Table 2). Varying frequencies of these three cell populations with distinct morphologies were dengue viral antigen–positive as evidenced by immunohistochemical staining (Fig. 5). Among these cells, virtually all of the CD61+CCR5− cell sorter–enriched subset of cells were dengue viral antigen–positive (Fig. 5C and Table 2). This subset lacked the presence of a visible nuclear contour and represented the lowest frequency of the three subsets enriched. The specific targeting of the DENV to this CD61+CCR5− subset was confirmed by co-culturing each of the subsets with Vero cells. Thus, as seen in Table 2, coculture of the CD61+CCR5− cells with Vero cells yielded the highest recoverable levels of infectious virus in the culture supernatant as compared to the coculture of the other two sorted cell subsets (Table 2). The detection of the nonstructural protein 1, a surrogate marker for dengue viral replication in Vero cells, further confirms the presence of infectious DENV in these cells (Fig. 5D). As a whole, although the number of CD61+CCR5− cells was significantly lower, the percentage of cocultured cells positive for dengue viral antigen was far higher (Fig. 5C and Table 2) than those of CD61−CCR5+ and CD61+CCR5+ populations (Fig. 5A and B, Table 2). However, to our surprise, even though dengue viral antigen (about 15% of the cells) was detected in the CD61−CCR5+ cells, infectious virus could not be recovered from the cells, despite several repeated attempts at coculturing them with the Vero cells (Table 2). These results suggest that cells with CCR5 phenotype maybe involved in viral clearance.
Table 2.
Properties of subsets of BM cells isolated by flow cytometry–assisted cell sorting from DENV–infected rhesus monkeys*
| RM#4 |
RM#5 |
RM#6 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Purity (%) | No. of cells |
Ag+ (%) | Coculture | Purity (%) | No. of cells |
Ag+ (%) | Coculture | Purity (%) | No. of cells |
Ag+ (%) | Coculture | |
| CD61+CCR5− | 82 | 101 | 100 | + | 88 | 168 | 100 | + | 91 | 176 | 100 | + |
| CD61+CCR5+ | 81 | 7640 | 45 | + | 83 | 9800 | 38 | + | 84 | 10170 | 25 | + |
| CD61−CCR5+ | 95 | 3120 | 20 | − | 94 | 5200 | 18 | − | 92 | 6400 | 10 | − |
Frozen and pooled BM mononuclear cells obtained on day 1, 3, 5, and 7 post infection from each animal were stained with appropriate monoclonal antibodies and highly enriched population of phenotypically defined subsets were isolated. The cell sorter strategy is described in Supplementary Figure E2 (online only, available at www.exphem.org). The purity and number of sorted cells are shown. The sorted cells were assessed for the expression of DENV antigen by immunofluorescent staining as well as for the presence of infectious DENV by coculture with Vero cells and subsequent qRT-PCR using culture supernatant. (+), recovery of infectious virus in the coculture as determined by real-time qRT-PCR; (−), no detectable virus was found in the coculture within a 7-day period of study.
Figure 5.
Staining of cells sorted from the BM of infected rhesus monkeys for dengue viral antigen and virus isolation. Mononuclear cells purified from BM aspirates of all infected monkeys were pooled and subjected to fluorescence-activated cell sorting as described in Supplementary Figure E2 (online only, available at www.exphem.org). Cells were then cytospinned down onto slides and immunostaining for DENV antigen was performed as described in the Materials and Methods. All three populations of cells were observed positive for dengue antigen, (A) CD61−CCR5+, (B) CD61+CCR5+, and (C) CD61+CCR5−, respectively. The surface of CD61+ cells characteristically shows grape-like or bleb appearance, indicating young platelets about to be detached from the cell body [35,36]. (D) Kinetics of dengue viral antigen, nonstructural protein 1 (NS1), in the supernatants of coculture assays. NS1 concentration was measured by enzyme-linked immunosorbent assay using purified NS1 antigen as a standard control.
Discussion
Results of the studies reported here, using biological BM specimens from the rhesus macaque model we described earlier, document the contribution of platelets and the megakaryocytic cell lineage as a potential target for DENV infection, consistent with the initial observation in human dengue patients [18]. Additionally, dengue viral RNA could be detected in both platelets and plasma isolated from the BM of the same animals. The amount of dengue viral RNA in the BM peaked at approximately days 1 to 3 post infection, which subsequently declined. The peak viral load in the peripheral blood from the same macaques, however, was observed at later time points, i.e., 3 to 5 days post infection [16]. These data suggest that DENV can reach the BM compartment within a short period of time after experimental infection and, by implication, shortly after the mosquito bite. We interpret these findings as evidence that DENV likely targets the BM compartment for its first round of replication, and then re-enters the circulating blood. Because the BM is the major source of hematopoietic cells, the production of which is tightly regulated, dysregulation within this microenvironment results in abnormalities in the BM mass, which is then reflected by a skewed profile of peripheral blood cells a couple of days later. Indeed, our results demonstrate a transient surge on day 1 and a subsequent drop in the BM mass on days 2 to 10 post infection, concurrent with the rapid decrease of viral load observed in the plasma at later time points post infection. Therefore, it is likely that the BM compartment plays a critical role in acute DENV production and in dengue pathogenesis. The data presentedhere support the previously accumulated data that have suggested a prominent role for hematopoietic progenitor cells in dengue pathogenesis. Whether the fluctuations in the cellular composition of the BM, which occurs as a function of age, can account for the differences in susceptibility to DENV, warrants further investigation.
Timing is a key factor that needs to be considered when attempting to obtain accurate information about the involvement of the BM in the context of DENV infection. The fact that virus infection occurs after accidental transfusion of DENV–infected BM indicates the early involvement of the BM cells in DENV infection, before development of clinical symptoms [28]. Because individuals usually do not seek medical help until clinical symptoms dictate it, the BM analysis from these patients presenting at this stage show either hypercellularity or a normal profile [1,29]. Even though transient BM suppression has been registered in dengue patients at early time points of infection, information about its early involvement in virus production and pathogenesis in acute dengue disease is largely unexplored. Lack of a suitable animal model that recapitulates all the cardinal features of the human dengue has hindered the progress of the investigation of BM in dengue pathogenesis. Thus, taking advantage of the nonhuman dengue primate model, our study focused on the utilization of immunohistological staining and cell-sorter techniques for the isolation and identification of highly enriched population of BM cell subsets from DENV–infected monkeys. Results from these studies revealed that the DENV permissive cells display a prototype megakaryocytic cell lineage characteristic, as evidenced by the expression of the cell surface marker(s), CD61 and/or CD41. The data presented here strongly suggest that megakaryocytes in the BM are the likely major target cell lineage of DENV, which could be one of the contributing factors to thrombocytopenia seen in dengue patients.
Further investigation on the phenotype of the cells positive for dengue viral antigen suggested that cells expressing CD61 and/or CCR5 may be involved in DENV infection in vivo. Although several of the subsets expressing CD61 were found to be positive for dengue viral antigen, a distinct difference was noted when attempting to recover infectious virus using coculture techniques. Dengue virus could be recovered upon coculture of the CD61+CCR5− and the CD61+CCR5+ subsets of cells with the Vero indicator cells, with the percentage of viral antigen–positive cells and level of infectious virus significantly higher in the former cell subset. Hence, the results indicate that DENV may preferentially target the CD61+CCR5− cell population in the BM. The precise reasons for the increased susceptibility of this sublineage of cells, however, remain to be defined. The inflammatory milieu caused by dengue infection of the BM may also result in the recruitment of immune cells, which may attempt to eliminate the infected cells through phagocytosis [30]. This explanation may account for the inability to recover infectious virus from the CD61−CCR5+ cells, despite the fact that some of these cells display dengue viral antigen. It is possible that some of the CD61+CCR5+ cells may have acquired DENV antigen via phagocytic activity as well as through direct infection. Interestingly, this view is consistent with previous reports of a failure to isolate DENV from liver tissues obtained immediately after the death of the patient, despite the presence of readily detectable levels of dengue viral antigen in liver Kupffer cells [31–33]. Thus, more research is warranted to determine the reasons for the failure to isolate infectious dengue virions from tissues that are clearly positive for dengue viral antigen using immunofluorescent type of techniques. Taken together, the data obtained from our nonhuman primate animal model indicate the potential contribution of megakaryocytes in the BM compartment to the life cycle of DENV infection. It has been suggested that megakaryocytes share several common features with hematopoietic stem cells [34]; thus, we could not rule out a possibility that the cells observed in our study are hematopoietic stem cells. Nevertheless, whether this cell type participates in disseminating virus to other parts of the human body and plays an important role in virus replication, transmission, and disease pathogenesis needs to be delineated.
Supplementary Material
Supplementary Figure E1. Two representative photographs of cells (A, B) showing platelet-like vesicles budding from the cell surface in freshly prepared blood smears from dengue-infected rhesus monkeys. Freshly prepared peripheral blood mononuclear cells were applied onto slides and subjected to periodic acid-Schiff assay.
Supplementary Figure E2. Flow cytometric strategy to sort cells expressing the cells surface markers CD61+ and/or CCR5+. BM aspirates were pooled and subjected to antibody staining and a two-step fluorescence-activated cell sorting was applied as described in the Materials and Methods. Figure at the bottom shows the purity of the sorted cells upon reanalysis.
Supplementary Figure E3. Representative photomicrograph depicting the immunostaining profile of post flow cytometry obtained enriched subset of cells that confirms the presence of dengue antigen within CD61+ cells. Cells collected from post enrichment were subjected to cytospin onto slides and subjected to immunostaining. (A) Immunohistochemical staining depicts dengue antigen (brown) and nucleus (light blue). (B) Immunofluorescence staining depicts CD61 (green), a megakaryocytic marker, dengue antigen (red) and nucleus, 4′,6-diamidino-2-phenylindole (blue).
Supplementary Table E1. Viral load in the BM of DENV-infected rhesus monkeys*
Acknowledgments
We thank the veterinary and research staff of the Yerkes National Primate Center at Emory for technical assistance as well as the Yerkes animal care staff for their excellent support and care of the animals involved in this study. The authors would like to appreciate the help, guidance, suggestions and discussions provided by Dr. Tristram Parslow and Dr. Shiyoung Li from the Department of Pathology and Laboratory Medicine at Emory University School of Medicine. The authors would like to thank your Dr. Hong Yi of the EM core facility of Emory University.
Funding disclosure The research was supported in part by the U19 Pilot Project Funds U19 AI057266 (RFA-AI-02-042), National Institutes of Health/Southeastern Regional Center of Excellence for Emerging Infections and Biodefense, Emory University Research Committee grants, Howard Hughes Medical Institute Med into Grad Initiative (KBC), and the NCRR p51 support to the Yerkes National Primate Research Center DRR000165.
Footnotes
Author contributions: Sansanee Noisakran discussed the experimental strategy, performed EM and fluorescence-activated cell sorting (FACS) experiments and analyses, and partially wrote the manuscript; Nattawat Onlamoon designed and performed the FACS experiments and analyses; Hui-Mien Hsiao performed the immunohistochemistry staining, real-time PCR and enzyme-linked immunosorbent assay; Kristina B. Clark discussed the experimental strategy, performed the subset of the EM and FACS assays, and edited the manuscript; Francois Villinger discussed the experimental strategy, directed the monkey experiments and edited the manuscript; Aftab A. Ansari discussed the experimental strategy, assisted in approval of Institutional Animal Care and Use Committee (IACUC) protocol, and edited the manuscript, and Guey Chuen Perng designed, defined and discussed the experiment strategy, wrote the IACUC protocol, and wrote the manuscript.
Conflict of interest disclosure No financial interest/relationships with financial interest relating to the topic of this article have been declared.
Supplementary data related to this article can be found online at doi:10. 1016/j.exphem.2011.11.011.
References
- 1.Bierman HR, Nelson ER. Hematodepressive virus diseases of Thailand. Ann Intern Med. 1965;62:867–884. doi: 10.7326/0003-4819-62-5-867. [DOI] [PubMed] [Google Scholar]
- 2.Nisalak A, Halstead SB, Singharaj P, Udomsakdi S, Nye SW, Vinijchaikul K. Observations related to pathogenesis of dengue hemorrhagic fever. 3. Virologic studies of fatal disease. Yale J Biol Med. 1970;42:293–310. [PMC free article] [PubMed] [Google Scholar]
- 3.Au WY, Ma ES, Kwong YL. Acute myeloid leukemia precipitated by dengue virus infection in a patient with hemoglobin H disease. Haematologica. 2001;86:E17. [PubMed] [Google Scholar]
- 4.Young NS. Flaviviruses and bone marrow failure. JAMA. 1990;263:3065–3068. [PubMed] [Google Scholar]
- 5.Srichaikul T, Punyagupta S, Kanchanapoom T, Chanokovat C, Likittanasombat K, Leelasiri A. Hemophagocytic syndrome in Dengue hemorrhagic fever with severe multiorgan complications. J Med Assoc Thai. 2008;91:104–109. [PubMed] [Google Scholar]
- 6.Nakao S, Lai CJ, Young NS. Dengue virus, a flavivirus, propagates in human bone marrow progenitors and hematopoietic cell lines. Blood. 1989;74:1235–1240. [PubMed] [Google Scholar]
- 7.Bente DA, Melkus MW, Garcia JV, Rico-Hesse R. Dengue fever in humanized NOD/SCID mice. J Virol. 2005;79:13797–13799. doi: 10.1128/JVI.79.21.13797-13799.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halstead SB, O’Rourke EJ, Allison AC. Dengue viruses and mononuclear phagocytes. II. Identity of blood and tissue leukocytes supporting in vitro infection. J Exp Med. 1977;146:218–229. doi: 10.1084/jem.146.1.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farrar J. In: Clinical features of dengue. Halstead SB, editor. Imperial College Press; Dengue. London: 2008. pp. 171–191. [Google Scholar]
- 10.Halstead SB. First ICMR Seminar on Dengue and Dengue Hemorrhagic Fever. ICMR; Kobe University School of Medicine, Kobe, Japan: 1980. Pathology and pathogenesis of DHF; p. 215. [Google Scholar]
- 11.World Health Organization . Dengue and dengue hemorrhagic fever. World Health Organization; Geneva: 2009. [Google Scholar]
- 12.Thisyakorn U, Nimmannitya S, Ningsanond V, Soogarun S. Atypical lymphocyte in dengue hemorrhagic fever: its value in diagnosis. Southeast Asian J Trop Med Public Health. 1984;15:32–36. [PubMed] [Google Scholar]
- 13.Chen RF, Liu JW, Yeh WT, et al. Altered T helper 1 reaction but not increase of virus load in patients with dengue hemorrhagic fever. FEMS Immunol Med Microbiol. 2005;44:43–50. doi: 10.1016/j.femsim.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 14.Rothwell SW, Putnak R, La Russa VF. Dengue-2 virus infection of human bone marrow: characterization of dengue-2 antigen-positive stromal cells. Am J Trop Med Hyg. 1996;54:503–510. doi: 10.4269/ajtmh.1996.54.503. [DOI] [PubMed] [Google Scholar]
- 15.Kuruvilla JG, Troyer RM, Devi S, Akkina R. Dengue virus infection and immune response in humanized RAG2(-/-)gamma(c)(-/-) (RAG-hu) mice. Virology. 2007;369:143–152. doi: 10.1016/j.virol.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 16.Onlamoon N, Noisakran S, Hsiao HM, et al. Dengue virus-induced hemorrhage in a nonhuman primate model. Blood. 2010;115:1823–1834. doi: 10.1182/blood-2009-09-242990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Institute of Laboratory Animal Research. Commission on Life Sciences. National Research Council . Guide for the care and use of laboratory animals. The National Academic Press; Washington, DC: 1996. [Google Scholar]
- 18.Noisakran S, Gibbons RV, Songprakhon P, et al. Detection of dengue virus in platelets isolated from dengue patients. Southeast Asian J Trop Med Public Health. 2009;40:253–262. [PubMed] [Google Scholar]
- 19.Yi H, Leunissen J, Shi G, Gutekunst C, Hersch S. A novel procedure for pre-embedding double immunogold-silver labeling at the ultra-structural level. J Histochem Cytochem. 2001;49:279–284. doi: 10.1177/002215540104900301. [DOI] [PubMed] [Google Scholar]
- 20.Wilde NT, Burgess R, Keenan DJ, Lucas GS. The effect of cardiopulmonary bypass on circulating megakaryocytes. Br J Haematol. 1997;98:322–327. doi: 10.1046/j.1365-2141.1997.2373055.x. [DOI] [PubMed] [Google Scholar]
- 21.Wang S, He R, Patarapotikul J, Innis BL, Anderson R. Antibody-enhanced binding of dengue-2 virus to human platelets. Virology. 1995;213:254–257. doi: 10.1006/viro.1995.1567. [DOI] [PubMed] [Google Scholar]
- 22.Honda S, Saito M, Dimaano EM, et al. Increased phagocytosis of platelets from patients with secondary dengue virus infection by human macrophages. Am J Trop Med Hyg. 2009;80:841–845. [PubMed] [Google Scholar]
- 23.Smith TJ, Brandt WE, Swanson JL, McCown JM, Buescher EL. Physical and biological properties of dengue-2 virus and associated antigens. J Virol. 1970;5:524–532. doi: 10.1128/jvi.5.4.524-532.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Junjhon J, Lausumpao M, Supasa S, et al. Differential modulation of prM cleavage, extracellular particle distribution, and virus infectivity by conserved residues at nonfurin consensus positions of the dengue virus pr-M junction. J Virol. 2008;82:10776–10791. doi: 10.1128/JVI.01180-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Junt T, Schulze H, Chen Z, et al. Dynamic visualization of thrombopoiesis within bone marrow. Science. 2007;317:1767–1770. doi: 10.1126/science.1146304. [DOI] [PubMed] [Google Scholar]
- 26.Kosaki G. Platelet production by megakaryocytes: protoplatelet theory justifies cytoplasmic fragmentation model. Int J Hematol. 2008;88:255–267. doi: 10.1007/s12185-008-0147-7. [DOI] [PubMed] [Google Scholar]
- 27.Mondal D, Williams CA, Ali M, Eilers M, Agrawal KC. The HIV-1 Tat protein selectively enhances CXCR4 and inhibits CCR5 expression in megakaryocytic K562 cells. Exp Biol Med (Maywood) 2005;230:631–644. doi: 10.1177/153537020523000905. [DOI] [PubMed] [Google Scholar]
- 28.Rigau-Perez JG, Vorndam AV, Clark GG. The dengue and dengue hemorrhagic fever epidemic in Puerto Rico, 1994-1995. Am J Trop Med Hyg. 2001;64:67–74. doi: 10.4269/ajtmh.2001.64.67. [DOI] [PubMed] [Google Scholar]
- 29.La Russa VF, Innis BL. Mechanisms of dengue virus-induced bone marrow suppression. Baillieres Clin Haematol. 1995;8:249–270. doi: 10.1016/s0950-3536(05)80240-9. [DOI] [PubMed] [Google Scholar]
- 30.Saito K, Hirokawa M, Inaba K, et al. Phagocytosis of codeveloping megakaryocytic progenitors by dendritic cells in culture with thrombopoietin and tumor necrosis factor-alpha and its possible role in hemophagocytic syndrome. Blood. 2006;107:1366–1374. doi: 10.1182/blood-2005-08-3155. [DOI] [PubMed] [Google Scholar]
- 31.Singharaj P, Nisalak A, Halstead SB. Attempts at virus recovery from patients dying of Thai haemorrhagic fever. Bull World Health Organ. 1966;35:57–58. [PMC free article] [PubMed] [Google Scholar]
- 32.Bhamarapravati N, Boonyapaknavik V. Pathogenetic studies on Thai haemorrhagic fever: immunofluorescent localization of dengue virus in human tissue. Bull World Health Organ. 1966;35:50–51. [PMC free article] [PubMed] [Google Scholar]
- 33.Jessie K, Fong MY, Devi S, Lam SK, Wong KT. Localization of dengue virus in naturally infected human tissues, by immunohistochemistry and in situ hybridization. J Infect Dis. 2004;189:1411–1418. doi: 10.1086/383043. [DOI] [PubMed] [Google Scholar]
- 34.Huang H, Cantor AB. Common features of megakaryocytes and hematopoietic stem cells: what’s the connection? J Cell Biochem. 2009;107:857–864. doi: 10.1002/jcb.22184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Djaldetti M, Fishman P, Bessler H, Notti I. SEM observations on the mechanism of platelet release from megakaryocytes. Thromb Haemost. 1979;42:611–620. [PubMed] [Google Scholar]
- 36.Polliack A, Leizerowitz R, Berrebi A, Gurfel D, Gamliel H. Surface features of leukaemic megakaryocytic precursors. A study of 5 cases of megakaryoblastic leukaemia with scanning electron microscopy. Scand J Haematol. 1983;30:145–150. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure E1. Two representative photographs of cells (A, B) showing platelet-like vesicles budding from the cell surface in freshly prepared blood smears from dengue-infected rhesus monkeys. Freshly prepared peripheral blood mononuclear cells were applied onto slides and subjected to periodic acid-Schiff assay.
Supplementary Figure E2. Flow cytometric strategy to sort cells expressing the cells surface markers CD61+ and/or CCR5+. BM aspirates were pooled and subjected to antibody staining and a two-step fluorescence-activated cell sorting was applied as described in the Materials and Methods. Figure at the bottom shows the purity of the sorted cells upon reanalysis.
Supplementary Figure E3. Representative photomicrograph depicting the immunostaining profile of post flow cytometry obtained enriched subset of cells that confirms the presence of dengue antigen within CD61+ cells. Cells collected from post enrichment were subjected to cytospin onto slides and subjected to immunostaining. (A) Immunohistochemical staining depicts dengue antigen (brown) and nucleus (light blue). (B) Immunofluorescence staining depicts CD61 (green), a megakaryocytic marker, dengue antigen (red) and nucleus, 4′,6-diamidino-2-phenylindole (blue).
Supplementary Table E1. Viral load in the BM of DENV-infected rhesus monkeys*