Abstract
Bdellovibrio bacteriovorus is a bacterial parasite with an unusual lifestyle. It grows and reproduces in the periplasm of a host prey bacterium. The complete genome sequence of B. bacteriovorus has recently been reported. We have reanalyzed the transport proteins encoded within the B. bacteriovorus genome according to the current content of the transporter classification database (TCDB). A comprehensive analysis is given on the types and numbers of transport systems that B. bacteriovorus has. In this regard, the potential protein secretory capabilities of at least 4 types of inner membrane secretion systems and 5 types for outer membrane secretion are described. Surprisingly, B. bacteriovorus has a disproportionate percentage of cytoplasmic membrane channels and outer membrane porins. It has far more TonB/ExbBD-type systems and MotAB-type systems for energizing outer membrane transport and motility than does E. coli. Analysis of probable substrate specificities of its transporters provides clues to its metabolic preferences. Interesting examples of gene fusions and of potentially overlapping genes were also noted. Our analyses provide a comprehensive, detailed appreciation of the transport capabilities of B. bacteriovorus. They should serve as a guide for functional experimental analyses.
Keywords: Bacterial parasitism, transport, genome analyses, vectorial metabolism, protein secretion
Bdellovibrio bacteriovorus is a Gram-negative δ-proteobacterium that preys on other Gram-negative bacteria [1]. B. bacteriovorus penetrates the outer membrane of its prey and grows intraperiplasmically [2]. There it differentiates from the attack phase cell into the growth phase cell [1,3]. It loses its flagellum and initiates growth. At this point, B. bacteriovorus modifies the host cell peptidoglycan [4,5] and converts the host cell into a spherical structure called a bdelloplast in a process dependent on glycanase [2,6]. Not until 45 minutes after initiating the growth phase does DNA replication begin. During the following 2-3 hours, B. bacteriovorus causes extensive host cell damage and grows into a long coiled filament [7]. Late steps in the differentiation cycle can be completed outside of the host cell [8]. Although wild-type B. bacteriovorus is an obligate parasite, it can be mutated to grow in culture [9].
The B. bacteriovorus developmental cycle has been divided into eight phases according to morphological and physiological observations [7,10,11]: (1) The attack phase: B. bacteriovorus swims rapidly and collides with its prey, remaining reversibly attached for a short “recognition” period [12]. (2) Irreversible attachment: Active adhesion, possibly involving multiple fimbriae, occurs at the pole opposite the flagellum. (3) Invasion: B. bacteriovorus forms a “penetration pore” in the host cell outer membrane and cell wall [13]. Invasion may involve retractive fimbriae pulling the prey through the pore [11]. Pore formation is believed to occur when B. bacteriovorus locally secretes hydrolytic enzymes to degrade outer envelope constituents. Before entry into the periplasm, B. bacteriovorus frequently sheds its flagellum [10]. The pore is ultimately resealed by the host cell. (4) Macromolecular synthesis: B. bacteriovorus initiates macromolecular (RNA, protein, lipid, polysaccharide) synthesis. The first round of DNA synthesis occurs [2,14]. Since B. bacteriovorus can synthesize only 11 amino acids, protein synthesis depends on the uptake of host degradation products [11]. (5) Bdelloplast formation: The rod-shaped host cell rounds up into the spherical bdelloplast [2,15], and cell growth continues. (6) Septation: After formation of a single long snake-like cell, B. bacteriovorus synchronously undergoes septation, generating multiple progeny cells. (7) Flagellation: The progeny cells synthesize flagella while in the exhausted host bdelloplast. (8) Exit phase: B. bacteriovorus secretes novel hydrolytic enzymes that cause bdelloplast lysis. Release of the progeny attack cells is achieved [16]. This progression of developmental events may be initiated and regulated by a set of sensor kinase/response regulator systems and orchestrated by a sigma factor cascade similar in principle to that established for Bacillus sporulation [11,17,18].
Throughout most of the growth phase, prey cytoplasmic and integral membrane proteins are degraded, as are other host cell macromolecules [19,20]. There is evidence that B. bacteriovorus secretes proteins, possibly porins [21] but definitely many degradative enzymes [20]. Many of these appear in the host cell cytoplasm although the mechanisms by which they get there are unknown. Upon completion of growth, the single filamentous cell septates, giving rise to multiple motile cells, their numbers depending on the size of the prey cell [22]. Extensive signaling between the predator and prey bacteria seems to be operative [23,24]. B. bacteriovorus has the potential of being a therapeutic agent for treatment of Gram-negative bacterial infections [11,25,26,27].
In 2004, the genome sequence of B. bacteriovorus was published [11], allowing prediction of its physiology from its gene content. The single circular chromosome contains 3.8 Mbp and includes an estimated 3,600 coding regions. Only 55% of the deducted proteins were assigned a putative function based on homology searches. The rest were of unknown function. The transport systems predicted by annotation of the genome sequence were shown to fall in the classes of ABC-type transporters and MFS permeases or secondary carriers belonging to other transporter families. In this analysis, we are updating the list of transport systems according to the content of the January 2006 version of the transporter classification database (TCDB; http://www.tcdb.org/). We propose possible substrates and functions for some of these transporters.
B. bacteriovorus exhibits a number of properties that suggest the need for a most unusual complement of transport proteins. Several of its metabolic pathways may be incomplete, based on the available gene function annotations [11]. Neither oxidation nor fermentation of carbohydrates, organic acids or alcohols has been demonstrated [1,28]. Biochemically, this organism lacks the phosphoenolpyruvate-dependent sugar transporting phosphotransferase system (PTS) [19]. It seems to depend primarily on non-carbohydrate macromolecular metabolism for carbon and energy [1,29,30]. It secretes many macromolecular degradative enzymes including carbohydrases, proteases, nucleases and lipases [1,19,20]. Thus, it must have a tremendous capacity for protein secretion across both of its membranes [20]. It appears to grow largely at the expense of host cell proteins, nucleic acids, and membrane and cell wall constituents [1,31]. For example, up to 80% of degraded host nucleic acids is incorporated into B. bacteriovorus DNA [32]. However, this is just a fraction of the nucleotides required for growth, so B. bacteriovorus must be capable of making all of the nucleotides required for DNA synthesis. In fact, complete pathways for purine and pyrimidine biosynthesis are encoded within the genome [11].
Only a few molecular transport activities in B. bacteriovorus have been characterized, and even fewer transport proteins have been associated with these activities. The energy-dependent uptake of intact nucleotides (UMP and ATP) has been demonstrated [33,34]. Uptake of nucleotides appears to be a rare trait for a bacterium, but at least two such systems appear to exist in B. bacteriovorus. Neither has been characterized in molecular terms.
We have reanalyzed the complement of recognizable transporters encoded within the B. bacteriovorus genome 2 years after its original annotation, as a plethora of new sequences have been made publicly available during this time. The B. bacteriovorus genome analyses reported by Rendulic et al. [11] identified transporters as either permeases or ABC-type transporters. Here we present a systematic classification of the B. bacteriovorus transporters based on the TC system [35], which facilitates a more detailed understanding of transport function and evolution [36]. The methodology used has been described previously [37].
The genome of B. bacteriovorus reveals the presence of potential efflux pumps for hydrophobic and amphipathic drugs and organic solvents as well as potential uptake systems for amino acids, peptides and inorganic anions. The general secretory (Sec) system, the twin arginine targeting protein translocation system, and ABC-type protein secretion systems are also discussed. B. bacteriovorus encodes complete flagellar and fimbrial protein export systems and probably a type II main terminal branch (MTB; TC #3.A.15) for secretion of proteins across the outer membrane [38]. Several other protein export systems are described as well. These systems must account for the unusual parasitic lifestyle of this bacterium.
Results
Overview of Transporter Types
Table 1 presents an overall summary of the classes of transporters found in B. bacteriovorus. The 406 transport proteins make up 172 transport systems. 11.3% of the genes encode recognizable transport proteins, corresponding to established entries in TCDB. An additional 161 (4.5%) of its genes encode potential transporters that, however, do not give good hits in TCDB (see below). The total potential percent of transport protein encoding genes is therefore 15.8%. Since in most free-living bacteria, 10-15% of the genes encode transport proteins [39], B. bacteriovorus may contain an unusually large number of transporters especially considering the fact that the genomes of most intracellular parasites encode lower proportions of transport proteins [39]. The B. bacteriovorus genome encodes higher numbers of ABC-type transporters than most other bacterial genomes analyzed [11].
Table 1.
Overview of the Bdellovibrio bacteriovorus transporter analyses.
| TC Classa | Class Description | No. of Transport Proteinsb | TC Subclass | Subclass Description | No. of Transport Proteinsb |
|---|---|---|---|---|---|
| 1. | Channels | 45 (29) | 1.A | α-type channels | 15 (12) |
| 1.B | β-barrel porins | 29 (16) | |||
| 1.C | Pore-forming toxins (proteins and peptides) | – | |||
| 1.D | Nonribosomally synthesized channels | – | |||
| 1.E | Holins | 1 (1) | |||
| 2. | Secondary carriers | 100 (70) | 2.A | Porters (uniporters, symporters, antiporters) | 73 (64) |
| 2.B | Nonribosomally synthesized porters | – | |||
| 2.C | Ion-gradient-driven energizers | 27 (6) | |||
| 3. | Primary active transporters | 227 (55) | 3.A | P-P-bond-hydrolysis-driven transporters | 202 (51) |
| 3.B | Decarboxylation-driven transporters | 4 (-) | |||
| 3.C | Methyltransfer-driven transporters | – | |||
| 3.D | Oxidoreduction-driven transporters | 21 (4) | |||
| 3.E | Light absorption-driven transporters | – | |||
| 4. | Group translocators | 0 | 4.A | Phosphotransfer-driven group translocators | - |
| 4.B | Nicotinamide ribonucleoside (NR) uptake permeases | - | |||
| 5. | Transmembrane electron carriers | 4 (2) | 5.A | Transmembrane 2-electron transfer carriers | 4 (2) |
| 5.B | Transmembrane 1-electron transfer carriers | – | |||
| 8. | Auxiliary transport proteins | 7 (-) | 8.A | Auxiliary transport proteins | 7 (-) |
| 9. | Poorly defined system | 23 (16) | 9.A | Recognized transporters of unknown biochemical mechanism | 4 (3) |
| 9.B | Putative uncharacterized transport proteins | 19 (13) | |||
| 9.C | Functionally characterized transporters lacking identified sequences | – | |||
| Total | 406 (172) | 406 (172) | |||
Transporter classes 6 & 7 have not been assigned in the TC system yet and therefore are absent from this table.
Numbers in parentheses represent the number of transport systems.
In most bacteria, approximately 3-8% of all the transport proteins encoded in the genome are channel-type transporters [39], B. bacteriovorus has a surprisingly large number of recognized channel proteins: 15 inner membrane channel proteins (3.7% of the total number of transport proteins) comprising 12 channels (7% of the transport systems), and 29 outer membrane porin-type channel-forming proteins (7.1% of the recognized transport proteins), corresponding to 16 systems, (9.3% of the transport systems). These numbers presumably reflect a need for rapid, low specificity uptake and export of ions and nutrients, consistent with the unusual lifestyle of B. bacteriovorus. Some of these channels may be expressed only at certain phases of the growth cycle (e.g., phase 4, see Introduction) or in response to specific stress conditions.
B. bacteriovorus has substantially more secondary carriers (70 systems, or 41% of the transport systems identified) than primary active transporters (55 systems, or 32%). The genome sequence gives no indication for group translocators of the phosphoenolpyruvate:sugar phosphotransferase system (PTS), confirming biochemical results of Romo et al. [19]. It has just two recognized transmembrane electron flow carrier, but 16 of its putative transporters fall into the TC class 9 category of poorly defined systems. Many (161) potential transport proteins (see below) have no counterpart in TCDB [35]. This is also an unusually large number for a bacterium with a genome of 3.8 Mbps. This observation may reflect a need for permeases of diverse function.
Transport Substrates
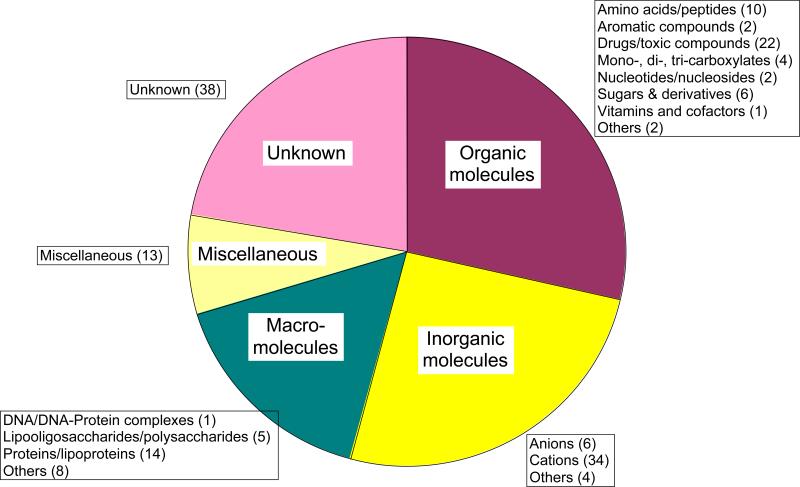
Figure 1 presents a breakdown of the transport systems according to substrate type. Forty-four (26%) of the recognized transporters are specific for inorganic molecules. Of these, the large majority transport cations (20%) while far fewer transport anions (3.5%). Nearly 2.5% are low specificity outer membrane porins.
Figure 1.
Pie chart of transporters in B. bacteriovorus according to predicted substrate specificity. Five different categories are shown. More detailed analyses, including numbers of systems found in each subcategory, specific for each substrate type are provided in the boxes adjacent to the five pieces of the pie. The chart is based on the data presented in Table 2 as discussed in detail in the text.
Small organic molecule transporters show a strong bias for drugs and toxic compounds, there being three times as many of these transporters as there are sugar or vitamin transporters. Systems specific for organic acids [40] are only half as plentiful as those for amino acids [14]. A few systems probably transport aromatic compounds and nucleosides. However, 22 transporters (13%) are drug/toxic compound efflux systems. This last percentage is comparable to, but on the high side relative to that found in many other bacteria.
Half of the macromolecular transporters are probably protein export systems, but lipid and polysaccharide exporters are also present. Nearly 30% of the identified transporters fall into the “miscellaneous” or “unknown” category. We suggest the probable substrates and transport mechanisms of several of these transporters. These results will be discussed in more detail below and on our website (http://www.biology.ucsd.edu/~msaier/supmat/Bba).
Distribution of Topological Types
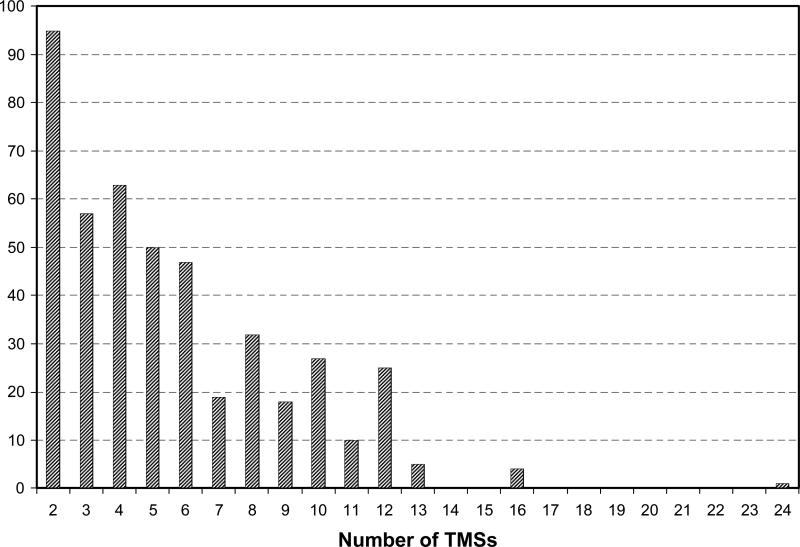
The TMHMM transmembrane helix prediction program [41,42] was used to predict the number of putative transmembrane segments (TMSs) in all the proteins encoded in the B. bacteriovorus genome. Of the 3587 recognized protein-encoding genes in B. bacteriovorus, 2736 (76%) are predicted to have 0 TMSs, and 849 (24%) are predicted to have 1 or more TMSs. Of the latter, 396 (11%) have only 1 TMS. Many of these will be secreted proteins such as periplasmic binding proteins, which require an N-terminal leader sequence to exit the cytoplasm via the general secretory (Sec) pathway. Remaining proteins with 2 or more TMSs (Fig. 2) include 453 proteins (13%), which have the greatest potential of being transporters. However, it should be noted some of these proteins are not involved in transport as discussed below. Further, only about half of the B. bacteriovorus proteins with 2 or more TMSs were classified into transporter families while a majority of the remainder were assigned as putative transporters with no functional information (see below). Of these, 152 (9.2%) have just 2 or 3 TMSs. Many of these are recognizable sensor histidine kinases, but those that function as transporters are likely to be oligomeric pore formers. No proteins with 3 or less TMSs per polypeptide chain have yet been identified that function as carriers [35,43]. Sixty-three proteins (1.8%) have 4 TMSs. These could be either carriers or channels. If carriers, they probably function as dimeric or tetrameric structures [44,45]. Proteins with 5 or more TMSs are likely to be secondary carriers or primary active transporters although some channel proteins are known to have 5 or more TMSs [46,47]. There are nearly equal numbers of predicted 5 and 6 TMS proteins (50 (1.4%) and 47 (1.3%), respectively) encoded within the B. bacteriovorus genome. Carriers of 5 or 6 TMSs generally function as dimers. Transmembrane proteins of 5 TMSs are mostly ABC transporters while those with 6 are primarily secondary carriers (see below). A total of 4.6% of the proteins in B. bacteriovorus have 4-9 TMSs; 7, 8 and 9 TMS carriers are also known [47]. Just 2% of the proteins have 10 or more TMSs. Of these, most are predicted to have 10 or 12 TMSs. As shown in Figure 2, large (≥10 TMSs) proteins with even numbers of TMSs predominate over those predicted to have odd numbers (77% even; 23% odd). We believe this has to do with the pathways taken for their evolutionary appearance [43]. The distribution of topological types is not strikingly different than from those of other bacteria [48,49]. The actual proportion of proteins with even numbers of TMSs may be even greater due to errors in topological prediction.
Figure 2.
Distribution of topological types of putative membrane proteins with 2-24 predicted TMSs. Number of proteins of a particular predicted topological structure is plotted on the X-axis versus the number of TMSs in that protein, plotted on the Y-axis. The plot illustrates the greater prevalence of proteins with even numbers of TMSs than odd numbers of TMSs with the sole exception of the putative 5 TMS protein. These 5 TMS proteins are common in secondary carriers and ATP-hydrolysis-driven uptake transporters of the ABC superfamily (see text). We believe this distribution reflects the evolutionary pathway taken for their appearance [43].
The five largest transmembrane proteins have 16 (4 proteins) and 24 (1 protein) TMSs. The 24 TMS protein (GI:42523740) is a member of the monovalent cation:proton antiporter-3 (CPA3) family (TC #2.A.63) and is a fusion of two previously recognized subunits of this multi-protein transporter complex [50,51]. Transporters of the CPA3 family include subunits that are homologous to subunits of the NADH dehydrogenase complex (TC #3.D.5). Of the four 16 TMS proteins, two (GI:42523556, GI:42523724) are of the CPA2 family (TC #2.A.37). Typical Na:H+ and K+:H+ antiporters of the CPA2 family have up to 14 TMSs (e.g., GmrA of Bacillus megaterium) [52]. The two B. bacteriovorus homologues proved to be fusion proteins where the integral membrane transporters are fused to soluble TrkA-like domains [53], and the two extra TMSs precede the TrkA domain, linking the transporter to this soluble regulatory domain.
The third 16 TMS protein (GI:42523211) is a member of the H+-translocating pyrophosphatase family (TC #3.A.10), most members of which have 16 TMSs [54]. Finally, the fourth 16 TMS protein (GI:42525209) is a homologue of subunit L of the proton-translocating NADH dehydrogenase complex which in other organisms has 16 TMSs.
Predicted Subcellular Localizations of B. bacteriovorus Proteins
We used the PSORTb program [55,56] to predict the subcellular localizations of the proteins in B. bacteriovorus. Of the assigned proteins, 860 proteins (23%) were predicted to be cytoplasmic, while 522 (14.1%) were predicted to be in the cytoplasmic membrane. Of the remaining, 56 (1.4%) may be in the periplasm, 100 (2.5%) may be in the outer membrane, and 31 (0.8%) may be extracellular. However, 2018 proteins (58%) could not be assigned a subcellular location using this program. It should be noted that the PSORTb program predicts slightly more proteins (14.1%) to be cytoplasmic membrane proteins than based merely on the number of proteins with 2 or more predicted transmembrane helices (13%) as noted above. The PSORTb program uses the HMMTOP algorithm to predict the number of transmembrane segments in a protein. Different prediction programs used to predict the topology and subcellular localization of proteins, often yield varying results [57].
Channels (TC #1.A)
As noted above, B. bacteriovorus has a large number of channel types. Three are homologous to known chloride channels of the ClC family (TC #1.A.11; Table 2). A fourth protein (GI:42524203) shows statistically significant sequence similarity to a central portion of an epithelial chloride channel (E-ClC, TC #1.A.13), but the rest of the protein does not resemble members of the E-ClC family. There is therefore no clear evidence that this protein functions in anion transport.
Table 2.
TC classification and functional predictions of putative transport proteins from Bdellovibrio bacteriovorus.
| TC Family | Family Name | Number of transport systemsa | Bba Protein ID | Size | # of TMSsb | Best blast-hit in TCDB and/or comments |
|---|---|---|---|---|---|---|
| 1.A.11 | Chloride Channel (ClC) Family | 3 | 42523302 | 442 | 8 | EriC [E. coli] |
| 42523636 | 406 | 10 | MJ0305 [M. jannaschii] | |||
| 42523824 | 442 | 10 | EriC [E. coli] | |||
| 1.A.22 | Large Conductance Mechanosensitive Ion Channel (MscL) Family | 1 | 42521798 | 144 | 2 | MscL [E. coli] |
| 1.A.23 | Small Conductance Mechanosensitive Ion Channel (MscS) Family | 4 | 42521793 | 396 | 5 | KefA [E. coli] |
| 42523536 | 351 | 4 | MscMJLR-like [M. jannaschii] | |||
| 42524113 | 380 | 4 | MscMJLR [M. jannaschii] | |||
| 42524226 | 316 | 2 | KefA [E. coli] | |||
| 1.A.30.1 | H+- or Na+-translocating Bacterial Flagellar Motor (Mot) Family | 3 | 42521781 | 262 | 4 | MotA [B. subtilis] |
| 42521782 | 249 | 1 | MotS [B. subtilis] | |||
| 42524415 | 319 | 1 | MotS [B. subtilis] | |||
| 42524416 | 293 | 4 | PomA [V. alginolyticus] | |||
| 42524629 | 318 | 0 | MotB [E. coli] | |||
| 42524630 | 265 | 3 | MotA [E. coli] | |||
| 1.A.35 | CorA Metal Ion Transporter (MIT) Family | 1 | 42522589 | 308 | 2 | CorA [M. jannaschii] |
| 1.B.3 | Sugar Porin (SP) Family | 1 | 42522760 | 447 | 0 | LamB [E. coli] |
| 1.B.6 | OmpA-OmpF Porin (OOP) Family | 4 | 42521994 | 171 | 0 | OmpA [E. coli] |
| 42522454 | 368 | 0 | OmpATb-like [M. tuberculosis] | |||
| 42524036 | 440 | 0 | OmpATb-like [M. tuberculosis] | |||
| 42524180 | 214 | 0 | OmpATb [M. tuberculosis] | |||
| 1.B.10 | Nucleoside-specific Channel-forming Outer Membrane Porin (Tsx) Family | 2 | 42522254 | 269 | 0 | OmpK [V. parahaemolyticus] |
| 42524633 | 253 | 1 | OmpK-like | |||
| 1.B.14 | Outer Membrane Receptor (OMR) Family | 4 | 42522910 | 698 | 0 | FhuA [E. coli] |
| 42523077 | 709 | 0 | ViuA [V. cholerae] | |||
| 42524068 | 687 | 0 | FecA [E. coli] | |||
| 42524920 | 610 | 0 | BtuB [E. coli] | |||
| 1.B.17 | Outer Membrane Factor (OMF) Family | 42522293 | 426 | 1 | NodT2 [R. leguminosarum] [with 3.A.1.115] | |
| 42522458 | 434 | 0 | TolC [E. coli] [with 2.A.6.2] | |||
| 42522925 | 462 | 1 | ToxI-like [2.A.6.2] [B. glumae] | |||
| 42523285 | 485 | 0 | TolC-like | |||
| 42523393 | 437 | 0 | PrtF [E. coli] | |||
| 42523686 | 404 | 0 | TolC [E. coli] | |||
| 42524451 | 509 | 0 | VceC-like [V. cholerae] | |||
| 1.B.22 | Outer Bacterial Membrane Secretin (Secretin) Family | 42522439 | 726 | 0 | PilQ [P. aeruginosa] | |
| 42524863 | 222 | 0 | XcpQ [P. aeruginosa] | |||
| 1.B.33 | Outer Membrane Protein Insertion Porin (OmpIP) Family | 4 | 42521763 | 245 | 0 | YfiO [E. coli] ComL |
| 42523001 | 799 | 0 | YaeT [E. coli] | |||
| 42523064 | 573 | 0 | D15-like [H. influenzae] | |||
| 42523505 | 392 | 0 | YfgL [E. coli] | |||
| 42523581 | 803 | 1 | Imp [E. coli] | |||
| 42523924 | 929 | 0 | YaeT-like [E. coli] | |||
| 42524276 | 423 | 0 | YfgL-like [E. coli] | |||
| 42524950 | 392 | 0 | Omp85-like [N. meningitidis] | |||
| 1.B.x | Unclassified outer membrane protein | 1 | 42525165 | 216 | 1 | LolA-like OMP |
| 1.E.14 | LrgA Holin (LrgA Holin) Family | 1 | 42524812 | 129 | 4 | LrgA [S. aureus] |
| 2.A.1 | MFS Superfamily | |||||
| 2.A.1.2 | Drug:H+ Antiporter-1 (12 Spanner) (DHA1) Family | 6 | 42522009 | 396 | 12 | Bcr [E. coli] |
| 42522069 | 405 | 10 | TetA [E. coli] | |||
| 42522244 | 367 | 11 | TetA [E. coli] | |||
| 42522883 | 393 | 12 | YdeA [E. coli] | |||
| 42523627 | 412 | 12 | PbuE/YdhL [B. subtilis] | |||
| 42525027 | 397 | 12 | NepI [E. coli] | |||
| 2.A.1.4 | Organophosphate:Pi Antiporter (OPA) Family | 2 | 42523618 | 513 | 13 | UhpC [E. coli] |
| 42524319 | 497 | 12 | Hpt [C. pneumoniae] | |||
| 2.A.1.6 | Metabolite:H+ Symporter (MHS) Family | 1 | 42523790 | 454 | 12 | KgtP [E. coli] |
| 2.A.1.24 | Unknown Major Facilitator-1 (UMF1) Family | 1 | 42522938 | 392 | 11 | YCL038c [S. cerevisiae] |
| 2.A.1.25 | Peptide-Acetyl-Coenzyme A Transporter (PAT) Family | 1 | 42521955 | 447 | 12 | AmpG [E. coli] |
| 2.A.1.30 | Putative Abietane Diterpenoid Transporter (ADT) Family | 1 | 42523759 | 398 | 12 | DitE [P. abietaniphila] |
| 2.A.1.36 | Acriflavin-sensitivity (YnfM) Family | 2 | 42522279 | 394 | 9 | YnfM [E. coli] |
| 42524997 | 408 | 12 | YnfM [E. coli] | |||
| 2.A.1.x | Member of the MFS Superfamily | 3 | 42522316 | 405 | 10 | Putative MFS |
| 42523216 | 418 | 12 | Putative MFS | |||
| 42523082 | 395 | 10 | Putative MFS | |||
| 2.A.2 | Glycoside-Pentoside-Hexuronide (GPH): Cation Symporter Family | 1 | 42524653 | 422 | 10 | MelB [E. coli] |
| 2.A.3 | Amino Acid/Polyamine/Organocation (APC) Superfamily | |||||
| 2.A.3.6 | Archaeal/Bacterial Transporter (ABT) Family | 1 | 42523390 | 412 | 12 | Cat-1 [A. fulgidus] |
| 2.A.4 | Cation Diffusion Facilitator (CDF) Family | 2 | 42521983 | 345 | 6 | YiiP [E. coli] |
| 42523674 | 310 | 5 | CzcD [A. eutrophus] | |||
| 2.A.5 | Zinc (Zn2+)-Iron (Fe2+) Permease (ZIP) Family | 1 | 42524041 | 251 | 7 | ZupT [E. coli] |
| 2.A.6 | RND Superfamily | |||||
| 2.A.6.1 | Heavy Metal Efflux (HME) Family | 1 | 42523684 | 1010 | 9 | CzcA [A. eutrophus] |
| 2.A.6.2 | (Largely Gram-negative Bacterial) Hydrophobe/Amphiphile Efflux-1 (HAE1) Family | 6 | 42522459 | 1032 | 12 | MdtC/YegO [E. coli] |
| 42522562 | 1050 | 10 | MdtC/YegO [E. coli] | |||
| 42522687 | 1033 | 10 | MdtC/YegO [E. coli] | |||
| 42523244 | 1039 | 12 | MdtC/YegO [E. coli] | |||
| 42523286 | 1033 | 11 | MdtC/YegO [E. coli] | |||
| 42524450 | 1070 | 12 | MdtC/YegO [E. coli] | |||
| 2.A.6.7 | (Largely Archaeal Putative) Hydrophobe/Amphiphile Efflux-3 (HAE3) Family | 1 | 42523469 | 1013 | 13 | MJ1562 [M. jannaschii] |
| 2.A.7 | DMT Superfamily | |||||
| 2.A.7.1 | 4 TMS Small Multidrug Resistance (SMR) Family | 2 | 42523246 | 145 | 4 | EmrE [E. coli] |
| 42523933 | 131 | 4 | SugE [E. coli] | |||
| 2.A.7.3 | 10 TMS Drug/Metabolite Exporter (DME) Family | 3 | 42521888 | 295 | 10 | PecM [E. chrysanthemi] |
| 42522043 | 285 | 10 | RhtA [E. coli] | |||
| 42524639 | 298 | 10 | RhtA [E. coli] | |||
| 2.A.7.x | Member of the DMT Superfamily | 2 | 42523696 | 309 | 9 | Putative DMT |
| 42523878 | 311 | 10 | Putative DME [2.A.7.3] | |||
| 2.A.9 | Cytochrome Oxidase Biogenesis (Oxa1) Family | 1 | 42525232 | 539 | 5 | YidC [E. coli] |
| 2.A.17 | Proton-dependent Oligopeptide Transporter (POT) Family | 1 | 42523325 | 451 | 11 | DtpT [L. lactis] |
| 2.A.19 | Ca2+:Cation Antiporter (CaCA) Family | 1 | 42523602 | 371 | 10 | ChaA [E. coli] |
| 2.A.22 | Neurotransmitter:Sodium Symporter (NSS) Family | 1 | 42523422 | 455 | 11 | MJ1319 [M. jannaschii] |
| 2.A.27 | Glutamate:Na+ Symporter (ESS) Family | 1 | 42524043 | 394 | 12 | GltS [E. coli] |
| 2.A.35 | NhaC Na+:H+ Antiporter (NhaC) Family | 1 | 42521884 | 427 | 9 | MleN [B. subtilis] |
| 2.A.37 | Monovalent Cation:Proton Antiporter-2 (CPA2) Family | 5 | 42521965 | 294 | 10 | GrmA [B. subtilis] |
| 42522245 | 650 | 11 | KefC [E. coli] | |||
| 42523556 | 746 | 16 | MagA [Magnetospirillum]+TrkA_C | |||
| 42523635 | 388 | 10 | NhaS3-like [Synechocystis] | |||
| 42523724 | 739 | 16 | GrmA [B. megaterium]+TrkA_C | |||
| 42525048 | 192 | 0 | YheR [E. coli] | |||
| 2.A.46 | Benzoate:H+ Symporter (BenE) Family | 1 | 42521934 | 387 | 12 | BenE [A. calcoaceticus] |
| 2.A.47 | Divalent Anion:Na+ Symporter (DASS) Family | 2 | 42523367 | 488 | 11 | SodiTl [S. oleracea] |
| 42523612 | 612 | 10 | SdrP [Synechocystis] | |||
| 2.A.50 | Glycerol Uptake (GUP) Family | 1 | 42523167 | 470 | 9 | GUP1 [S. cerevisiae] |
| 2.A.51 | Chromate Ion Transporter (CHR) Family | 1 | 42523317 | 378 | 9 | SrpC [Synechococcus] |
| 2.A.58 | Phosphate:Na+ Symporter (PNaS) Family | 1 | 42523679 | 535 | 8 | YjbB [E. coli] |
| 2.A.63 | Monovalent Cation (K+ or Na+):Proton Antiporter-3 (CPA3) Family | 1 | 42523736 | 109 | 3 | MnhG [S. aureus] |
| DNA | NA | MnhF [S. aureus] | ||||
| 42523737 | 112 | 2 | MnhE [S. aureus] | |||
| 42523738 | 486 | 13 | MnhD [S. aureus] | |||
| 42523739 | 98 | 3 | MnhC [S. aureus] | |||
| 42523740 | 853 | 24 | MnhAB [S. aureus] | |||
| 2.A.64 | Twin Arginine Targeting (Tat) Family | 1 | 42523658 | 79 | 1 | TatE [E. coli] |
| 42523995 | 81 | 0 | TatA [E. coli] | |||
| 42525186 | 253 | 6 | TatC [E. coli] | |||
| 42525187 | 118 | 0 | TatB [E. coli] | |||
| 2.A.66 | MOP Superfamily | |||||
| 2.A.66.1 | Multi Antimicrobial Extrusion (MATE) Family | 1 | 42523788 | 404 | 11 | NorM [B. vietnamiensis] |
| 2.A.66.2 | Polysaccharide Transport (PST) Family | 1 | 42523185 | 422 | 12 | MTH347 [M. thermautotrophicus] |
| 2.A.66.4 | Mouse Virulence Factor (MVF) Family | 2 | 42523761 | 520 | 11 | MviN [S. typhimurium] |
| 42522981 | 523 | 12 | MviN [S. typhimurium] | |||
| 2.A.68 | p-Aminobenzoyl-glutamate Transporter (AbgT) Family | 2 | 42524987 | 514 | 12 | MtrF/AbgT |
| 42521870 | 454 | 10 | Putative AtoE | |||
| 2.A.72 | K+ Uptake Permease (KUP) Family | 1 | 42523474 | 669 | 12 | KUP [E. coli] |
| 2.A.85 | The Aromatic Acid Exporter (AraE) Family | 1 | 42524263 | 344 | 8 | YccS [E. coli] |
| 2.C.1 | TonB-ExbB-ExbD/TolA-TolQ-TolR (TonB) Family of Auxiliary Proteins for Energization of Outer Membrane Receptor (OMR)-mediated Active Transport | 6 | 42521812 | 461 | 0 | TolB [E. coli] |
| 42521813 | 286 | 1 | TonB-like [E. coli] | |||
| 42521814 | 138 | 1 | ExbD/TolR [E. coli] | |||
| 42521815 | 245 | 3 | TolQ [E. coli] | |||
| 42522010 | 316 | 1 | TonB-like ? | |||
| 42522032 | 165 | 1 | ExbD/TolR-like ? | |||
| 42522033 | 149 | 1 | ExbD-like [E. coli] | |||
| 42522034 | 248 | 3 | ExbB [E. coli] | |||
| 42522085 | 209 | 3 | TolQ [E. coli] | |||
| 42522086 | 134 | 1 | TolR-like [E. coli] | |||
| 42522223 | 168 | 0 | Pal [E. coli] | |||
| 42522225 | 223 | 0 | YbgF-like [E. coli] | |||
| 42522342 | 295 | 1 | TonB-like ? | |||
| 42522410 | 221 | 3 | TolQ [E. coli] | |||
| 42522411 | 158 | 1 | TolR [E. coli] | |||
| 42522990 | 167 | 1 | ExbD/TolR-like [E. coli] | |||
| 42522991 | 185 | 0 | TolR-like [E. coli] | |||
| 42522992 | 236 | 3 | TolQ-like [E. coli] | |||
| 42523572 | 224 | 1 | YbgF-like [E. coli] | |||
| 42523821 | 175 | 1 | ExbD/TolR-like ? | |||
| 42523822 | 151 | 1 | ExbD-like [E. coli] | |||
| 42523823 | 244 | 3 | ExbB [E. coli] | |||
| 42523907 | 175 | 0 | YbgF-like [E. coli] | |||
| 42524267 | 244 | 0 | TonB-like ? | |||
| 42524795 | 341 | 0 | TolB-like ? | |||
| 42524919 | 209 | 1 | TonB-like [E. coli] | |||
| 42525096 | 471 | 1 | TonB-like ? | |||
| 3.A.1 | ABC Superfamily c | |||||
| 3.A.1.1 | Carbohydrate Uptake Transporter-1 (CUT1) Family | 1 | 42522756 | 347 | 0 | [C] MalK |
| 42522757 | 272 | 6 | [M] MalG2 | |||
| 42522758 | 733 | 7 | [R+M] MalE1+MalF1 | |||
| 3.A.1.2 | Carbohydrate Uptake Transporter-2 (CUT2) Family | 1 | 42522051 | 291 | 8 | [M] |
| 42522052 | 342 | 8 | [M] FrcC-like [R. meliloti] | |||
| 42522053 | 494 | 0 | [C] | |||
| 42522054 | 339 | 0 | [R] | |||
| 3.A.1.3 | Polar Amino Acid Uptake Transporter (PAAT) Family | 1 | 42522399 | 247 | 0 | BgtA [C] |
| 42522400 | 247 | 4 | BgtB_C [M] | |||
| 42522401 | 262 | 0 | BgtB_N [R] | |||
| 3.A.1.4 | Hydrophobic Amino Acid Uptake Transporter (HAAT) Family | 1 | 42524744 | 373 | 0 | LivK [R] |
| 42524745 | 300 | 8 | LivH [M] | |||
| 42524746 | 323 | 8 | LivM [M] | |||
| 42524747 | 270 | 0 | LivG [C] | |||
| 42524748 | 237 | 0 | LivF [C] | |||
| 3.A.1.5 | Peptide/Opine/Nickel Uptake Transporter (PepT) Family | 3 | 42521975 | 540 | 0 | DppE [R] |
| 42521976 | 343 | 6 | DppB [M] | |||
| 42521977 | 339 | 6 | DppC [M] | |||
| 42523449 | 487 | 0 | MppA [R] | |||
| 42523651 | 322 | 0 | OppD [C] | |||
| 42523652 | 329 | 0 | OppF [C] | |||
| 42523653 | 564 | 0 | OppA [R] | |||
| 42523654 | 323 | 6 | OppB [M] | |||
| 42523655 | 404 | 6 | OppC [M] | |||
| 42524127 | 274 | 6 | DppC [M] | |||
| 42524128 | 310 | 7 | DppB [M] | |||
| 3.A.1.7 | Phosphate Uptake Transporter (PhoT) Family | 1 | 42523158 | 333 | 0 | PstS-like [R] |
| 42523159 | 312 | 6 | PstC [M] | |||
| 42523160 | 305 | 8 | PstA [M] | |||
| 42523161 | 255 | 0 | PstB [C] | |||
| 3.A.1.9 | Phosphonate Uptake Transporter (PhnT) Family | 1 | 42524492 | 322 | 7 | PhnE [M] |
| 42524493 | 262 | 0 | PhnC [C] | |||
| 42524494 | 297 | 0 | PhnD [R] | |||
| 3.A.1.11 | Polyamine/Opine/Phosphonate Uptake Transporter (POPT) Family | 1 | 42522934 | 344 | 0 | PotD [R] |
| 42522935 | 292 | 0 | PotA [C] | |||
| 42522936 | 277 | 6 | PotB [M] | |||
| 42522937 | 277 | 6 | PotC [M] | |||
| 3.A.1.15 | Manganese/Zinc/Iron Chelate Uptake Transporter (MZT) Family (Similar to 3.A.1.12 and 3.A.1.16) | 1 | 42524734 | 291 | 0 | ZnuA [R] |
| 42524735 | 189 | 0 | ZnuC [C] | |||
| 42524736 | 270 | 7 | ZnuB [M] | |||
| 3.A.1.16 | Nitrate/Nitrite/Cyanate Uptake Transporter (NitT) Family | 1 | 42523495 | 310 | 0 | [R] |
| 42523496 | 253 | 5 | [M] CmpB-like [Synechococcus] | |||
| 42523497 | 266 | 0 | [C] | |||
| 3.A.1.17 | Taurine Uptake Transporter (TauT) Family (Similar to 3.A.1.12 and 3.A.1.16) | 1 | 42524162 | 266 | 0 | TauB [C] |
| 42524163 | 275 | 6 | TauC [M] | |||
| 42524164 | 333 | 1 | TauA [R] | |||
| 3.A.1.19 | Thiamin Uptake Transporter (ThiT) Family (Most similar to 3.A.1.10, 3.A.1.6 and 3.A.1.8 in that order) | 1 | 42522187 | 346 | 1 | ThiB [R] |
| 42522188 | 487 | 11 | ThiP [M] | |||
| 42522189 | 208 | 0 | ThiQ [C] | |||
| 3.A.1.102 | Lipooligosaccharide Exporter (LOSE) Family | 1 | 42521775 | 308 | 0 | NodI [C] |
| 42521776 | 262 | 6 | NodJ [M] | |||
| 3.A.1.106 | Lipid Exporter (LipidE) Family | 3 | 42521794 | 573 | 5 | MsbA [M+C] |
| 42523036 | 588 | 4 | MsbA [M+C] | |||
| 42524487 | 567 | 5 | MsbA [M+C] | |||
| 3.A.1.115 | Na+ Exporter (NatE) Family | 1 | 42523356 | 250 | 0 | [C] |
| 42523357 | 246 | 0 | [C] | |||
| 42523358 | 368 | 6 | [M] NatB-like | |||
| 42523359 | 358 | 6 | [M] | |||
| 3.A.1.122 | Macrolide Exporter (MacB) Family | 1 | 42522295 | 650 | 4 | MacB [C+M] |
| 3.A.1.125 | Lipoprotein Translocase (LPT) Family | 2 | 42522999 | 405 | 4 | LolE [M] |
| 42523000 | 220 | 0 | LolD [C] | |||
| 42523263 | 416 | 4 | LolC [M] | |||
| 42524549 | 402 | 4 | LolE [M] | |||
| 42524550 | 231 | 0 | LolD [C] | |||
| 3.A.1.208 | Conjugate Transporter Family (ABCC) | 2 | 42523967 | 597 | 5 | AtMRP2_N [A. thaliana] |
| 42523968 | 620 | 6 | AtMRP2_C [A. thaliana] | |||
| 42524005 | 1228 | 10 | MRP3 [H. sapiens] | |||
| 3.A.1.x | Orphan members of the ABC Superfamily | 15 | 42521819 | 252 | 0 | [C] |
| 42521882 | 251 | 0 | [R] [3.A.1.3 ?] | |||
| 42521925 | 305 | 0 | [C] | |||
| 42521962 | 221 | 0 | [C] | |||
| 42521963 | 849 | 10 | [M] [duplicated LolE-like] | |||
| 42522096 | 254 | 0 | [R] [3.A.1.3; TcyA B. subtilis] | |||
| 42522111 | 273 | 1 | [R] [3.A.1.3; ArtJ E. coli] | |||
| 42522191 | 246 | 0 | [R] [3.A.1.3 ?] | |||
| 42522255 | 496 | 0 | [R] [duplicated 3.A.1.3 ?] | |||
| 42522275 | 274 | 1 | [R] [3.A.1.3; ArtJ-like E. coli] | |||
| 42522313 | 243 | 1 | [R] [3.A.1.3; ArgT-like E. coli] | |||
| 42522356 | 242 | 0 | [R] [3.A.1.3 ?] | |||
| 42522414 | 240 | 0 | [C] | |||
| 42522416 | 274 | 0 | [R] [3.A.1.3; GlnH E. coli] | |||
| 42522431 | 296 | 0 | [R] [3.A.1.9; PhnD E. coli] | |||
| 42522432 | 247 | 0 | [C] | |||
| 42522433 | 253 | 6 | [M] [NosY-like] [PliI ?] | |||
| 42522485 | 252 | 0 | [C] | |||
| 42522556 | 603 | 0 | [C+C] | |||
| 42522582 | 314 | 0 | [C] | |||
| 42522583 | 257 | 6 | [M] ? | |||
| 42522584 | 511 | 4 | [M] [NA; ABC ?] | |||
| 42522612 | 292 | 0 | [R] [3.A.1.3 ?] | |||
| 42522628 | 330 | 0 | [r] [3.A.1.10; FutA1 Synechocystis] | |||
| 42522659 | 229 | 0 | [C] | |||
| 42522660 | 421 | 3 | [M] [LolE-like] | |||
| 42522753 | 569 | 0 | [C] | |||
| 42522767 | 245 | 0 | [R] [3.A.1.3; HisJ S. typhimurium] | |||
| 42522834 | 285 | 0 | [R] [3.A.1.3 ?] | |||
| 42522852 | 556 | 0 | [C+C] | |||
| 42522853 | 277 | 5 | [M] [NA; ABC ?] | |||
| 42522868 | 247 | 0 | [R] [3.A.1.3 ?] | |||
| 42522895 | 582 | 0 | [R] [3.A.1.2+3; RbsB E. coli + HisJ S. typhimurium] | |||
| 42522931 | 543 | 0 | [C+C] | |||
| 42522945 | 254 | 1 | [R] [3.A.1.3; ArtJ-like E. coli] | |||
| 42523223 | 248 | 6 | [M] [3.A.1.105] | |||
| 42523224 | 287 | 0 | [C] [3.A.1.105] | |||
| 42523258 | 274 | 6 | [M] [9.B. 11; Tgd1 A. thaliana] | |||
| 42523259 | 236 | 0 | [C] [3.A.5 FtsE-like ATPase] | |||
| 42523260 | 317 | 1 | [R] [Ttg2C] | |||
| 42523261 | 261 | 0 | [R] [3.A.1.3 ?] | |||
| 42523262 | 265 | 0 | [R] [3.A.1.3 ?] | |||
| 42523274 | 247 | 0 | [R] [3.A.1.3; ArgT-like E. coli] | |||
| 42523464 | 237 | 1 | [R] [3.A.1.3 ?] | |||
| 42523514 | 246 | 1 | [R] [3.A.1.3; GlnH-like E. coli] | |||
| 42523547 | 600 | 1 | [R] [duplicated; 3.A.1.2 ?] | |||
| 42523561 | 257 | 0 | [R] [3.A.1.3; ArgT E. coli] | |||
| 42523595 | 257 | 6 | [M] [3.A.1.102] | |||
| 42523596 | 305 | 0 | [C] [3.A.1.102] | |||
| 42523831 | 267 | 0 | [R] [3.A.1.13; ButF S. typhimurium] | |||
| 42523888 | 239 | 0 | [R] [3.A.1.3 ?] | |||
| 42524071 | 246 | 0 | [R] [3.A.1.3; CysX E. coli] | |||
| 42524132 | 347 | 0 | [R] [3.A.1.2] | |||
| 42524161 | 149 | 4 | [M] [NA; ABC ?] | |||
| 42524193 | 260 | 0 | [R] [3.A.1.3; TcyA-like B. subtilis] | |||
| 42524262 | 287 | 0 | [R] [3.A.1.3] | |||
| 42524284 | 254 | 0 | [R] [3.A.1.3 ?] | |||
| 42524507 | 459 | 1 | [R] [Ttg2C] | |||
| 42524508 | 254 | 0 | [C] | |||
| 42524509 | 275 | 5 | [M] [9.B.11; Tgd1 A. thaliana] | |||
| 42524523 | 264 | 4 | [M] [NA; ABC ?] | |||
| 42524770 | 262 | 5 | [M] [9.B.11; Tgd1 A. thaliana] | |||
| 42524771 | 248 | 0 | [C] | |||
| 42524772 | 272 | 1 | [R] [Ttg2C] | |||
| 42524780 | 474 | 0 | [R] [3.A.1.3 ? + SLT] | |||
| 42524915 | 622 | 0 | [C+C] | |||
| 42524918 | 515 | 0 | [C+C] | |||
| 42524922 | 265 | 0 | [R] [3.A.1.3; ArtI E. coli] | |||
| 42524935 | 276 | 0 | [C] | |||
| 42524936 | 270 | 7 | [M] ? | |||
| 42524937 | 266 | 6 | [M] [NA; ABC ?] | |||
| 42524980 | 554 | 0 | [C+C] | |||
| 42525129 | 258 | 1 | [R] [Ttg2C] | |||
| 42525130 | 239 | 5 | [M] [9.B.11; Tgd1 A. thaliana] | |||
| 42525131 | 259 | 3 | [M] [9.B. 11; Tgd1 A. thaliana] | |||
| 42525132 | 243 | 0 | [C] | |||
| 42525133 | 252 | 0 | [C] | |||
| 42525140 | 237 | 0 | [C] | |||
| 42525141 | 786 | 9 | [M] ? | |||
| 42525194 | 556 | 0 | [R] [3.A.1.4; ? NatB Synechocystis] | |||
| 3.A.2 | H+ or Na+ translocating F-type, V-type and A-type ATPase (F-ATPase) Superfamily | 1 | 42521659 | 229 | 6 | F0-A |
| 42521660 | 105 | 2 | F0-C | |||
| 42525217 | 140 | 0 | F1-ε | |||
| 42525218 | 468 | 0 | F1-β | |||
| 42525219 | 295 | 0 | F1-γ | |||
| 42525220 | 507 | 0 | F1-α | |||
| 42525221 | 182 | 0 | F1-δ | |||
| 42525222 | 186 | 2 | F0-B | |||
| 42525223 | 144 | 1 | F0-B | |||
| 3.A.3 | P-type ATPase (P-ATPase) Superfamily | 5 | 42522486 | 825 | 9 | pMA1, Ca2+-ATPase |
| 42523248 | 141 | 2 | KdpA_N, K+-ATPase | |||
| 42523249 | 160 | 3 | KdpA_C, K+-ATPase | |||
| 42523682 | 724 | 8 | CopA, Cu2+-ATPase | |||
| 42523748 | 692 | 8 | CopB, Cu2+, Cu+, Ag+ ATPase | |||
| 42524029 | 798 | 8 | CopA, Cu2+-ATPase | |||
| 3.A.5 | General Secretory Pathway (Sec) Family | 1 | 42521801 | 220 | 0 | FtsE |
| 42521895 | 889 | 0 | SecA | |||
| 42522594 | 161 | 2 | SecG | |||
| 42522713 | 402 | 1 | FtsY | |||
| 42523591 | 450 | 0 | Ffh | |||
| 42523691 | 312 | 6 | SecF | |||
| 42523692 | 564 | 5 | SecD | |||
| 42523693 | 117 | 1 | YjaC | |||
| 42524357 | 442 | 10 | SecY | |||
| 42524389 | 125 | 3 | SecE | |||
| 3.A.6 | Type III (Virulence-related) Secretory Pathway (IIISP) Family | 1 | 42524690 | 699 | 7 | FlhA |
| 42524691 | 353 | 3 | FlhB | |||
| 42524692 | 259 | 7 | FliR | |||
| 42524693 | 90 | 2 | FliQ | |||
| 42524694 | 252 | 5 | FliP | |||
| 42524695 | 224 | 1 | FliO | |||
| 42524696 | 122 | 0 | FliN | |||
| 42524697 | 333 | 0 | FliM | |||
| 42524760 | 442 | 0 | FliI | |||
| 42524761 | 261 | 0 | FliH | |||
| 42524763 | 549 | 2 | FliF | |||
| 3.A.10 | H+ translocating Pyrophosphatase (H+PPase) Family | 1 | 42523211 | 688 | 16 | V-PPase [A. thaliana] |
| 3.A.12 | Septal DNA Translocator (S-DNA-T) Family | 1 | 42521689 | 797 | 5 | SpoIIIE of [B. subtilis] |
| 3.A.15 | Outer Membrane Protein Secreting Main Terminal Branch (MTB) Family | 2 | 42522434 | 259 | 6 | PilD |
| 42522813 | 190 | 1 | PilA | |||
| 42523016 | 566 | 0 | PilB | |||
| 42523017 | 347 | 0 | PilT | |||
| 42523018 | 405 | 3 | PilC | |||
| 42523092 | 411 | 0 | PulK ? | |||
| 42523093 | 294 | 1 | PulJ ? | |||
| 42523094 | 166 | 1 | PulI ? | |||
| 42523095 | 194 | 1 | FimT ? /PulH ? | |||
| 42523096 | 136 | 1 | PulG | |||
| 42523100 | 405 | 3 | PulF | |||
| 42523101 | 564 | 0 | PulE | |||
| 42523102 | 765 | 0 | PulD [1.B.22] | |||
| 42523103 | 304 | 1 | PulC | |||
| 42523986 | 289 | 1 | FimU ? | |||
| 42524677 | 433 | 0 | PilQ-like [1.B.22] | |||
| 42525175 | 367 | 0 | PilU/PilT-like | |||
| 3.B.1 (?) | Na+ transporting Carboxylic Acid Decarboxylase (NaT-DC) Family | 42524925 | 127 | 0 | Gamma [A. fermentans] | |
| 42524928 | 535 | 0 | Alpha [P. abyssi] | |||
| 42525181 | 522 | 0 | Alpha [V. parvula] | |||
| 42525183 | 175 | 0 | Gamma [P. abyssi] | |||
| 3.D.1 | Proton-translocating NADH Dehydrogenase (NDH) Family | 1 | 42524471 | 174 | 1 | Nqo9 |
| 42524472 | 504 | 0 | Nqo3 | |||
| 42524473 | 431 | 0 | Nqo1 | |||
| 42524474 | 159 | 0 | Nqo2 | |||
| 42524475 | 560 | 0 | Nqo5-Nqo4 | |||
| 42524476 | 187 | 0 | Nqo6 | |||
| 42525207 | 486 | 12 | Nqo14 | |||
| 42525208 | 517 | 13 | Nqo13 | |||
| 42525209 | 643 | 16 | Nqo12 | |||
| 42525210 | 107 | 3 | Nqo11 | |||
| 42525211 | 178 | 5 | Nqo10 | |||
| 42525212 | 385 | 8 | Nqo8 | |||
| 42525213 | 127 | 3 | Nqo7 | |||
| 3.D.4 | Proton-translocating Cytochrome Oxidase (COX) Superfamily | 3 | 42521909 | 318 | 2 | Cox2 |
| 42521910 | 535 | 12 | Cox1 | |||
| 42521911 | 221 | 5 | Cox3 | |||
| 42521912 | 112 | 3 | Cox4 | |||
| 42521913 | 292 | 8 | CoxX | |||
| 42524021 | 441 | 12 | Cox1-like | |||
| 42524022 | 215 | 0 | Cox2-like | |||
| 42524031 | 706 | 13 | CcoN-CcoO | |||
| 5.A.1 | Disulfide Bond Oxidoreductase D (DsbD) Family | 1 | 42524631 | 653 | 9 | DsbD [E. coli] |
| 5.A.3 | The Prokaryotic Molybdopterin-containing Oxidoreductase (PMO) Family | 1 | 42523112 | 1033 | 0 | DmsAB [H. salinarium] |
| 42523113 | 453 | 10 | DmsC [H. salinarium] | |||
| 42524233 | 151 | 1 | TorC_N [E. coli] | |||
| 8.A.1 | Membrane Fusion Protein (MFP) Family | 42522294 | 321 | 1 | MacA [for 3.A.1.122] | |
| 42523355 | 204 | 0 | MFP [for 3.A.1.115] | |||
| 42522395 | 582 | 1 | Probably with [NA SapB-like 9.B.20] | |||
| 42525142 | 297 | 1 | AcrA-like [for 3.A.1.x] | |||
| 8.A.21 | Epithelial Na+ Channel (ENaC) Family | 42523755 | 250 | 0 | Stomatin homolog [P. horikoshii] | |
| 42523756 | 424 | 5 | NfeD protease [P. horikoshii] | |||
| 42524093 | 307 | 1 | Stomatin homolog-like protein | |||
| 9.A.8 | Ferrous Iron Uptake (FeoB) Family | 1 | 42523346 | 76 | 0 | [FeoA] FeoB2_N [P. gingivalis] |
| 42523347 | 638 | 8 | FeoB [L. biflexa] | |||
| 9.A.19 | Mg2+ Transporter-E (MgtE) Family | 1 | 42523969 | 447 | 4 | MgtE [B. firmus] |
| 9.A.23 | Ferroportin (FP) Family | 1 | 42523494 | 440 | 8 | Fpn1 [M. musculus] Putative MFS? |
| 9.B.3 | Putative Bacterial Murein Precursor Exporter (MPE) Family | 2 | 42523896 | 374 | 9 | RodA [E. coli] |
| 42524580 | 380 | 9 | FtsW [E. coli] | |||
| 9.B.17 | Putative Fatty Acid Transporter (FAT) Family | 42521902 | 498 | 0 | FadD [E. coli] | |
| 42522110 | 562 | 0 | CaiC [E. coli] | |||
| 42522828 | 645 | 0 | CaiC [E. coli] | |||
| 42523290 | 554 | 0 | FadD [E. coli] | |||
| 42524534 | 593 | 0 | FadD [E. coli] | |||
| 42524536 | 805 | 0 | X+FadD [E. coli] | |||
| 9.B.22 | Putative Permease (PerM) Family | 2 | 42523616 | 345 | 7 | Yct2 [B. subtilis] |
| 42523150 | 371 | 8 | PerM-like ? [E. coli] | |||
| 9.B.26 | PF27 (PF27) Family | 2 | 42523213 | 90 | 2 | Y615-like [C-half] |
| 42524601 | 183 | 5 | Y615 [Synechocystis] | |||
| 9.B.27 | YdjX-Z (YdjX-Z) Family | 1 | 42521725 | 224 | 4 | YdjZ [E. coli] |
| 9.B.30 | Hly III (Hly III) Family | 2 | 42522243 | 215 | 7 | HlyIII [B. cereus] |
| 42525120 | 208 | 7 | HlyIII [B. cereus] | |||
| 9.B.37 | HlyC/CorC (HCC) Family | 2 | 42522543 | 346 | 3 | YrkA [B. subtilis] |
| 42523633 | 343 | 3 | YrkA [B. subtilis] | |||
| 9.B.53 ? | Unknown IT-6 (UIT6) Family | 1 | 42524007 | 429 | 12 | Putative transporter ? [L. interrogans] |
| 9.B.63 | 9 TMS Putative Metabolite Efflux (9-PME) Family | 1 | 42522953 | 325 | 8 | YeiH [E. coli] |
Proteins of certain families are known to function either as part of multi-component transport systems or are accessory proteins. Therefore this information is considered when calculating the number of transport systems.
The numbers of putative α-helical transmembrane segments (TMSs) were calculated using the TMHMM program. Unfortunately, in the case of outer membrane porins (TC #1.B), the numbers do not reflect the numbers of β-strands and therefore are of limited value.
The various protein components of ABC transporters are labeled as [M]: Integral membrane protein, [C]: Cytoplasmic ATP-hydrolyzing protein, and [R]: Extracytoplasmic (periplasmic) solute-binding receptor. Duplication of domains (e.g., [C+C]) or fusion of two or more protein domains (e.g., [R+M]) are also indicated.
B. bacteriovorus possesses five mechanosensitive channels, one of the MscL-type (TC #1.A.22) and four of the MscS-type (TC #1.A.23). Both types of channels are known to function in hypoosmotic stress adaptation [58]. Only one of these five channel proteins had been discussed previously [11] although the NCBI Genbank records do include annotations noting that these proteins are putative mechanosensitive ion channels.
As reported previously [11], B. bacteriovorus encodes 3 MotA and 3 MotB homologues (TC #1.A.30.1). These occur within three operons that each contains a single motA and a single motB gene. This observation is surprising since B. bacteriovorus encodes only a single flagellum. Possibly the three flagellar “torque generators” act on a single flagellum under different conditions. For example, for swimming vs. swarming motility as is the case of Bacillus subtilis [59,60]. However, these Mot proteins are distantly related to gliding motility genes in Myxococcus xanthus [61], and gliding motility may be a characteristic of B. bacteriovorus [11]. One or more of these MotAB pairs may therefore function to energize gliding motility rather than flagellar rotation.
Finally, B. bacteriovorus has a single divalent metal ion channel of the MIT or CorA family (TC #1.A.35). CorA family members can be specific for a single divalent cation or can allow entry of several [62]. This homologue may provide a primary mechanism for divalent cation (Mg2+, Co2+, etc.) uptake in this organism.
Outer Membrane Porins (TC #1.B)
B. bacteriovorus has a fair complement of outer membrane β-structured porins. These include a single member of the 16 TMS sugar porin family (TC #1.B.3), 4 paralogues of the 8 TMS OmpA-type porin family (TC #1.B.6) and two members of the 12 TMS Tsx nucleoside-specific porin family (TC #1.B.10). Four outer membrane receptors (TC #1.B.14) probably function in the energy-dependent uptake of Fe-siderophore complexes (3 systems) and vitamin B12 (1 system). There are also seven outer membrane factors (TC #1.B.17) that presumably function in conjunction with inner membrane efflux pumps. One of these resembles PtrF of E. coli and probably acts with an ABC-type protease exporter; a second most closely resembles NodT2 and may therefore catalyze oligosaccharide export; several others probably act with RND-type drug efflux pumps. TolC of E. coli can function with multiple transporters from different families, and three of the OMF family members in B. bacteriovorus proved most similar to TolC. Consequently, these proteins may be multifunctional. Surprisingly, we could identify only four membrane fusion proteins (MFP, TC #8.A.1) [63]. These proteins probably function with ABC- (3) and RND-type (1) drug exporters. All MFPs characterized to date function with a single efflux transporter, so at least 7 might be expected to be present. Since these proteins are sequence divergent, some MFPs encoded in the B. bacteriovorus genome may not have been identified by our current search and annotation techniques.
Two outer membrane secretins (TC #1.B.22) were found. One, a PilQ homologue, may function as a “porthole” in the export of type IV pilus subunits [64,65]. The other, an XcpQ homologue, is likely to serve as the porthole for a type II protein secretion system of the main terminal branch [66,67].
B. bacteriovorus has four homologues of E. coli YaeT (TC #1.B.33) and one homologue of E. coli Imp (OstA). It also has one homologue of YfiO and two of YfgL (none of NlpB) [68]. These E. coli proteins are known to function as a complex for the assembly and insertion of outer membrane macromolecules, proteins, lipids and/or lipopolysaccharides [68]. Other subunits of the E. coli complex may exist but have not yet been identified. One such candidate is encoded by a gene in an operon that also encodes several homologues of other constituents of the E. coli outer membrane biogenesis complex. It seems clear that B. bacteriovorus assembles its outer membrane as does E. coli. However, the presence of four YaeT homologues and two YfgL homologues suggests a level of complexity greater than observed for E. coli. There may be four distinct systems corresponding to the four YaeT homologues, and these may share the other components of these systems. Alternatively, more than one YaeT homologue may participate in the formation of a single complex. Interestingly, B. bacteriovorus has a LolA-like outer membrane protein that may function in lipoprotein export [11].
Finally B. bacteriovorus encodes a single holin of the LrgA family (TC #1.E.14), not discussed previously [11], although the Genbank record does contain annotation suggesting homology to the LrgA family. This protein is encoded by a gene that is downstream of and within the same operon as a putative autolysin, a murine hydrolase. It would seem that B. bacteriovorus encodes a chromosomal holin/autolysin system that may function in programmed cell death [69]. A penicillin-binding protein, possibly also involved in cell wall metabolism, is encoded within the same operon.
Secondary Carriers (TC #2.A)
B. bacteriovorus has substantial representation of transporters of the Major Facilitator Superfamily (MFS, TC #2.A.1). These include 6 drug exporters of the DHA1 family (TC #2.A.1.2) and two sequence divergent members of the OPA family (TC #2.A.1.4) that may function in sugar-phosphate:inorganic phosphate antiport. One member of the MFS (TC #2.A.1.6) shows greatest sequence similarity to the KgtP α-ketoglutarate uptake transporter of E. coli (see TCDB). Two more show greatest similarity with a putative acriflavine uptake transporter (TC # 2.A.1.36). Such systems probably have some other aromatic compounds as their natural substrates. One putative MFS transporter closely resembles the AmpG transporter of E. coli (TC #2.A.1.25) which takes up cell wall degradation products [70,71]. Three MFS members in B. bacteriovorus could not be assigned a substrate type due to their low BLAST scores with functionally characterized transporters. They may be members of uncharacterized families not yet in TCDB.
Some other families in TCDB have been shown to be distantly related to the MFS [72,73,74]. Of these, B. bacteriovorus has one member in the GPH family (TC #2.A.2) of glycoside permeases which might be a melibiose uptake system, and one member of the POT family (TC #2.A.17) of peptide uptake systems. The presence of these two transporters expands the limited repertoire of transporters B. bacteriovorus has for taking up sugars and amino acid derivatives. A single member of the APC superfamily (TC #2.A.3) of transporters for amino acids and their derivatives is present in B. bacteriovorus, fewer than in most bacteria with a genome of comparable size. This transporter is a member of the ABT family (TC #2.A.3.6) for which no functionally characterized members are available.
Two members of the Cation Diffusion Facilitator (CDF) family (TC #2.A.4) are also present. CDF carriers function in prokaryotes as heavy metal (Co2+, Cd2+, Zn2+, Ni2+, Cu2+ and Hg2+) efflux pumps probably using a Me2+/H+ antiport mechanism [75,76]. These pumps can exhibit broad or narrow specificities, so the two CDF carriers in B. bacteriovorus may be a broad and a narrow specificity system like the two proteins in TCDB (YiiP and CzcD, respectively) that they most closely resemble.
A single member of the ZIP family (TC #2.A.5) of heavy metal uptake carriers occurs in B. bacteriovorus. These carriers can be Zn2+-specific or broad specificity (Fe2+, Co2+, Mn2+, etc.) uptake systems. The B. bacteriovorus homologue most resembles ZupT, a broad specificity system of E. coli [77].
Three families within the RND superfamily (TC #2.A.6) are represented in B. bacteriovorus. The first family is the heavy metal efflux (HME) family (TC #2.A.6.1). The single B. bacteriovorus homologue in this family is most similar to the CzcA protein of Ralstonia eutropha, a Co2+, Zn2+, Cd2+ efflux system. The second family is the Hydrophobe/Amphiphile Exporter (HAE1) family of drug efflux pumps (TC #2.A.6.2). Six of these paralogues are present in B. bacteriovorus. All of them most closely resemble MdtC (YegO) of E. coli, a broad specificity drug/detergent/organic solvent/lipid exporter [78]. These systems might protect the parasite against defense mechanisms of the host bacterium. The last B. bacteriovorus RND homologue is a member of the mostly archaeal HAE3 family (TC #2.A.6.7) which is still poorly characterized.
Within the Drug/Metabolite Transporter (DMT) superfamily (TC #2.A.7) are two members of the SMR family (TC #2.A.7.1) of small multidrug resistance systems. One most closely resembles the E. coli EmrE broad specificity cationic drug exporter, while the other resembles the E. coli SugE narrow specificity cationic drug exporter [44,79]. Three members of the DME family (TC #2.A.7.3) are probably metabolite efflux pumps. Two of these most closely resemble the RhtA protein of E. coli which is a threonine/homoserine exporter that may also be able to accommodate other semipolar amino acids [80]. Two more distantly related putative 10 TMS members of the DMT superfamily could not be assigned membership to an established family within the DMT superfamily. They may be members of new families.
B. bacteriovorus encodes a single Ca2+:cation antiporter of the CaCA family (TC #2.A.19). All characterized members of this family function in Ca2+ extrusion from the cytoplasm using a monovalent cation antiport mechanism. Therefore, this is likely to be its function in B. bacteriovorus. Prokaryotic members of the NSS family (TC #2.A.22) are amino acid uptake systems, and the one from B. bacteriovorus most resembles the tryptophan uptake system of Symbiobacterium thermophilum, TnaT [81]. There is also a single member of the glutamate:sodium symporter (ESS) family (TC #2.A.27) represented in B. bacteriovorus, a protein resembling GltS of E. coli which transports both D- and L-glutamate as well as various glutamate derivatives.
B. bacteriovorus encodes within its genome many putative cation:proton antiporters, one of the NhaC family (TC #2.A.35). NhaC-type systems can function as Na+:H+ antiporters or malate • 2H+:lactate • Na+ antiporters [82,83]. Thus, members of this family may merely act as cation exchangers, but they may also be capable of electroneutral transport of organic anions.
Five members of the CPA2 family (TC #2.A.37) of monovalent cation transporters are encoded within the B. bacteriovorus genome, and they most closely match four different transporters in TCDB. Two resemble Bacillus GrmA, a spore germination protein of unknown transport specificity, which, however, closely resembles the GerN Na+/H+-K+ antiporter of Bacillus cereus; the second resembles the KefC glutathione-regulated K+ efflux protein of E. coli; the third looks like the MagA iron-regulated transporter of a magnetotactic bacterium; and the fourth is like NhaS3 of Synechocystis, a Na+:H+ antiporter. It seems likely that each of these B. bacteriovorus homologues will prove to catalyze a different reaction, but always acting on monovalent cations.
Two additional monovalent cation transporters are found within the B. bacteriovorus genome. The first is a multicomponent Na+ or K+:H+ antiporter of the CPA3 family (TC #2.A.63). All seven characteristic constituents of these systems were identified in B. bacteriovorus although only 6 had been annotated. One of them (GI:42523740) is a fusion protein of two previously recognized subunits of these systems. The system in B. bacteriovorus most closely resembles the Na+:H+ antiporter of the Gram-positive bacterium, Staphylococcus aureus, rather than the K+:H+ antiporter of the Gram-negative bacterium, Rhizobium meliloti [50,84]. It is therefore likely that the B. bacteriovorus homologue is a Na+:H+ antiporter. These complex Na+:H+ antiporters, with subunits homologous to subunits in NADH dehydrogenase, are possibly dependent on NADH2. Such a Na+:H+ antiporter has not been characterized in a Gram-negative bacterium before.
The K+ uptake (KUP) permease (TC #2.A.72) of E. coli may use a K+:H+ symport mechanism, allowing a 106-fold accumulation of K+ over the external medium [85,86]. This protein has an N-terminal 12 TMS topology (residues 1-450) followed by a hydrophilic domain of unknown function (residues 450-622). The same structure is observed for the B. bacteriovorus protein that it closely resembles. This suggests that the B. bacteriovorus homologue may function, and may be regulated, like the E. coli homologue.
Four members of the MOP superfamily (TC #2.A.66), from three different constituent families, were identified in B. bacteriovorus. One is probably an MDR efflux pump of the MATE family (TC #2.A.66.1) of drug:Na+ antiporters; the second is likely to be a polysaccharide exporter of the PST family (TC #2.A.66.2); and the third and fourth are members of the “Mouse Virulence family” (TC #2.A.66.4) with no functionally characterized member.
B. bacteriovorus encodes one or two members of each of several small solute carrier families. One resembles the BenE benzoate:H+ symporter of Acinetobacter calcoaceticus (TC #2.A.46) [87]; a second and third are members of the DASS family (TC #2.A.47) of divalent anion:Na+ symporters within the IT superfamily [88]; the fourth is a full-sized chromate/sulfate transporter of the CHR family (TC #2.A.51) [89,90,91,92]; the fifth is a putative phosphate:Na+ uptake symporter of the PNaS family (TC #2.A.58); the sixth is a member of the ArAE family (TC #2.A.85) which may export one or more aromatic acids [93] and functions with a Membrane Fusion Protein (TC #8.A.1) [94]; and the seventh and eighth are putative AbgT family (TC #2.A.68) homologues which might be peptide, p-aminobenzoyl-glutamate and/or drug uptake porters. One of these proteins (GI:42521870) had been annotated as a short chain fatty acid transporter like AtoE of E. coli (TC #2.A.73.1.1).
Two remaining pmf-dependent systems listed in Table 2 under the 2.A category of TCDB are involved in protein trafficking. One resembles the YidC protein of the Oxa1 family (TC #2.A.9). This protein probably catalyzes protein insertion into the cytoplasmic membrane [95]. The other is the Twin Arginine Targeting and Translocation (TAT) protein secretion system (TC #2.A.69) [96]. All bacteria with a Tat system have at least 1 TatC constituent and at least 1 TatA constituent. E. coli and B. bacteriovorus have three TatA homologues, TatA, TatB and TatE [96]. B. bacteriovorus has a single TatC as does E. coli, and it is encoded within a bicistronic operon that also encodes a TatB homologue. Unlike the gene arrangement in E. coli, TatA and TatE are both encoded elsewhere on the chromosome.
Outer Membrane Receptor (OMR) Energizers for Active Transport Across the Outer Membrane
Category 2.C.1 in TCDB includes a single family of multicomponent, pmf-dependent transporter energizers. Two such systems are present in E. coli, and at least three constituents show sequence similarity between these two systems. The TonB/ExbB/ExbD system shows sequences and functions similar to the TolA/TolQ/TolR system. The latter system has additional auxiliary proteins called Pal (a lipoprotein), TolB and YbgF. B. bacteriovorus encodes at least one complete TolA-type system with minimally one copy of each of the recognized E. coli auxiliary constituents (6 non-homologous proteins). However, encoded in this B. bacteriovorus genome are 6 TonB/TolA homologues, six ExbB/TolQ-like constituents, nine ExbD/TolR-like proteins, three YbgF homologues, two TolB homologues and one Pal lipoprotein. Operon analyses revealed that one set of TolA, TolB, TolQ, and TolR are encoded together in a single operon. Five separate TolQ/TolR pairs are encoded in five other distinct operons, and three of these also encode an extra ExbD/TolR-like homologue. Pal, TolB and YbgF homologues, present in 1, 2 and 3 copies, respectively (Table 2), are encoded at sites distant from each other and the other Tol genes, with the single exception noted above where a TolB homologue is encoded within an operon with TolA, TolQ and TolR homologues.
The operon structures were examined in the proximity of these genes. In brief, there are four OMR receptors in Bba; three are probably specific for complex iron; and one is specific for vitamin B12. Six TolQ/TolR homologues are present, but there are fewer TolB, Pal and YbgF homologues. It is probable that these last mentioned proteins either can function with multiple co-transcribed pairs of TolQR proteins or are not required. This might suggest that each OMR interacts with its own TolQ/R pair, any of which can use the same TolB/Pal/YbgF complex. It is interesting to recall that TolQ/R homologues (MotA/B homologues) may function in adventurous gliding motility in Myxococcus xanthus [61](see acc. #AAO22857). The possibility that B. bacteriovorus is capable of gliding motility using type four pili has not been demonstrated [11].
Primary Active Transporters – ABC Superfamily (TC #3.A.1)
The ABC superfamily of ATP-driven transporters is the largest transporter superfamily represented in the B. bacteriovorus genome. Fourteen potential uptake systems and 20 potential efflux systems were identified, and all of these systems appear to be complete, having all of the expected constituents. One maltose-type system of the CUT1 family (TC #3.A.1.1) and one ribose-type system of the CUT2 family (TC #3.A.1.2) were identified. In the former system, the MalE binding protein and the MalF protein constituent are fused in a single polypeptide chain derived from a single fused gene, an unusual arrangement. In the latter system, one cytoplasmic (C) ATP-hydrolyzing protein (RbsA), one periplasmic receptor (R) and two membrane (M) constituents were found. This arrangement resembles that of a minority of CUT-2 transporters. Others have a single membrane constituent and thus have only three constituents, one C, one M and one R. The E. coli ribose system has the equivalent four gene products, RsbABCD, where A is the cytoplasmic ATPase, B is the periplasmic receptor, and C and D are the channel-forming membrane proteins.
One system for uptake of polar amino acids (PAAT family, TC #3.A.1.3) and one system for uptake of hydrophobic amino acids (HAAT family, TC #3.A.1.4) were found. Both systems appear to be complete with three constituents in the PAAT family system (1R, 1C and 1M) and five in the HAAT family system (1R, 2Cs and 2Ms).
There is a complete oligopeptide uptake system (TC #3.A.1.5) like that in E. coli with two Rs, two Cs and two Ms. However, a strange situation is observed for the dipeptide (Dpp) system where there is a single receptor (R) and two pairs of membrane proteins (M) but no ATPase (C). Possibly these two systems use the sequence similar OppD and OppF to energize transport. Complete ABC uptake transporters specific for (1) phosphate (resembling PstABC/PstS of E. coli; 4 constituents; TC #3.A.1.7), (2) phosphonates (most like PhnCDE of E. coli; 3 constituents; TC #3.A.1.9), (3) polyamines (PotABCD of E. coli; 4 constituents; TC #3.A.1.11), (4) zinc (ZnuABC of E. coli; 3 constituents; TC #3.A.1.15), (5) inorganic anions (nitrate, nitrite, cyanide and bicarbonate; 3 constituents; TC #3.A.1.16), taurine (3 constituents; TC #3.A.1.17), and (6) thiamin (3 constituents; TC #3.A.1.19) were found.
ABC efflux systems include (1) a single system specific for lipooligosaccharides (2 constituents; TC #3.A.1.102), (2) three systems specific for lipids and/or drugs (one constituent each; TC #3.A.1.106), (3) either one 4-component or two 2-component Na+ exporter(s) (2 constituents each; TC #3.A.1.115), (4) a macrolide exporter (1 constituent; TC #3.A.1.122), (5) two probable lipoprotein exporters (2 or 3 constituents each; TC #3.A.1.125), and (6) two eukaryotic-like MDR pumps (TC #3.A.1.208), one resembling the plant AtMRP2 system and the other resembling the human MRP3 system. There are also about a dozen functionally unassigned ABC systems or orphan proteins. We have made functional predictions for some of these proteins when warranted (see Table 2). The Genbank records also include annotations of recognizable domains for many of these proteins.
Primary Active Transporters – Other Cation-transporting ATPases
B. bacteriovorus has one complete H+-translocating F-type ATPase (TC #3.A.2) and one H+-translocating pyrophosphatase (TC #3.A.10). Both enzymes can reversibly synthesize pyrophosphate bonds using the proton electrochemical gradient (the proton motive force, pmf) as the driving force. B. bacteriovorus also has five P-type ATPases (TC #3.A.3), one likely to be specific for Ca2+ (efflux), one for K+ (uptake), and three heavy metal systems that could be either uptake or efflux systems. No other cation-translocating ATPases appear to be encoded within the B. bacteriovorus genome.
Primary Active Transporters – ATP-dependent Protein Secretion Systems
As reported previously [11], B. bacteriovorus has a complete (11 component) general secretory (Sec) system (TC #3.A.5) including SecYEG, SecA, SecDF, FtsY and Ffh, YjaC, the 4.5 S RNA and FtsE. A very sequence-divergent FtsX was also identified (see below). B. bacteriovorus has a complete (11 component) flagellar protein export system (TC #3.A.6) [11] and possesses a single member of the septal DNA translocator (S-DNA-T) family (TC #3.A.12), essential for DNA translocation after septum formation in many bacteria. This protein may be required to complete DNA translocation after synchronous cell division of B. bacteriovorus snakes (phase 6 in the developmental cycle) (see Introduction).
B. bacteriovorus has protein constituents resembling those of two related types of outer membrane protein secretion systems (TC #3.A.15). One of these is the type II protein secretion system or main terminal branch (MTB), like the PulC-O,S system found in Klebsiella pneumoniae (14 constituents) [67], and the other is the pilin secretion/fimbrial assembly system like the PilA-EQTU FimTU system of Pseudomonas aeruginosa (10 constituents) [97,98]. The system(s) found in B. bacteriovorus has(have) at least 13 constituents, 7 most closely resembling the Pul system, and 6 most closely resembling the Pil system. However, there are many other pil (pilus) and fim (fimbrium) genes present on the B. bacteriovorus chromosome. Rendulic et al. [11] suggested that these proteins comprise a single system, a Pil-type rather than an MTB-type system. If, as proposed, B. bacteriovorus has a type 4 pilus-driven system for passage through the host cell envelope, then the suggestion that it is a Pil-type system is valid. Indeed, Schwudke et al. [99] have shown that a pilus gene, flp1, shows increased expression in the attack phase of Bba, compared to the intracellular replication phase. However, it is known that B. bacteriovorus secretes many proteins across its two-membrane envelope, and consequently, a Pul-type system would be expected to be useful. The nine pul genes in B. bacteriovorus occur within two operons, and these are distant in sequence from any of the pil genes recognized using TC-BLAST [36]. Other pil annotated genes not listed in Table 2 are present in the B. bacteriovorus genome, but these genes are not homologous to genes encoding the proteins of the P. aeruginosa pil system in TCDB (TC #3.A.15.2.1). They also localized to regions of the chromosome distant from the two pil operons in B. bacteriovorus that encode five of the pil genes listed in Table 3. On a purely bioinformatic basis, it is difficult to distinguish the Pul-from the Pil-type systems with certainty. However, based on their degrees of sequence similarity, we propose that B. bacteriovorus has both a complete Pil biogenesis system and a functional Pul-type (type II) protein secretion system.
Table 3.
Unusual compositions of protein complexes in Bba.
| Complex | Component | # of Homologues in Bba | # of Homologues in E. coli |
|---|---|---|---|
| A. Bacterial Motor Complexes | |||
| MotA | 3 | 1 | |
| MotB | 3 | 1 | |
| B. OMR Energizers | |||
| TolA/TonB | 6 | 2 | |
| TolQ/ExbB (like MotA) | 6 | 2 | |
| TolR/ExbD (like MotB) | 9 | 2 | |
| TolB | 2 | 1 | |
| Pal | 1 | 1 | |
| YbgF | 3 | 1 | |
| C. Outer Membrane Assembly Complex | |||
| YaeT | 4 | 2 | |
| OstA-L | 1 | 1 | |
| OstA-S | 0 | 1 | |
| YfgL | 2 | 1 | |
| OfiO | 1 | 1 | |
| NlpB | 0 | 1 |
Many prokaryotes have Na+-transporting organic acid decarboxylases (TC #3.B.1) which include α-, β- and γ-subunits [100,101]. B. bacteriovorus has two copies of both the α- and γ-subunits, but we could not identify a β-subunit. The β-subunit is the actual transporter, while α is the decarboxylase (often present without β), and γ, a Zn2+-binding protein of catalytic importance [102,103] is thought to be the linker connecting α and β. The absence of a recognizable β leaves the question open as to whether B. bacteriovorus can couple decarboxylation to Na+ expulsion.
Primary Active Transporters – Cation-translocating Electron Transfer Complexes
Many bacteria possess Na+-translocating NADH dehydrogenase complexes of 14 dissimilar subunits [104,105,106]. In B. bacteriovorus, 13 proteins comprise the NADH dehydrogenase (TC #3.D.1), encoded within two operons. One of the 13 proteins is a fusion protein (Nqo5-Nqo4). Thus, the system is complete and is presumed to be functional. B. bacteriovorus also has a complete proton pumping cytochrome oxidase complex (TC #3.D.4) with all five expected proteins (Cox1-4 and X) encoded within a single operon. Surprisingly, it also has several homologues of cytochrome oxidase subunits that map elsewhere on the chromosome. A Cox1-like homologue and a Cox2-like homologue are encoded within a single operon and show low sequence similarity to the Cox subunits. They have high sequence identity with the two subunits of nitric oxide reductases [107,108], and therefore presumably serve this function. These enzyme complexes may be capable of coupling proton export to electron flow [107].
Transmembrane Electron Flow Carriers (TC #5.A)
B. bacteriovorus possesses two transmembrane electron flow systems that can influence cellular energetics. One is disulfide bond oxidoreductase D (DsbD; TC #5.A.1) in which electrons from an electron donor such as NADH in the cytoplasm are transferred sequentially via thioredoxin reductase, thioredoxin and DsbD to a periplasmic disulfide-containing protein electron acceptor. Several such periplasmic proteins (DsbC, DsbE and DsbG) can be reduced via the DsbD pathway, and some of them can further reduce other disulfide-containing periplasmic proteins in Gram-negative bacteria [109]. The DsbD pathway in B. bacteriovorus undoubtedly facilitates proper folding of and disulfide bond formation in periplasmic proteins as is the case for E. coli [110,111].
The second transmembrane electron flow carrier is a single member of the Prokaryotic Molybdopterin-containing Oxidoreductase (PMO) family (TC #5.A.3), probably a dimethylsulfoxide (DMSO) oxidoreductase. One large protein (1033 aas) is a DmsAB fusion protein including the equivalent of the α- and β-subunits while the other is a DmsC subunit (the γ-subunit) [112,113]. Both proteins show greatest sequence similarity with an archaeal enzyme in TCDB (Table 2).
(Putative) Transporters of Unknown Function or Mechanism
In the 9A category of incompletely characterized transporters, we find one FeoAB iron uptake system (TC #9.A.8), one MgtE magnesium uptake porter (TC #9.A.19), and one putative iron transporter of the Ferroportin family (TC #9.A.23), as reported previously [11]. Our preliminary analyses suggest that this last B. bacteriovorus protein may be distantly related to members of the Major Facilitator Superfamily in agreement with the Genbank annotation.
In the 9B series of putative permeases, we find two homologues of Bacterial Murein Precursor Exporters of the MPE family (TC #9.B.30) which are found in many, if not all, bacteria. These porters probably serve the function of exporting precursors essential for bacterial cell wall synthesis [114,115]. Members of the Putative Fatty Acid Transporter family (TC #9.B.17) are acyl CoA synthetases that may in some cases function in fatty acid uptake coupled to esterification with cytoplasmic coenzyme A [116,117,118,119].
B. bacteriovorus encodes four potential hemolysins that could function in host cell lysis or pore formation in the cytoplasmic membrane. The two members of the HlyIII family (TC #9.B.30) are homologous to the B. cereus hemolysin III [120,121]. The two members of the HlyC/CorC family (TC #9.B.37) include one protein believed to be a hemolysin [122] and one protein believed to be a component (CorB) of the Ca2+/Mg2+ uptake transporter [123]. All proteins that show homology with members of the 9B class are homologous to TC entries of unestablished functions (Table 2).
Updated annotations and distant homologues of established transporters
Table S1 on our website (http://www.biology.ucsd.edu/~msaier/supmat/Bba) lists the proteins originally annotated as ABC-type proteins (8) or permeases (10) for which we now suggest new annotations. In addition to identifying the proteins and providing the original annotations, this table presents the protein sizes and the predicted numbers of TMSs. In some cases where we suspected erroneous annotations, we noted that several homologues in the NCBI database also appeared to be annotated in the same way, thus providing an explanation for discrepancies noted in this analysis. This allowed us to identify erroneous annotations in the current databases as well as surprising examples of gene overlap in the B. bacteriovorus genome. These findings are described in our website (http://www.biology.ucsd.edu/~msaier/supmat/Bba).
Putative Transporters Lacking Functional Data or Close Homologues in TCDB
One hundred and nine putative transporters were identified on the basis of their predicted transmembrane α-helical topologies, PSORTb localizations, and operon associations (Table S2). Like some of the proteins mentioned in the previous section and on our website, some of the proteins included in Table S2 had been annotated as permease-like constituents, and these were also investigated. The results of these studies are presented on our website.
Discussion and overview
B. bacteriovorus is an organism with an unusual lifestyle. It grows only in the intraperiplasmic space of another host Gram-negative bacterium (see Introduction). It derives its nutrients by degrading host macromolecules and hence secretes degradative enzymes from its own cytoplasm to that of its host cell [20]. Such mechanisms are not yet understood.
We have analyzed the transporters in this organism to determine what systems might confer upon B. bacteriovorus its unique physiological characteristics. We identified several additional putative transport proteins than reported in the original genome annotation effort [11]. Our most interesting and provocative findings will be summarized here, and the potential physiological importance of some of our observations will be considered.
Monovalent Ion Transport
B. bacteriovorus has an unusually large complement of transporters for monovalent cations (H+, K+ and Na+). Some of these are primary active transporters that promote the generation of electrochemical gradients (pmf and smf) that in part comprise the membrane potential. B. bacteriovorus probably uses respiratory function primarily for this purpose, NADH dehydrogenase to generate Na+ gradients, and both cytochrome oxidase and nitric oxide reductase to create the proton electrochemical gradient. These are then used to make high-energy phosphodiester bonds in ATP and pyrophosphate via the F-type ATPase and the H+-translocating pyrophosphatase, respectively. The DsbD oxidoreductase and the dimethylsulfoxide (DMSO) reductase both catalyze transmembrane electron flow from the cytoplasm to the periplasm and thus tend to dissipate the pmf. The Kdp-like P-type K+ uptake system uses ATP to bring K+ into the cell, so if this is an electrogenic system, it should dissipate the pmf. This system is not likely to influence the membrane potential appreciably except under highly selective conditions [124]. The ABC-type Na+ exporter(s) (NatE) also use ATP to pump Na+ out of the cell, so this process is expected to enhance the pmf. If ATP or pyrophosphate is available, then the F-type ATPase or the H+-translocating pyrophosphatase, respectively, can generate a pmf using these high-energy compounds.
All other recognized monovalent ion transporters are cation uniporters, antiporters and symporters, but these derive from many independent families as well as different subfamilies within the primary families. One NhaC cation/proton (or malate•2H+/lactate Na+) antiporter, and five CPA2 cation/proton antiporters, are encoded in the B. bacteriovorus genome. However, four of the 5 proteins from the latter family most closely resemble phylogenetically divergent TC entries within the NhaC family, and may therefore serve dissimilar functions. These homologous systems may, of course, be developmentally regulated, being activated only during specific stages in the B. bacteriovorus life cycle. Single members from (1) the multicomponent CPA3 family, which may use NADH to drive Na+ efflux and (2) a KUP-type K+/H+ symporter for K+ uptake are also present in B. bacteriovorus.
Divalent Cation Transporters
B. bacteriovorus appears to have three or four types of divalent cation uptake systems. One is an ABC-type system similar to the E. coli ZnuABC zinc transport system; a second is represented by three P-type copper ion/heavy metal uptake and/or efflux ATPases [125]; a third is a CorA-type divalent ion-specific channel-type system used primarily for cation uptake; and the fourth is a Zip family carrier-type system responsible for Zn2+, Fe2+ and/or Mn2+ uptake. While the former two types of systems (1 and 2) are driven by ATP-hydrolysis, both of these last two mentioned systems (3 and 4) are probably driven by the membrane potential. These four types of systems could exhibit overlapping but distinct specificities and affinities, and be regulated in response to different stimuli.
Other systems are probably efflux systems. These include (1) a P-type Ca2+-ATPase, (2) the three heavy metal P-type ATPase efflux systems mentioned above, (3) a Ca2+/H+ antiporter (for Ca2+ extrusion), (4) an RND-type heavy metal efflux system, and (5) two CDF-type heavy metal efflux systems, one possibly of broad specificity and the other of narrow specificity (see Results and TCDB). While (1 and 2) are ATP hydrolysis-driven, (3-5) are pmf-driven. These systems probably interact functionally to maintain essential concentrations while preventing the accumulation of toxic levels of these substances.
Anion Transporters
Many systems in B. bacteriovorus transport organic and inorganic solutes with inwardly directed polarity, and in some cases we cannot be sure if the natural substrates are one or the other or both. Of the potential organic anion transporters, there is one system for aromatic acid uptake and one for aromatic acid efflux belonging to the Major Facilitator (TC #2.A.1.15) and ArAE (TC #2.A.85) families, respectively. There is also a member of a family of ABC systems for the uptake of organic phosphonates. Members of the DASS family in the IT superfamily may take up either organic and/or inorganic anions.
At least one member of each of 5 different families is likely to transport inorganic anions. There are two transporters that are probably specific for sulfate (the CHR and SulP families), two likely to be specific for phosphate (the PNaS family of phosphate:Na+ symporters and the PP family of ATP-dependent phosphate uptake permeases in the ABC superfamily), and one specific for various anions (NO3-, NO2-, CN- and HCO3-) in the NNP family within the ABC superfamily.
Nutrient Transport
B. bacteriovorus appears to have numerous uptake systems for amino acids and peptides but relatively few for sugars. The three probable sugar uptake permeases are (1) a member of the GPH family, probably specific for glycosides such as melibiose, (2) an ABC Cut1 family system specific for maltose and malto-oligosaccharides, and (3) a Cut2 family system specific for ribose. In contrast, numerous systems are present for the uptake (and efflux) of amino acids and their derivatives. Five potential peptide uptake permeases derive from four TC families: (1) a potential peptide transporter of the AbgT family in the IT superfamily, (2) an AmpG-like carrier, probably specific for cell wall degradation products in the MF superfamily, (3) a peptide transporting permease within another family of the MFS, the POT family [72], and (4) an oligopeptide (OPP) system and either one or two dipeptide (DPP) system(s), all within the ABC superfamily. In this last case, only a single pair of ATPase subunits was identified, although receptors and membrane proteins were found for two or three systems. Consequently, we postulate that the former ATPase subunits may be shared by both or all three systems. Although we know of no precedence for such a postulate in the ABC superfamily, the sharing of energy-coupling proteins by multiple transporters has been amply documented in the bacterial phosphotransferase system (PTS) superfamily [126].
Uptake systems for amino acids include: (1) a member of the BAT family within the APC superfamily (of unknown specificity, but almost certainly specific for amino acids and their derivatives as are all members of this superfamily), (2) a member of the NSS family, possibly a tryptophan uptake system, (3) a member of the ESS family, likely to transport acidic amino acids such as D- and L-glutamate and their derivatives, and (4) two ABC-type systems, one specific for polar amino acids (the PAAT family) and one specific for hydrophobic amino acids (the HAAT family).
Candidates for the functionally characterized ATP/ADP antiporter(s) in B. bacteriovorus [33,34] include two sequence-divergent members of the organophosphate/phosphate antiporter (OPA) family in the MF superfamily and a sequence divergent member of the phosphate transporter family (PNaS; TC #2.A.58; see our website). If these do in fact prove to be the “missing” antiporters, they will represent new specificities within these families. Other members of the MFS, and members of other families, distantly related to any of the functionally characterized members, potentially could also fulfill this role.
Drugs and Hydrophobic Substances
B. bacteriovorus encodes drug/amphiphile exporters from all superfamilies that are known to include MDR pumps. Thus, there are (1) six MFS (H+ antiport) systems, (2) six RND (H+ antiport) systems, (3) five ABC (ATP-dependent) systems from three different ABC families, one from a predominantly eukaryotic family, (4) two cationic drug exporters of the SMR family within the DMT H+-antiporter superfamily, one probably of narrow specificity and one of broad specificity, and (5) one Na+ antiporter from the MATE family within the MOP superfamily. Altogether there are 20 putative drug/amphiphile exporters, more than 10% of the total number of transporters identified. These pumps may protect B. bacteriovorus against efforts of the host cell to defend itself against bacterial predation using toxic chemicals.
Macromolecular Exporters
Protein secretion and membrane insertion must play a major role in the predatory lifestyle of B. bacteriovorus. It has been suggested that this parasite somehow exports hydrolytic enzymes directly from its own cytoplasm to that of its prey across three membranes (see ref. [20] for a review). The only systems known to achieve this feat in a single step are the type III [127] and type IV [64] secretion systems of Gram-negative bacteria, but these systems are lacking in B. bacteriovorus. An alternative mechanism must therefore be proposed. We suggest that B. bacteriovorus secretes these enzymes into the host periplasm, and that the host cell then takes them up by retrotranslocation [128]. Although such a process has not been documented in prokaryotes, it is well documented in eukaryotes where other mechanisms may also be operative [129].
All living organisms studied to date have a general secretory pathway for exporting unfolded polypeptide chains across the cytoplasmic membrane. However, not all constituents are common to all organisms. Only the SecYEG, Ffh and FtsY proteins are found ubiquitously. B. bacteriovorus has these proteins plus all of the auxiliary constituents found in E. coli [130] that are thought to increase the efficiency of protein export. It also possesses a 4 component Tat system for secreting folded proteins across this membrane. Many organisms have simplified 2 or 3 component Tat systems [96]. Thus, the B. bacteriovorus Sec and Tat systems are just as complicated as those in E. coli.
Potential hemolysins that could destabilize the host cell membrane if exported to the host periplasm and a holin that probably exports an autolysin across the B. bacteriovorus cytoplasmic membrane were also detected. But how does B. bacteriovorus export proteins across its own outer membrane? At least four types of such systems were positively identified. (1) Type I ABC systems export proteins across both membranes in a single energy-coupled step. These systems are usually substrate protein specific and are therefore limited in scope. (2) The flagellar export machinery has been shown to be capable of exporting pathogenicity-related proteins in addition to flagellar proteins, but again, this process appears to be of limited physiological significance [131,132]. (3) The same arguments apply to pilus (Pil) and fimbrial (Fim) protein secretion systems: they appear to be specific for the subunits of the pilus or fimbrium they assemble. (4) B. bacteriovorus also has several “autotransporters” that have C-terminal domains that insert into the outer membrane and translocate their N-terminal domains across this structure. However, each autotransporter is usually specific for its own N-terminal protein domain [133].
What system, then, is responsible for secreting the majority of the many exported proteins that B. bacteriovorus encodes in its genome across the outer membrane? We identified many and perhaps all of the essential components of the Main Terminal Branch (MTB) (Type II) pathway (TC #3.A.15) [67,134]. In many Gram-negative bacteria, such systems provide the principal pathway for protein secretion across the outer membrane. Although these systems are distantly related to Pil-type systems, the sequence similarity of the putative B. bacteriovorus “Pul” proteins to authentic Pul proteins of Klebsiella was much greater than that of the characterized Pil homologues. Also, the putative pul genes of B. bacteriovorus were found in operons that did not include and mapped distantly from any of the pil genes. These observations lead us to suggest that the MTB provides the primary route for the export of secreted protein across the outer membrane of B. bacteriovorus. It is noteworthy that E. coli lacks such a system.
The genome also encodes other potential macromolecular (polysaccharide, lipid and lipopolysaccharide) exporters including several of the ABC, RND and MOP superfamilies [135,136,137]. Many other observations suggested an unusual degree of complexity in B. bacteriovorus that is lacking in most well studied bacteria such as E. coli and B. subtilis. For example, we noted unusual numbers of homologues of the flagellar motor (3 each of MotA and MotB), of the TolAQR energizers of outer membrane OMR-type importers and of the outer membrane assembly complexes (see Table 3). In each case, the numbers of homologues suggest that although the molecular mechanisms are similar in the two organisms, the degree of functional complexity is much greater in B. bacteriovorus than in E. coli. These pmf- or smf-dependent energizers may also provide novel functions such as serving as the motor for gliding motility [61].
An interesting observation relates to the relatively large proportion of fusion proteins in B. bacteriovorus that in most bacteria are encoded by two distinct genes. Why B. bacteriovorus has a disproportionate number of such fusion proteins is not clear. Fusion often facilitates complex formation and increases both stability and efficiency of the assembled complex [138,139,140], but why B. bacteriovorus as compared to other bacteria would prefer to use this mechanism remains to be determined. Possibly B. bacteriovorus is subject to a wider range of stress conditions by virtue of its parasitic lifestyle. This possibility might also explain the large proportion of stress-relieving channel proteins and multidrug efflux pumps.
Supplementary Material
Acknowledgments
This work was supported by NIH grants GM077402, GM55434 and GM64368. We thank Mary Beth Hiller for her assistance in the preparation of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Stolp H. The bdellovibrios: bacterial parasites of bacteria. Ann. Rev. Phytopathol. 1973;11:53–76. [Google Scholar]
- 2.Tudor JJ, McCann MP, Acrich IA. A new model for the penetration of prey cells by bdellovibrios. J. Bacteriol. 1990;172:2421–2426. doi: 10.1128/jb.172.5.2421-2426.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Starr MP, Huang JC. Physiology of the bdellovibrios. Adv. Microb. Physiol. 1972;8:215–261. doi: 10.1016/s0065-2911(08)60191-5. [DOI] [PubMed] [Google Scholar]
- 4.Ruby EG. Cell-envelope modifications accompanying intracellular growth of Bdellovibrio bacteriovorus. In: Moulder JW, editor. Intracellular Parasitism. CRC Press; Boca Raton, FL: 1989. pp. 17–34. Ch. 2. [Google Scholar]
- 5.Ruby EG, Rittenberg SC. Attachment of diaminopimelic acid to bdelloplast peptidoglycan during intraperiplasmic growth of Bdellovibrio bacteriovorus 109J. J. Bacteriol. 1984;158:597–602. doi: 10.1128/jb.158.2.597-602.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Araki Y, Ruby EG. A soluble enzyme activity that attaches free diaminopimelic acid to bdelloplast peptidoglycan. Biochemistry. 1988;27:2624–2629. doi: 10.1021/bi00407a053. [DOI] [PubMed] [Google Scholar]
- 7.Rittenberg SC, Shilo M. Early host damage in the infection cycle of Bdellovibrio bacteriovorus. J. Bacteriol. 1970;102:149–160. doi: 10.1128/jb.102.1.149-160.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruby EG, Rittenberg SC. Differentiation after premature release of intraperiplasmically growing Bdellovibrio bacteriovorous. J. Bacteriol. 1983;154:32–40. doi: 10.1128/jb.154.1.32-40.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diedrich DL, Denny CF, Hashimoto T, Conti SF. Facultatively parasitic strain of Bdellovibrio bacteriovorus. J. Bacteriol. 1970;101:989–996. doi: 10.1128/jb.101.3.989-996.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lambert C, Evans KJ, Till R, Hobley L, Rendulic S, Schuster SC, Aizawa SI, Sockett RE. Characterising the flagellar filament and the role of motility in bacterial prey-penetration by Bdellovibrio bacteriovorus. Mol. Microbiol. 2006;60:274–286. doi: 10.1111/j.1365-2958.2006.05081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rendulic S, Jagtap P, Rosinus A, Eppinger M, Baar C, Lanz C, Keller H, Lambert C, Evans KJ, Goesmann A, Meyer F, Sockett RE, Schuster SC. A predator unmasked: life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science. 2004;303:689–692. doi: 10.1126/science.1093027. [DOI] [PubMed] [Google Scholar]
- 12.Thomashow LS, Rittenberg SC. Waveform analysis and structure of flagella and basal complexes from Bdellovibrio bacteriovorus 109J. J. Bacteriol. 1985;163:1038–1046. doi: 10.1128/jb.163.3.1038-1046.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hasegawa H, Naganuma K, Nakagawa Y, Matsuyama T. Membrane filter (pore size, 0.22-0.45 μm; thickness, 150 μm) passing-through activity of Pseudomonas aeruginosa and other bacterial species with indigenous infiltration ability. FEMS Microbiol. Lett. 2003;223:41–46. doi: 10.1016/S0378-1097(03)00327-6. [DOI] [PubMed] [Google Scholar]
- 14.Rittenberg SC. Bdellovibrios–intraperiplasmic growth. In: Burns RG, Slater JH, editors. Experimental Microbial Ecology. Blackwell Scientific; Oxford, UK: 1982. pp. 379–391. [Google Scholar]
- 15.Thomashow MF, Rittenberg SC. Intraperiplasmic growth of Bdellovibrio bacteriovorus 109J: solubilization of Escherichia coli peptidoglycan. J. Bacteriol. 1978;135:998–1007. doi: 10.1128/jb.135.3.998-1007.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fackrell HB, Robinson J. Purification and characterization of a lytic peptidase produced by Bdellovibrio bacteriovorus 6-5-S. Can. J. Microbiol. 1973;19:659–666. doi: 10.1139/m73-107. [DOI] [PubMed] [Google Scholar]
- 17.Dworkin J, Losick R. Developmental commitment in a bacterium. Cell. 2005;121:401–409. doi: 10.1016/j.cell.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 18.Stragier P, Losick R. Cascades of sigma factors revisited. Mol. Microbiol. 1990;4:1801–1806. doi: 10.1111/j.1365-2958.1990.tb02028.x. [DOI] [PubMed] [Google Scholar]
- 19.Romo AJ, Ruby EG, Saier MH., Jr. Effect of Bdellovibrio bacteriovorus infection on the phosphoenolpyruvate:sugar phosphotransferase system in Escherichia coli: evidence for activation of cytoplasmic proteolysis. Res. Microbiol. 1992;143:5–14. doi: 10.1016/0923-2508(92)90029-n. [DOI] [PubMed] [Google Scholar]
- 20.Saier MH., Jr. Protein uptake into E. coli during Bdellovibrio infection: A process of reverse secretion? FEBS Lett. 1994;337:14–17. doi: 10.1016/0014-5793(94)80620-9. [DOI] [PubMed] [Google Scholar]
- 21.Tudor JJ, Karp MA. Translocation of an outer membrane protein into prey cytoplasmic membranes by bdellovibrios. J. Bacteriol. 1994;176:948–952. doi: 10.1128/jb.176.4.948-952.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eksztejn M, Varon M. Elongation and cell division in Bdellovibrio bacteriovorus. Arch. Microbiol. 1977;114:175–181. doi: 10.1007/BF00410781. [DOI] [PubMed] [Google Scholar]
- 23.Gray KM, Ruby EG. Prey-derived signals regulating duration of the developmental growth phase of Bdellovibrio bacteriovorus. J. Bacteriol. 1990;172:4002–4007. doi: 10.1128/jb.172.7.4002-4007.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomashow MF, Cotter TW. Bdellovibrio host dependence: the search for signal molecules and genes that regulate the intraperiplasmic growth cycle. J. Bacteriol. 1992;174:5767–5771. doi: 10.1128/jb.174.18.5767-5771.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hampton T. Researchers eye “predatory” bacterium for novel antimicrobial strategies. JAMA. 2004;291:1188–1189. doi: 10.1001/jama.291.10.1188. [DOI] [PubMed] [Google Scholar]
- 26.Moore A. Finding my enemy's enemies. EMBO Rep. 2004;5:754–757. doi: 10.1038/sj.embor.7400216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sockett RE, Lambert C. Bdellovibrio as therapeutic agents: a predatory renaissance? Nat. Rev. Microbiol. 2004;2:669–675. doi: 10.1038/nrmicro959. [DOI] [PubMed] [Google Scholar]
- 28.Seidler RJ, Starr MP. Isolation and characterization of host-independent Bdellovibrios. J. Bacteriol. 1969;100:769–785. doi: 10.1128/jb.100.2.769-785.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruby EG, McCabe JB. Metabolism of periplasmic membrane-derived oligosaccharides by the predatory bacterium Bdellovibrio bacteriovorus 109J. J. Bacteriol. 1988;170:646–652. doi: 10.1128/jb.170.2.646-652.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Starr MP, Seidler RJ. The Bdellovibros. Annu. Rev. Microbiol. 1971;25:649–678. doi: 10.1146/annurev.mi.25.100171.003245. [DOI] [PubMed] [Google Scholar]
- 31.Barel G, Sirota A, Volpin H, Jurkevitch E. Fate of predator and prey proteins during growth of Bdellovibrio bacteriovorus on Escherichia coli and Pseudomonas syringae prey. J. Bacteriol. 2005;187:329–335. doi: 10.1128/JB.187.1.329-335.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matin A, Rittenberg SC. Kinetics of deoxyribonucleic acid destruction and synthesis during growth of Bdellovibrio bacteriovorus strain 109D on Pseudomonas putida and Escherichia coli. J. Bacteriol. 1972;111:664–673. doi: 10.1128/jb.111.3.664-673.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruby EG, McCabe JB. An ATP transport system in the intracellular bacterium, Bdellovibrio bacteriovorus 109J. J. Bacteriol. 1986;167:1066–1070. doi: 10.1128/jb.167.3.1066-1070.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruby EG, McCabe JB, Barke JI. Uptake of intact nucleoside monophosphates by Bdellovibrio bacteriovorus 109J. J. Bacteriol. 1985;163:1087–1094. doi: 10.1128/jb.163.3.1087-1094.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Busch W, Saier MH., Jr. The Transporter Classification (TC) System. CRC Crit. Rev. Biochem. Mol. Biol. 2002;37:287–337. doi: 10.1080/10409230290771528. 2002. [DOI] [PubMed] [Google Scholar]
- 36.Saier MH, Jr., Tran CV, Barabote RD. TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucl. Acids Res. 2006;34:D181–D186. doi: 10.1093/nar/gkj001. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lorca GL, Barabote RD, Zlotopolski V, Tran C, Winnen B, Hvorup RN, Stonestrom AJ, Nguyen E, Huang LW, Kim DS, Saier MH., Jr. Transport capabilities of eleven gram-positive bacteria: Comparative genomic analyses. Biochim. Biophys. Acta. 2007;1768:1342–1366. doi: 10.1016/j.bbamem.2007.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saier MH., Jr. Protein secretion systems in Gram-negative bacteria. Microbe. 2006;1:414–419. doi: 10.1007/s00232-006-0049-7. [DOI] [PubMed] [Google Scholar]
- 39.Ren Q, Paulsen IT. Comparative analyses of fundamental differences in membrane transport capabilities in prokaryotes and eukaryotes. PLoS Comput. Biol. 2005;3:e27. doi: 10.1371/journal.pcbi.0010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barel G, Jurkevitch E. Analysis of phenotypic diversity among host-independent mutants of Bdellovibrio bacteriovorus 109J. Arch. Microbiol. 2001;176:211–216. doi: 10.1007/s002030100312. [DOI] [PubMed] [Google Scholar]
- 41.Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- 42.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 43.Saier MH., Jr. Tracing pathways of transport protein evolution. Mol. Microbiol. 2003;48:1145–1156. doi: 10.1046/j.1365-2958.2003.03499.x. [DOI] [PubMed] [Google Scholar]
- 44.Chung Y-J, Saier MH., Jr. SMR-type multidrug resistance pumps. Curr. Opin. Drug Discov. Dev. 2001;4:237–245. [PubMed] [Google Scholar]
- 45.Pornillos O, Chang G. Inverted repeat domains in membrane proteins. FEBS Lett. 2006;580:358–362. doi: 10.1016/j.febslet.2005.12.054. [DOI] [PubMed] [Google Scholar]
- 46.Saier MH., Jr. Families of proteins forming transmembrane channels. J. Membr. Biol. 2000a;175:165–180. doi: 10.1007/s00232001065. [DOI] [PubMed] [Google Scholar]
- 47.Saier MH., Jr. A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol. Mol. Biol. Rev. 2000b;64:354–411. doi: 10.1128/mmbr.64.2.354-411.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paulsen IT, Chauvaux S, Choi P, Saier MH., Jr. Characterization of glucose-specific catabolite repression-resistant mutants of Bacillus subtilis: identification of a novel Hexose:H+ symporter. J. Bacteriol. 1998;180:498–504. doi: 10.1128/jb.180.3.498-504.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paulsen IT, Nguyen L, Sliwinski MK, Rabus R, Saier MH., Jr. Microbial genome analyses: comparative transport capabilities in eighteen prokaryotes. J. Mol. Biol. 2000;301:75–100. doi: 10.1006/jmbi.2000.3961. [DOI] [PubMed] [Google Scholar]
- 50.Hiramatsu T, Kodama K, Kuroda T, Mizushima T, Tsuchiya T. A putative multisubunit Na+/H+ antiporter from Staphylococcus aureus. J. Bacteriol. 1998;180:6642–6648. doi: 10.1128/jb.180.24.6642-6648.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ito M, Guffanti AA, Wang W, Krulwich TA. Results of non-polar mutations in each of the seven Bacillus subtilis mrp genes suggest complex interactions among the gene products in support of Na+- and Alkali- but not cholate-resistance. J. Bacteriol. 2000;182:5663–5670. doi: 10.1128/jb.182.20.5663-5670.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tani K, Watanabe T, Matsuda H, Nasu M, Kondo M. Cloning and sequencing of the spore germination gene of Bacillus megaterium ATCC 12872: similarities to the NaH-antiporter gene of Enterococcus hirae. Microbiol. Immunol. 1996;40:99–105. doi: 10.1111/j.1348-0421.1996.tb03323.x. [DOI] [PubMed] [Google Scholar]
- 53.Roosild TP, Miller S, Booth IR, Choe S. A mechanism of regulating transmembrane potassium flux through a ligand-mediated conformational switch. Cell. 2002;109:781–791. doi: 10.1016/s0092-8674(02)00768-7. [DOI] [PubMed] [Google Scholar]
- 54.Au KM, Barabote RD, Hu KY, Saier MH., Jr. Evolutionary appearance of H+-translocating pyrophosphatases. Microbiology. 2006;152:1243–1247. doi: 10.1099/mic.0.28581-0. [DOI] [PubMed] [Google Scholar]
- 55.Gardy JL, Laird MR, Chen F, Rey S, Walsh CJ, Ester M, Brinkman FS. PSORTb v.2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics. 2005;21:617–623. doi: 10.1093/bioinformatics/bti057. [DOI] [PubMed] [Google Scholar]
- 56.Rey S, Acab M, Gardy JL, Laird MR, deFays K, Lambert C, Brinkman FS. PSORTdb: a protein subcellular localization database for bacteria. Nucleic Acids Res. 2005;33:D164–168. doi: 10.1093/nar/gki027. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kall L, Sonnhammer EL. Reliability of transmembrane predictions in whole-genome data. FEBS Lett. 2002;532:415–418. doi: 10.1016/s0014-5793(02)03730-4. [DOI] [PubMed] [Google Scholar]
- 58.Pivetti CD, Yen M-R, Miller S, Busch W, Tseng Y-H, Booth IR, Saier MH., Jr. Two families of prokaryotic mechanosensitive channel proteins. Microbiol. Mol. Biol. Rev. 2003;67:66–85. doi: 10.1128/MMBR.67.1.66-85.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Calvio C, Celandroni F, Ghelardi E, Amati G, Salvetti S, Ceciliani F, Galizzi A, Senesi S. Swarming differentiation and swimming motility in Bacillus subtilis are controlled by swrA, a newly identified dicistronic operon. J. Bacteriol. 2005;187:5356–5366. doi: 10.1128/JB.187.15.5356-5366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ito M, Hicks DB, Henkin TM, Guffanti AA, Powers BD, Zvi L, Uematsu K, Krulwich TA. MotPS is the stator-force generator for motility of alkaliphilic Bacillus, and its homologue is a second functional Mot in Bacillus subtilis. Mol. Microbiol. 2004;53:1035–1049. doi: 10.1111/j.1365-2958.2004.04173.x. [DOI] [PubMed] [Google Scholar]
- 61.Youderian P, Burke N, White DJ, Hartzell PL. Identification of genes required for adventurous gliding motility in Myxococcus xanthus with the transposable element mariner. Mol. Microbiol. 2003;49:555–570. doi: 10.1046/j.1365-2958.2003.03582.x. [DOI] [PubMed] [Google Scholar]
- 62.Kehres DG, Lawyer CH, Maguire ME. The CorA magnesium transporter gene family. Microb. Comp. Genomics. 1998;3:151–169. doi: 10.1089/omi.1.1998.3.151. [DOI] [PubMed] [Google Scholar]
- 63.Dinh T, Paulsen IT, Saier MH., Jr. A family of extracytoplasmic proteins that allow transport of large molecules across the outer membranes of Gram-negative bacteria. J. Bacteriol. 1994;176:3825–3831. doi: 10.1128/jb.176.13.3825-3831.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cao TB, Saier MH., Jr. Conjugal Type IV macromolecular transfer systems of Gram-negative bacteria: Organismal distribution, structural constraints and evolutionary conclusions. Microbiology. 2001;147:3201–3214. doi: 10.1099/00221287-147-12-3201. [DOI] [PubMed] [Google Scholar]
- 65.Collins RF, Ford RC, Kitmitto A, Olsen RO, Tønjum T, Derrick JP. Three-dimensional structure of the Neisseria meningitidis secretin PilQ determined from negative-stain transmission electron microscopy. J. Bacteriol. 2003;185:2611–2617. doi: 10.1128/JB.185.8.2611-2617.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chami M, Guilvout I, Gregorini M, Remigy HW, Muller SA, Valerio M, Engel A, Pugsley AP, Bayan N. Structural insights into the secretin PulD and its trypsin-resistant core. J. Biol. Chem. 2005;280:37732–33741. doi: 10.1074/jbc.M504463200. [DOI] [PubMed] [Google Scholar]
- 67.Sandkvist M. Biology of type II secretion. Mol. Microbiol. 2001;40:271–283. doi: 10.1046/j.1365-2958.2001.02403.x. [DOI] [PubMed] [Google Scholar]
- 68.Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, Kahne D. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 69.Bayles KW. Are the molecular strategies that control apoptosis conserved in bacteria? Trends Microbiol. 2003;11:306–311. doi: 10.1016/s0966-842x(03)00144-6. [DOI] [PubMed] [Google Scholar]
- 70.Chahboune A, Decaffmeyer M, Brasseur R, Joris B. Membrane topology of the Escherichia coli AmpG permease required for recycling of cell wall anhydromuropeptides and AmpC beta-lactamase induction. Antimicrob. Agents Chemother. 2005;49:1145–1149. doi: 10.1128/AAC.49.3.1145-1149.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cheng Q, Park JT. Substrate specificity of the AmpG permease required for recycling of cell wall anhydro-muropeptides. J. Bacteriol. 2002;184:6434–6436. doi: 10.1128/JB.184.23.6434-6436.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chang AB, Lin R, Studley WK, Tran CV, Saier MH., Jr. Phylogeny as a guide to structure and function of membrane transport proteins. Mol. Membrane Biol. 2004;21:171–181. doi: 10.1080/09687680410001720830. [DOI] [PubMed] [Google Scholar]
- 73.Pao SS, Paulsen IT, Saier MH., Jr. The major facilitator superfamily. Microbiol. Mol. Biol. Rev. 1998;62:1–32. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Saier MH, Jr., Beatty JT, Goffeau A, Harley KT, Heijne WHM, Huang S-C, Jack DL, Jahn PS, Lew K, Liu J, Pao SS, Paulsen IT, Tseng T-T, Virk PS. The major facilitator superfamily. J. Mol. Microbiol. Biotechnol. 1999;1:257–279. [PubMed] [Google Scholar]
- 75.Chao Y, Fu D. Kinetic study of the antiport mechanism of an Escherichia coli zinc transporter, ZitB. J. Biol. Chem. 2004;279:12043–12050. doi: 10.1074/jbc.M313510200. [DOI] [PubMed] [Google Scholar]
- 76.Guffanti AA, Wei Y, Rood SV, Krulwich TA. An antiport mechanism for a member of the cation diffusion facilitator family: divalent cations efflux in exchange for K+ and H+ Mol. Microbiol. 2002;45:145–153. doi: 10.1046/j.1365-2958.2002.02998.x. [DOI] [PubMed] [Google Scholar]
- 77.Grass G, Franke S, Taudte N, Nies DH, Kucharski LM, Maguire ME, Rensing C. The metal permease ZupT from Escherichia coli is a transporter with a broad substrate spectrum. J. Bacteriol. 2005;187:1604–1611. doi: 10.1128/JB.187.5.1604-1611.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yu EW, Aires JR, Nikaido H. AcrB multidrug efflux pump of Escherichia coli: composite substrate-binding cavity of exceptional flexibility generates its extremely wide substrate specificity. J. Bacteriol. 2003;185:5657–5664. doi: 10.1128/JB.185.19.5657-5664.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chung YJ, Saier MH., Jr. Overexpression of the Escherichia coli sugE gene confers resistance to a narrow range of quaternary ammonium compounds. J. Bacteriol. 2002;184:2543–2545. doi: 10.1128/JB.184.9.2543-2545.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Livshits VA, Zakataeva NP, Aleshin BB, Vitushkina MV. Identification and characterization of the new gene rhtA involved in threonine and homoserine efflux in Escherichia coli. Res. Microbiol. 2003;154:123–135. doi: 10.1016/S0923-2508(03)00036-6. [DOI] [PubMed] [Google Scholar]
- 81.Androutsellis-Theotokis A, Goldberg NR, Ueda K, Beppu T, Beckman ML, Das S, Javitch JA, Rudnick G. Characterization of a functional bacterial homologue of sodium-dependent neurotransmitter transporters. J. Biol. Chem. 2003;278:12703–12709. doi: 10.1074/jbc.M206563200. [DOI] [PubMed] [Google Scholar]
- 82.Ito M, Guffanti AA, Zemsky J, Ivey DM, Krulwich TA. Role of the nhaC-encoded Na+/H+ antiporter of alkaliphilic Bacillus firmus OF4. J. Bacteriol. 1997;179:3851–3857. doi: 10.1128/jb.179.12.3851-3857.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wei Y, Guffanti AA, Ito M, Krulwich TA. Bacillus subtilis YqkI is a novel malic/Na+-lactate antiporter that enhances growth on malate at low protonmotive force. J. Biol. Chem. 2000;275:30287–30292. doi: 10.1074/jbc.M001112200. [DOI] [PubMed] [Google Scholar]
- 84.Putnoky P, Kereszt A, Nakamura T, Endre G, Grosskopf E, Kiss P, Kondorosi A. The pha gene cluster of Rhizobium meliloti involved in pH adaptation and symbiosis encodes a novel type of K+ efflux system. Mol. Microbiol. 1998;28:1091–1101. doi: 10.1046/j.1365-2958.1998.00868.x. [DOI] [PubMed] [Google Scholar]
- 85.Trchounian A, Kobayashi H. Kup is the major K+ uptake system in Escherichia coli upon hyper-osmotic stress at a low pH. FEBS Lett. 1999;447:144–148. doi: 10.1016/s0014-5793(99)00288-4. [DOI] [PubMed] [Google Scholar]
- 86.Zakharyan E, Trchounian A. K+ influx by Kup in Escherichia coli is accompanied by a decrease in H+ efflux. FEMS Microbiol. Lett. 2001;204:61–64. doi: 10.1111/j.1574-6968.2001.tb10863.x. [DOI] [PubMed] [Google Scholar]
- 87.Neidle EL, Hartnett C, Ornston LN, Bairoch A, Rekik M, Harayama S. Nucleotide sequences of the Acinetobacter calcoaceticus benABC genes for benzoate 1,2-dioxygenase reveal evolutionary relationships among multicomponent oxygenases. J. Bacteriol. 1991;173:5385–5395. doi: 10.1128/jb.173.17.5385-5395.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Prakash S, Cooper G, Singhi S, Saier MH., Jr. The ion transporter superfamily. Biochim. Biophys. Acta. 2003;1618:79–92. doi: 10.1016/j.bbamem.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 89.Alvarez AH, Moreno-Sánchez R, Cervantes C. Chromate efflux by means of the ChrA chromate resistance protein from Pseudomonas aeruginosa. J. Bacteriol. 1999;181:7398–7400. doi: 10.1128/jb.181.23.7398-7400.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nicholson ML, Laudenbach DE. Genes encoded on a cyanobacterial plasmid are transcriptionally regulated by sulfur availability and CysR. J. Bacteriol. 1995;177:2143–2150. doi: 10.1128/jb.177.8.2143-2150.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nies D, Koch S, Wachi S, Peitzsch N, Saier MH., Jr. CHR, a novel family of prokaryotic proton motive force-driven transporters probably containing chromate/sulfate antiporters. J. Bacteriol. 1998;180:5799–5802. doi: 10.1128/jb.180.21.5799-5802.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pimentel BE, Moreno-Sanchez R, Cervantes C. Efflux of chromate by Pseudomonas aeruginosa cells expressing the ChrA protein. FEMS Microbiol. Lett. 2002;212:249–254. doi: 10.1111/j.1574-6968.2002.tb11274.x. [DOI] [PubMed] [Google Scholar]
- 93.Van Dyk TK, Templeton LJ, Cantera KA, Sharpe PL, Sariaslani FS. Characterization of the Escherichia coli AaeAB efflux pump: a metabolic relief valve? J. Bacteriol. 2004;186:7196–7204. doi: 10.1128/JB.186.21.7196-7204.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Harley KT, Saier MH., Jr. A novel ubiquitous family of putative efflux transporters. J. Mol. Microbiol. Biotechnol. 2000;2:195–198. [PubMed] [Google Scholar]
- 95.Yen M-R, Harley KT, Tseng Y-H, Saier MH., Jr. Phylogenetic and structural analyses of the Oxa1 family of putative protein translocation constituents. FEMS Microbiol. Lett. 2001;204:223–231. doi: 10.1111/j.1574-6968.2001.tb10889.x. [DOI] [PubMed] [Google Scholar]
- 96.Yen M-R, Tseng Y-H, Nguyen EH, Wu L-F, Saier MH., Jr. Sequence and phylogenetic analyses of the twin arginine targeting (Tat) protein export system. Arch. Microbiol. 2002;177:441–450. doi: 10.1007/s00203-002-0408-4. [DOI] [PubMed] [Google Scholar]
- 97.Darzins A, Russell MA. Molecular genetic analysis of type-4 pilus biogenesis and twitching motility using Pseudomonas aeruginosa as a model system–a review. Gene. 1997;192:109–115. doi: 10.1016/s0378-1119(97)00037-1. [DOI] [PubMed] [Google Scholar]
- 98.Hobbs M, Mattick JS. Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol. Microbiol. 1993;10:233–243. doi: 10.1111/j.1365-2958.1993.tb01949.x. [DOI] [PubMed] [Google Scholar]
- 99.Schwudke D, Bernhardt A, Beck S, Madela K, Linscheid MW, Appel B, Strauch E. Transcriptional activity of the host-interaction locus and a putative pilin gene of Bdellovibrio bacteriovorus in the predatory life cycle. Curr. Microbiol. 2005;51:310–316. doi: 10.1007/s00284-005-0030-1. [DOI] [PubMed] [Google Scholar]
- 100.Buckel W. Sodium ion-translocating decarboxylases. Biochim. Biophys. Acta. 2001;1505:15–27. doi: 10.1016/s0005-2728(00)00273-5. [DOI] [PubMed] [Google Scholar]
- 101.Dimroth P, Jockel P, Schmid M. Coupling mechanism of the oxaloacetate decarboxylase Na+ pump. Biochim. Biophys. Acta. 2001;1505:1–14. doi: 10.1016/s0005-2728(00)00272-3. [DOI] [PubMed] [Google Scholar]
- 102.Schmid M, Wild MR, Dahinden P, Dimroth P. Subunit gamma of the oxaloacetate decarboxylase Na+ pump: interaction with other subunits/domains of the complex and binding site for the Zn2+ metal ion. Biochemistry. 2002;41:1285–1292. doi: 10.1021/bi015764l. [DOI] [PubMed] [Google Scholar]
- 103.Dahinden P, Pos KM, Dimroth P. Identification of a domain in the α-subunit of the oxaloacetate decarboxylase Na+ pump that accomplishes complex formation with the γ-subunit. FEBS J. 2005;272:846–855. doi: 10.1111/j.1742-4658.2004.04524.x. [DOI] [PubMed] [Google Scholar]
- 104.Hirst J. The dichotomy of complex I: a sodium ion pump or a proton pump. Proc. Natl. Acad. Sci. USA. 2003;100:773–775. doi: 10.1073/pnas.0330050100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sapra R, Bagramyan K, Adams MWW. A simple energy-conserving system: proton reduction coupled to proton translocation. Proc. Natl. Acad. Sci. USA. 2003;100:7545–7550. doi: 10.1073/pnas.1331436100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Steuber J, Schmid C, Rufibach M, Dimroth P. Na+ translocation by complex I (NADH: quinone oxidoreductase) of Escherichia coli. Mol. Microbiol. 2000;35:428–434. doi: 10.1046/j.1365-2958.2000.01712.x. [DOI] [PubMed] [Google Scholar]
- 107.Flock U, Reimann J, Adelroth P. Proton transfer in bacterial nitric oxide reductase. Biochem. Soc. Trans. 2006;34(Pt 1):188–190. doi: 10.1042/BST0340188. [DOI] [PubMed] [Google Scholar]
- 108.Meakin GE, Jepson BJ, Richardson DJ, Bedmar EJ, Delgado MJ. The role of Bradyrhizobium japonicum nitric oxide reductase in nitric oxide detoxification in soya bean root nodules. Biochem. Soc. Trans. 2006;34(Pt 1):195–196. doi: 10.1042/BST0340195. [DOI] [PubMed] [Google Scholar]
- 109.Kimball RA, Martin L, Saier MH., Jr. Reversing transmembrane electron flow: The DsbD and DsbB protein families. J. Mol. Microbiol. Biotechnol. 2003;5:133–149. doi: 10.1159/000070263. [DOI] [PubMed] [Google Scholar]
- 110.Katzen F, Beckwith J. Transmembrane electron transfer by the membrane protein DsbD occurs via a disulfide bond cascade. Cell. 2000;103:769–779. doi: 10.1016/s0092-8674(00)00180-x. [DOI] [PubMed] [Google Scholar]
- 111.Krupp R, Chan C, Missiakas D. DsbD-catalyzed transport of electrons across the membrane of Escherichia coli. J. Biol. Chem. 2001;276:3696–3701. doi: 10.1074/jbc.M009500200. [DOI] [PubMed] [Google Scholar]
- 112.Hille R. Molybdenum and tungsten in biology. Trends Biochem. Sci. 2002;27:360–367. doi: 10.1016/s0968-0004(02)02107-2. [DOI] [PubMed] [Google Scholar]
- 113.McEwan AG, Hanson GR, Bailey S. Dimethylsulphoxide reductase from purple phototrophic bacteria: structures and mechanism(s) Biochem. Soc. Trans. 1998;26:390–396. doi: 10.1042/bst0260390. [DOI] [PubMed] [Google Scholar]
- 114.Boyle DS, Khattar MM, Addinall SG, Lutkenhaus J, Donachie WD. ftsW is an essential cell-division gene in Escherichia coli. Mol. Microbiol. 1997;24:1263–1273. doi: 10.1046/j.1365-2958.1997.4091773.x. [DOI] [PubMed] [Google Scholar]
- 115.Gérard P, Vernet T, Zapun A. Membrane topology of the Streptococcus pneumoniae FtsW division protein. J. Bacteriol. 2002;184:1925–1931. doi: 10.1128/JB.184.7.1925-1931.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Faergeman NJ, Black PN, Zhao XD, Knudsen J, DiRusso CC. The acyl-CoA synthetases encoded within FAA1 and FAA4 in Saccharomyces cerevisiae function as components of the fatty acid transport system linking import, activation, and intracellular utilization. J. Biol. Chem. 2001;276:37051–37059. doi: 10.1074/jbc.M100884200. [DOI] [PubMed] [Google Scholar]
- 117.Hall AM, Wiczer BM, Herrmann T, Stremmel W, Bernlohr DA. Enzymatic properties of purified murine fatty acid transport protein 4 and analysis of acyl-CoA synthetase activities in tissues from FATP4 null mice. J. Biol. Chem. 2005;280:11948–11954. doi: 10.1074/jbc.M412629200. [DOI] [PubMed] [Google Scholar]
- 118.Hirsch D, Stahl A, Lodish HF. A family of fatty acid transporters conserved from mycobacterium to man. Proc. Natl. Acad. Sci. USA. 1998;95:8625–8629. doi: 10.1073/pnas.95.15.8625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zou Z, DiRusso CC, Ctrnacta V, Black PN. Fatty acid transport in Saccharomyces cerevisiae. Directed mutagenesis of FAT1 distinguishes the biochemical activities associated with Fat1p. J. Biol. Chem. 2002;277:31062–31071. doi: 10.1074/jbc.M205034200. [DOI] [PubMed] [Google Scholar]
- 120.Baida G, Budarina ZI, Kuzmin NP, Solonin AS. Complete nucleotide sequence and molecular characterization of hemolysin II gene from Bacillus cereus. FEMS Microbiol. Lett. 1999;180:7–14. doi: 10.1111/j.1574-6968.1999.tb08771.x. [DOI] [PubMed] [Google Scholar]
- 121.Baida GE, Kuzmin NP. Cloning and primary structure of a new hemolysin gene from Bacillus cereus. Biochim. Biophys. Acta. 1995;1264:151–154. doi: 10.1016/0167-4781(95)00150-f. [DOI] [PubMed] [Google Scholar]
- 122.ter Huurne AA, Muir S, van Houten M, van der Zeijst BA, Gaastra W, Kusters JG. Characterization of three putative Serpulina hyodysenteriae hemolysins. Microb. Pathog. 1994;16:269–282. doi: 10.1006/mpat.1994.1028. [DOI] [PubMed] [Google Scholar]
- 123.Gibson MM, Bagga DA, Miller CG, Maguire ME. Magnesium transport in Salmonella typhimurium: the influence of new mutations conferring Co2+ resistance on the CorA Mg2+ transport system. Mol. Microbiol. 1991;5:2753–2762. doi: 10.1111/j.1365-2958.1991.tb01984.x. [DOI] [PubMed] [Google Scholar]
- 124.Epstein W. The roles and regulation of potassium in bacteria. Prog. Nucleic Acid Res. Mol. Biol. 2003;75:293–320. doi: 10.1016/s0079-6603(03)75008-9. [DOI] [PubMed] [Google Scholar]
- 125.Odermatt A, Suter H, Krapf R, Solioz M. Primary structure of two P-type ATPases involved in copper homeostasis in Enterococcus hirae. J. Biol. Chem. 1993;268:12775–12779. [PubMed] [Google Scholar]
- 126.Tchieu JH, Norris V, Edwards JS, Saier MH., Jr. The complete phosphotransferase system in Escherichia coli. J. Mol. Microbiol. Biotechnol. 2001;3:329–346. [PubMed] [Google Scholar]
- 127.Nguyen L, Paulsen IT, Tchieu J, Hueck CJ, Saier MH., Jr. Phylogenetic analyses of the constituents of type III protein secretion systems. J. Mol. Microbiol. Biotechnol. 2000;2:125–144. [PubMed] [Google Scholar]
- 128.Ahner A, Brodsky JL. Checkpoints in ER-associated degradation: excuse me, which way to the proteasome? Trends Cell Biol. 2004;14:474–478. doi: 10.1016/j.tcb.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 129.Schmitz A, Herzog V. Endoplasmic reticulum-associated degradation: exceptions to the rule. Eur. J. Cell Biol. 2004;83:501–509. doi: 10.1078/0171-9335-00412. [DOI] [PubMed] [Google Scholar]
- 130.Cao TB, Saier MH., Jr. The general protein secretory pathway: Phylogenetic analyses leading to evolutionary conclusions. Biochim. Biophys. Acta. 2003;1609:115–125. doi: 10.1016/s0005-2736(02)00662-4. [DOI] [PubMed] [Google Scholar]
- 131.Saier MH., Jr. Evolution of bacterial type III protein secretion systems. Trends Microbiol. 2004;12:113–115. doi: 10.1016/j.tim.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 132.Song YC, Jin S, Louie H, Ng D, Lau R, Zhang Y, Weerasekera R, Al Rashid S, Ward LA, Der SD, Chan VL. FlaC, a protein of Campylobacter jejuni TGH9011 (ATCC43431) secreted through the flagellar apparatus, binds epithelial cells and influences cell invasion. Mol. Microbiol. 2004;53:541–553. doi: 10.1111/j.1365-2958.2004.04175.x. [DOI] [PubMed] [Google Scholar]
- 133.Loveless BJ, Saier MH., Jr. A novel family of channel-forming, autotransporting, bacterial virulence factors. Mol. Membr. Biol. 1997;14:113–123. doi: 10.3109/09687689709048171. [DOI] [PubMed] [Google Scholar]
- 134.Ma Q, Zhai Y, Schneider CJ, Ramseier TM, Saier MH., Jr. Protein secretion systems of Pseudomonas aeruginosa and P. fluorescens. Biochim. Biophys. Acta. 2003;1611:223–233. doi: 10.1016/s0005-2736(03)00059-2. [DOI] [PubMed] [Google Scholar]
- 135.Hvorup RN, Winnen B, Chang AB, Jiang Y, Zhou XF, Saier MH. The multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) exporter superfamily. Eur. J. Biochem. 2003;270:799–813. doi: 10.1046/j.1432-1033.2003.03418.x. [DOI] [PubMed] [Google Scholar]
- 136.Reizer J, Reizer A, Saier MH., Jr. A new subfamily of bacterial ABC-type transport systems catalyzing export of drugs and carbohydrates. Prot. Sci. 1992;1:1326–1332. doi: 10.1002/pro.5560011012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Tseng T-T, Gratwick KS, Kollman J, Park D, Nies DH, Goffeau A, Saier MH., Jr. The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development proteins. J. Mol. Microbiol. Biotechnol. 1999;1:107–125. [PubMed] [Google Scholar]
- 138.Amar P, Ballet P, Barlovatz-Meimon G, Benecke A, Bernot G, Bouligand Y, Bourguine P, Delaplace F, Delosme JM, Demarty M, Fishov I, Fourmentin-Guilbert J, Fralick J, Giavitto JL, Gleyse B, Godin C, Incitti R, Kepes F, Lange C, Le Sceller L, Loutellier C, Michel O, Molina F, Monnier C, Natowicz R, Norris V, Orange N, Pollard H, Raine D, Ripoll C, Rouviere-Yaniv J, Saier M, Jr., Soler P, Tambourin P, Thellier M, Tracqui P, Ussery D, Vincent JC, Vannier JP, Wiggins P, Zemirline A. Hyperstructures, genome analysis and I-cells. Acta Biotheor. 2002;50:357–373. doi: 10.1023/a:1022629004589. [DOI] [PubMed] [Google Scholar]
- 139.Liu N, Caderas G, Deillon C, Hoffmann S, Klauser S, Cui T, Gutte B. Fusion proteins from artificial and natural structural modules. Curr. Protein Pept. Sci. 2001;2:107–121. doi: 10.2174/1389203013381161. [DOI] [PubMed] [Google Scholar]
- 140.Norris V, Gascuel P, Guespin-Michel J, Ripoll C, Saier MH., Jr. Metabolite-induced metabolons: the activation of transporter-enzyme complexes by substrate binding. Mol. Microbiol. 1999;31:1592–1595. doi: 10.1046/j.1365-2958.1999.01275.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.