Abstract
Purpose:
The failure of colorectal cancer treatments is partly due to overexpression of CXCR4 by tumor cells, which plays a critical role in cell metastasis. Moreover, serum alkaline phosphatase (ALP) levels are frequently elevated in patients with metastatic colorectal cancer. A polysaccharide, dextran, was chosen as the vector of siRNA. Spermine was conjugated to oxidized dextran by reductive amination process to obtain cationized dextran, so-called dextran-spermine, in order to prepare CXCR4-siRNAs/dextran-spermine nanoparticles. The fabricated nanoparticles were used in order to investigate whether downregulation of CXCR4 expression could affect serum ALP in mouse models of colorectal cancer.
Methods:
Colorectal cancer was established in BALB/C mice following injection of mouse colon carcinoma cells CT.26WT through the tail vein. CXCR4 siRNA for two sites of the target gene was administered following injection of naked siRNA or siRNA encapsulated into nanoparticles.
Results:
In vivo animal data revealed that CXCR4 silencing by dextran-spermine nanoparticles significantly downregulated CXCR4 expression compared with naked CXCR4 siRNA. Furthermore, there was correlation between CXCR4 expression and serum ALP.
Conclusion:
CXCR4 siRNA/dextran-spermine nanoparticles appear to be highly effective, and may be suitable for further in vivo applications. Further research evaluation will be needed to determine the effect of CXCR4 silencing on serum ALP levels, which may be a useful marker to predict liver metastasis in colorectal cancer.
Keywords: nanoparticles, cationized dextran, colorectal cancer, serum ALP enzyme, CXCR4, siRNA
Introduction
Liver metastasis is the main cause of colorectal cancer-related mortality. Death due to colorectal cancer is often a result of liver metastases. Despite extensive research into the biology of cancer progression, the molecular mechanisms involved in colorectal cancer metastasis are not well characterized.1 Several studies have demonstrated that CXCR4 and its ligand stromal cell-derived factor 1a (SDF-1) are highly expressed in tissues of metastatic growth, such as the lung, liver, and lymph nodes. CXCR4 is necessary for the outgrowth of colon cancer micrometastasis and significantly correlates with survival and liver metastasis.2–6
Alkaline phosphatase (ALP) has isoenzymes mainly derived from the leukocytes, bones, colon, placenta, kidneys, and liver.7,8 Elevated serum ALP levels are frequently associated with a variety of diseases, such as in patients with metastatic colorectal cancer.9 Kohne et al10 suggested that to stratify metastatic colorectal cancer patients in clinical trials, it is necessary to measure ALP, white blood cells, hemoglobin, and platelets. However, the significance of ALP in terms of detecting hepatic metastasis or prognosis is not well established.
RNA interference was first identified as a defense mechanism against the invasion of foreign genes in the nematode Caenorhabditis elegans and has subsequently been discovered in diverse eukaryotes such as fungi, insects, plants, and vertebrates. Researchers demonstrated that synthetic siRNAs are able to induce RNAi in mammalian cells.11 There are two types of small RNA molecules, microRNA or miRNA and small interfering RNA, sometimes known as siRNA or silencing RNA, which are central to RNA interference.12
Researchers demonstrated that inhibition of CXCR4 and its ligand CXCL12 signaling by siRNA knockdown has been found to reduce metastasis breast cancer.13 Delivery of therapeutic siRNA to specific tissues is a major challenge for systemic siRNA delivery. One reason is that the backbone of RNA contains ribose, which has a hydroxyl (OH) group in the 2′ position of the pentose ring instead of a hydrogen (H) atom. This extra hydroxyl group makes the RNA backbone more sensitive to hydrolysis.14 Encapsulation generally provides much better protection of siRNAs against serum degradation than chemical modification.15,16
Azzam et al17,18 Hosseinkhani et al19 Eliyahu et al20 and Hosseinkhani et al21 showed that dextran engrafted spermine capable of complexing and transfecting various genes to different cell lines in vitro and in vivo. Dextran is a water-soluble polysaccharide with multiple hydroxyl groups applicable to chemical modification. Furthermore, it has been extensively used for pharmaceutical applications to demonstrate feasibility in gene expression and cancer gene therapy.22,23 Dextran-spermine can form nanoparticles when mixed with siRNA, and the physicochemical properties are changed by the molecular weight of dextran.24
The aim of this study was to investigate the effect of CXCR4 gene silencing on serum ALP levels in colorectal cancer metastasis to the liver by using CXCR4 siRNAs/dextran-spermine nanoparticles.
Materials and methods
Mouse colon carcinoma cell line (CT26.WT) was purchased from American Type Culture Collection (Manassas, VA). SiGENOME-ON-TARGET plus SMART pool duplex CXCR4 in vitro (Genbank Accession No: NM_00 3467) was:
siRNA1: Sense, G.A.A.G.C.A.U.G.A.C.G.G.A.C.A. A.G.U.A.U.U
siRNA2: Sense, G.G.C.C.U.U.A.U.C.C.U.G.C.C.U.G.G. U.A.U.U
siRNA3: Sense, U.A.A.C.U.A.C.A.C.C.G.A.G.G.A. A.U.G.U.U
siRNA4: Sense, C.A.A.G.C.A.A.G.G.G.U.G.U.G.A. G.U.U.U.U.U
In vivo individual siRNA duplex CXCR4 I (Genbank Accession No: BC098322) (Sense: A.C.C.A.A.C.A.G.U. C.A.G.A.G.G.C.C.A.A.U.U) Cat No: LEEIG-000534, siRNA duplex CXCR4 II (Sense: G.G.U.C.A.U.G.G.G.U.U. A.C.C.A.G.A.A.G.U.U) Cat No: LEEIG-000536 and CXCR4 ON-TARGET plus siCONTROL nontargeting pool were purchased from Dharmacon RNA Technologies (Lafayette, CO). Dextran-spermine (FI-67) was prepared as a gift from University-Hadassah Medical School, Jerusalem, Israel. Other chemical reagents that were used in our study were RPMI (Sigma, Dorset, UK), fetal bovine serum (PAA, Somerset, UK), trypsin-EDTA (1X) (PAA), MTS kit (Promega, Madison, WI), Trizol reagent (Invitrogen, Carls-bad, CA), DNaseI kit (Sigma-Aldrich, St Louis, MO), first strand cDNA synthesis kit (K1612 Fermentas), Quantitect SYBR green reverse transcriptase polymerase chain reaction (RT-PCR) one-step kit (Qiagen, Valencia, CA), rabbit polyclonal to CXCR4 (ab2074) (Abcam, Cambridge, UK), goat antirabbit immunoglobulin (Ig)G (H + L), FITC-labeled (Anaspec, Fremont, CA), LDH optimized assay kit (D-68298, Roche Diagnostics GmbH, Indianapolis, IN), and Triton X-100 (Sigma-Aldrich).
Preparation of dextran-spermine
Dextran-spermine with molecular weight (GPC): Mw = 48400, 20% monoquaternary spermine, elemental analysis of N(%): 9.01 and primary amine (TNBS method): 0.58 mmol/g was prepared as described elsewhere.17 Briefly, a solution of oxidized polysaccharide (2 g) in 200 mL double-distilled water was slowly added over 5–7 hours to a basic solution containing 1.5 equimolar amount of the corresponding oligoamine dissolved in 100 mL of borate buffer (0.1 M, pH 11). The mixture was gently stirred at room temperature for 24 hours. The amine-based conjugates (reduced) were obtained after reducing the amine conjugates with excess NaBH4 (2 g), which was added to the mixture at room temperature for 48 hours. The reduction was repeated with the additional portion of NaBH4 and stirred for 24 hours in the same conditions. The mixture was dialyzed against double-distilled water (6 × 5 L) at 4°C with 3500 cutoff cellulose tubing (Membrane Filtration Products, Inc, San Antonio, TX). The resulting light yellow solution was dialyzed as described previously followed by freeze-drying to obtain yellowish reduced amine-based conjugates in 50% overall yield.
Preparation and characterization of CXCR4 siRNAs nanoparticles
In this study, two kinds of siRNA were chosen specifically for the CXCR4 mRNA to ensure no similarity with other genes. The formation of CXCR4 siRNA I, II/dextran-spermine nanoparticles was performed by simply mixing of CXCR4 siRNA and dextran-spermine at a weight ratio of 1:5 (siRNA I, II/dextran-spermine) in aqueous solution. Calculation of the concentration of the siRNA sample was done by Beer’s law, A260 = (ε)(C)(L), where ε is the extinction coefficient (from the Product Transfer Form), C is the siRNA concentration, and L is the path length of the cuvette. The final concentration of the resuspended siRNA could be done by solving for C and multiplying by the dilution factor. The equation was used to convert between nmol to μg of siRNA: (X nmoL)(Y g/moL)(moL/109 nmoL)(106 μg/g) = Z μg. Briefly, 5 μg of CXCR4 siRNAs I, II was added to 25 μL of RNase free water and the solution was pipetted up and down three to five times and was placed on an orbital mixer/shaker for 10 minutes at room temperature. The same volume of RNase free water containing 25 μg of dextran-spermine was placed on an orbital mixer/shaker for 10 minutes at room temperature. The solution was gently agitated for 30 minutes to form self-assembled siRNAs/dextran-spermine nanoparticles separately.
Size measurements, morphology, and zeta potential of nanoparticles
Morphology of nanoparticles was visualized by transmission electron microscopy model LEO 912AB with Omega energy filter. Nanoparticle size and Fractional volume density distribution (q= Fractional density in the size class) was analyzed using a particle size analyzer (Nanophox/Sympatecs gMBh) at 25°C. Zeta potentials of the nanoparticles were measured using Zetasizer analysis (Malvern Instruments, Malvern, UK) at 25°C with clear disposable zeta cell. The data represent the average ± standard deviations.
Animal experiments
Animal study was done on 7–8-week-old balb/c (Charles River, MA) female mice, which were divided into six groups with six mice per group. The protocol was approved by the animal care committee of Universiti Putra Malaysia (UPM/FPS/00265). Colorectal cancer models were established in balb/c mice by different injections of mouse colon carcinoma cell line (CT26.WT): (1) intravenous (IV) injection through the tail vain and (2) subcuteanous injection. In Group A, the animals were given IV injections of 1 × 105 CT26.WT cells transfected with nonspecific control siRNA duplexes (150 ng/g body weight). The animals were given postinjections of the control siRNA twice weekly through the tail vein. In group B, the animals were given IV injections of 1 × 105 CT26.WT cells (transfected with CXCR4 siRNAI, II/dextran-spermine, 150 ng/g body weight) through the tail vein. In group C, the animals were given IV injections of 1 × 105 CT26.WT cells (transfected with naked CXCR4 siRNAI, II, 150 ng/g body weight) through the tail vein. In group D, the animals were given IV injections of 1 × 105 CT26.WT cells (transfected with naked CXCR4 siRNAI, II, 150 ng/g body weight) through the tail vein. Postinjection of naked CXCR4 siRNAI, II was done twice weekly (150 ng/g body weight) through the tail vein. In group E, the animals were given IV injections of 1 × 105 CT26.WT cells (transfected with CXCR4 siRNAI, II/dextran-spermine, 150 ng/g body weight) through the tail vein. Postinjection of CXCR4 siR-NAI, II/dextran-spermine was done twice weekly (150 ng/g body weight) through the tail vein. In group F, the animals in group F were given nonspecific control siRNA duplexes twice weekly via injection through the tail vein without any injection of CT26.WT cells. Animals were sacrificed after 35 days.
RNA preparation and real-time reverse transcription-polymerase chain reaction analyses
Total RNA was isolated from six groups of mice frozen liver tissues (50 mg) using Trizol reagent, according to the manufacturer’s instruction. Real-time quantitative RT-PCR was performed in the real-time PCR machine (tubes) RotorGene 3000 (Corbett) with Quantitect SYBR green RT-PCR one-step kit as described by the manufacturer. Briefly, 500 ng of RNA was placed into a 20 μL reaction volume containing 0.2 μM of each primer, 10 μL of SYBER green I RT-PCR one-step master mix, and 0.2 μL of reverse transcriptase. A typical protocol included reverse transcription at 50°C for 30 minutes and a denaturation step at 95°C for 15 minutes, followed by 40 cycles with 94°C denaturation for 15 seconds, 57°C annealing for 30 seconds, and 72°C extension for 30 seconds. Melt curve analysis: melt data acquiring to Cycling A (FAM/Sybr), ramp from 72°C to 95°C, hold 45 seconds on the first step, hold 5 seconds on the next step. Two equations was used to calculate relative changes in CXCR4 and β-actin expression from real-time quantitative RT-PCR experiment.25
| (1) |
whereas ΔCttarget = Ctcontrol – Cttreatment;
ΔCtreference = Ctcontrol – Cttreatment
| (2) |
whereas ΔΔCt = ΔCtreference – ΔCttarget
The one-way analysis of variance and least significant difference post hoc test were used to determine whether there were any significant differences among the means of CXCR4 expression of groups A, B, C, D, and E.
Serum alkaline phosphatase measurement
Serum ALP of animal serum was measured in animals by Hitachi (902 Automatic Analyzer, Berlin, Germany) after 35 days. The blood samples were obtained by heart puncture and were allowed to clot. ALP activity was carried out using an ALP optimized assay kit (Roche Diagnostics GmbH, Basel, Switzerland), according to the manufacturer’s instructions. ALP activities were expressed as units per liter (U/L).
Immunohistochemistry staining of colon and distal ileum
Immunohistochemistry was performed on groups A, B, E, and F of the distal ileum and colon to confirm the translation of CXCR4 mRNA to protein. Specimens were routinely fixed in 10% formalin in the immediate postoperative period and paraffin-embedded within the first 6 hours after procurement. After sectioning samples (5 μm), slides were dried overnight at 37°C and were deparaffinized with xylene. The sections were treated with sodium citrate buffer (10 mM sodium citrate, 0.05% Tween 20, pH 6.0) at 98°C for 20 minutes, and the slides were washed 2 × 5 minutes in TBS plus 0.025% Triton X-100 with gentle agitation. Blocking was done in 10% normal serum with 1% bovine serum albumin in TBS for 2 hours at room temperature. Samples were incubated overnight at 4°C with a primary antibody diluted in TBS with 1% bovine serum albumin at a dilution of 1:100. The next day, sections were rinsed 2 × 5 minutes with tris-buffered saline (TBS) 0.025% Triton with gentle agitation. The slides were incubated in 0.3% H2O2 in TBS for 15 minutes. Sections were labeled with a goat antirabbit IgG horseradish peroxides (HRP), 200 μg/0.5 mL (Santa Cruz Biotechnology) and developed with 3,3′-diaminobenzidine (brown-colored staining) and counterstained with hematoxylin (blue color staining).
Results
Characterization of CXCR4 siRNAI, II/dextran-spermine nanoparticles
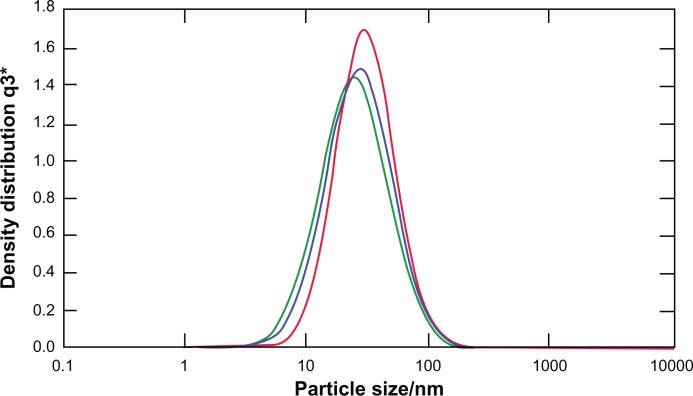
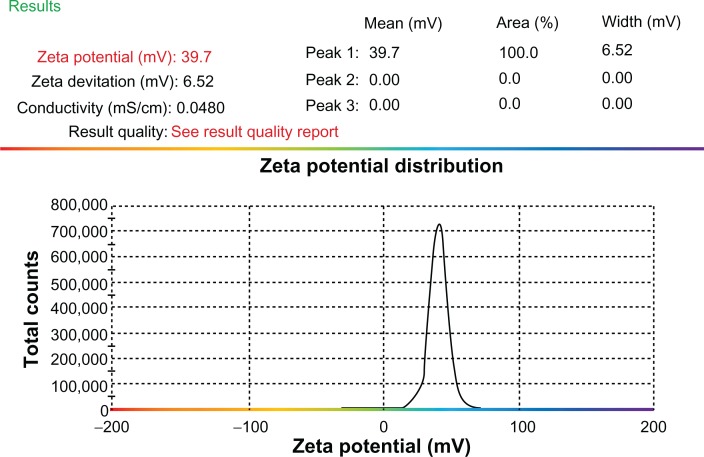
In this study, transmission electron microscopy was applied to determine the morphology of nanoparticles 8 hours after preparation. Nanoparticles were formed by mixing CXCR4 siRNAI, II with dextran-spermine at various weight-mixing ratios (siRNA/dextran-spermine) from 1:1 to 1:5. Nanoparticles formed at a ratio of 1:5 repelled one another and did not form aggregation. CXCR4 siRNAs/dextran-spermine nanoparticles formed small, round particles with good repelling properties against each other (data not shown). The particle size of nanoparticles was automatically calculated by particle size analysis NANOPHOX (0128 P) (Nanophox, Clausthal-Zellerfeld, Germany). The average size of nanoparticles was 54.74 ± 0.00 nm. This experiment was done three times and each time was repeated 40 times (Figure 1). The zeta potential or surface charge of nanoparticles (mV) was automatically calculated and found to be 39.7 ± 0.2, and zeta deviation (mV) was found to be 6.52 ± 0.6 (Figure 2).
Figure 1.
Particles size distribution of CXCR4 siRNAI, II/dextran-spermin q3 = Fractional volume density distribution.
Notes: The size of the particles was calculated at a weight-mixing ratio of 1:5 (CXCR4 siRNAI, II to dextran-spermine). The average of size of particles was below 100 nm.
Abbreviation: q3*, density distribution by volume.
Figure 2.
Zeta potential of CXCR4 siRNAI, II/dextran-spermine.
Notes: The zeta potential of particles was calculated at a weight-mixing ratio of 1:5 (CXCR4 siRNAI, II to dextran-spermine). The average of zeta potential was 39.7 ± 0.2 mV.
CXCR4 expression by RT-PCR one-step
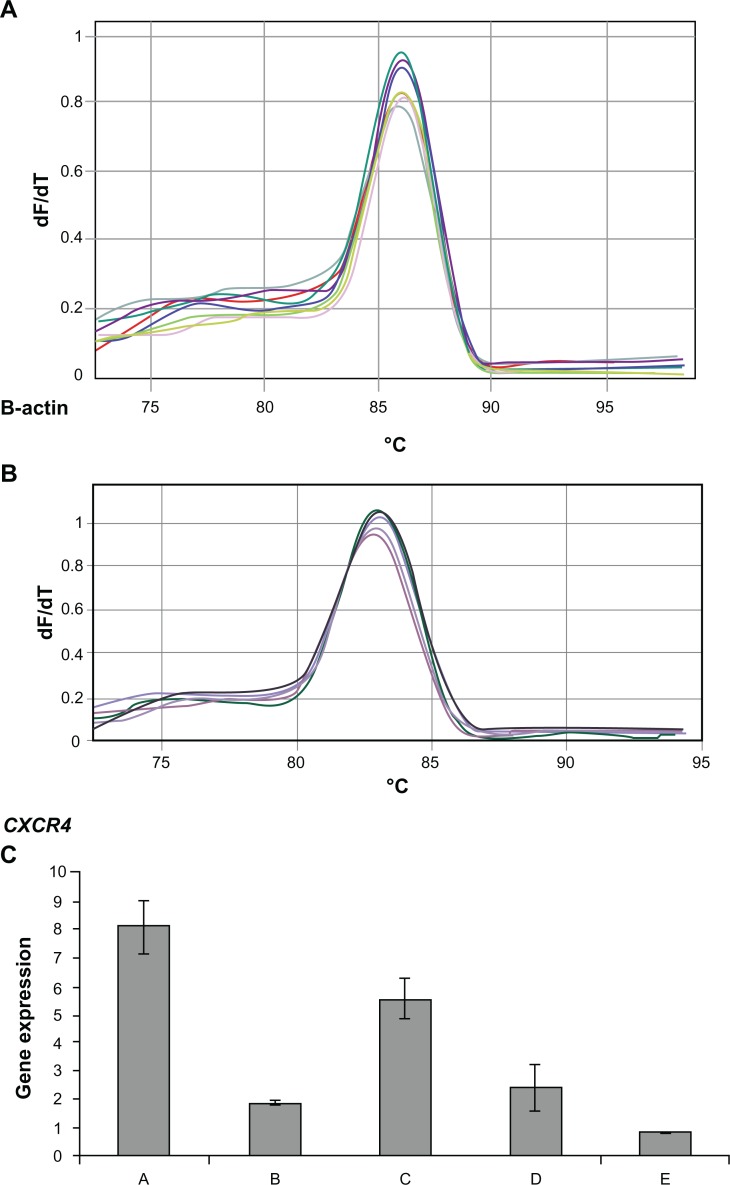
Total RNA was isolated from the liver of six groups of animals 35 days after CT26.WT cell injection. RT-PCR was performed on the real-time PCR machine RotorGene 3000 (Corbett), and delta-delta Ct method was used to analyze CXCR4 and housekeeping gene expression. Standard curve was done with one to two dilutions. CXCR4 efficiency was 1.34 and R^2 value: 0.99 and B-actin efficiency 1.04 and R^2 value: 0.99. Based on one-way analysis of variance and least significant difference post hoc test, there were significant differences among groups at the level F = 85.36 (P = 0.000) (Figure 3A–C).
Figure 3.
Melting curve for standard curve samples in real-time reverse transcription-polymerase chain reaction (RT-PCR) for β-actin endogeneous control: fluorescence melting curve obtained from real-time amplification of β-actin gene within the range of 72°C–97°C (A). Melting curve for standard curve samples in real-time RT-PCR for CXCR4 expression: fluorescence melting curve obtained from real-time amplification of CXCR4 gene within the range of 72°C–97°C (B). CXCR4 expression among groups A, B, C, D, and E: real-time RT-PCR was performed on the real-time PCR machine (tubes) Rotor-Gene 3000, and data analysis of CXCR4 and ß-actin expression was performed with the delta-delta Ct method. Standard curve was constructed with dilution (1:2) (C).
Serum ALP levels
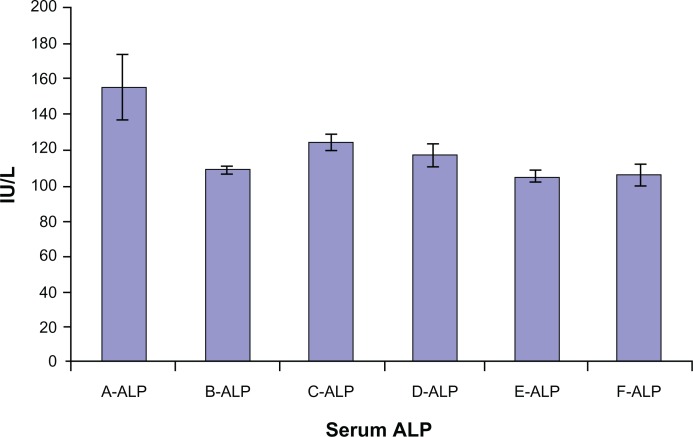
Animal study demonstrated that self-assembled siRNAs I, II/dextran-spermine nanoparticles achieved better inhibition of CXCR4 expression along with lowering serum ALP level. Among groups A to E, the highest level of ALP enzyme was 155 ± 18 IU/L in group A. On the contrary, the lowest level of CXCR4 serum ALP enzyme was 105 ± 4 IU/L in group E (Figure 4).
Figure 4.
Serum alkaline phosphatase (ALP) level in mice groups A, B, C, D, E, and F after 30 days of treatment.
Immunohistochemistry staining of colon and distal ileum
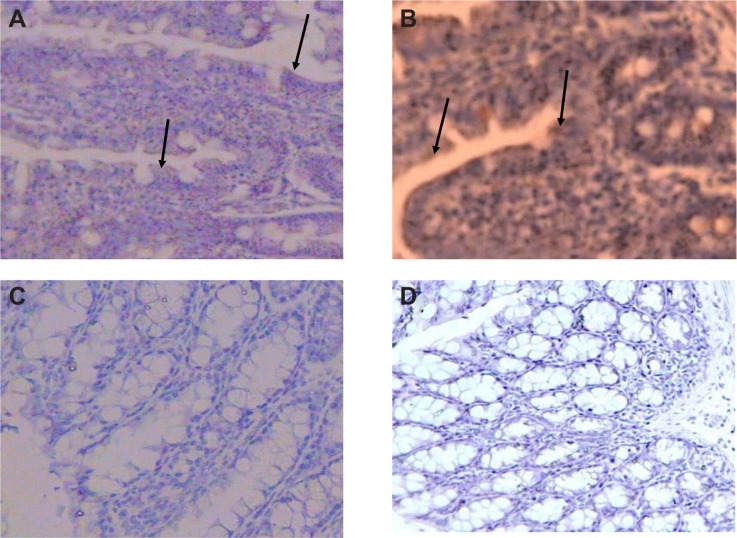
Specimens of mouse colon and ileum were studied immunohistochemically for the presence of CXCR4. The chemokine receptor CXCR4 is a seven-transmembrane G protein-coupled receptor (integral membrane proteins) for stromal cell-derived factor (SDF-1,3 also named CXCL12).2,6 Our observation revealed that sections labeled with a goat antirabbit IgG-HRP and developed with 3,3′-diaminoben-zidine produced a brown color during the enzymatic degradation of H2O2 by HRP. Results demonstrated that strong immunoreactivity was detected in the colon and distal ileum (group A). In group D, moderate immunoreactivity was detected in the colon and distal ileum. In group E, faint immunoreactivity was detected in the colon. In group F, faint to negative immunoreactivity was detected in the colon (Figure 5A–D). It is important to note that after injecting mouse colon carcinoma cells (CT26.WT) through the tail vein, most of the cells were localized in Peyer’s patches of colon and distal ileum (data not shown).
Figure 5.
Immunohistochemical staining of the colon and distal ileum. Specimens of mouse colon and ileum were studied immunohistochemically for the presence of CXCR4 (integral membrane proteins). Sections labeled with goat antirabbit immunoglobulin G horseradish peroxides and developed with 3,3′-diaminobenzidine produced a brown color and were counterstained with hematoxylin (blue color). Immunohistochemical staining of the colon and distal ileum showed strong immunoreactivity in the colon and distal ileum in group A. Arrows show CXCR4 protein (magnification 40×) (A). Moderate immunoreactivity was detected in the colon and distal ileum in group D. Arrows show CXCR4 protein (magnification 40×) (B). Faint immunoreactivity was detected in the colon in group E (magnification 40×) (C). Faint to negative immunoreactivity was detected in the colon in group F (magnification 40×) (D).
Discussion
The ideal gene delivery system must protect the nucleic acid from degradation and should be nontoxic, nonimmunogenic, and biodegradable.21 Reports have shown that adequate size and positive potential are essential for enhanced delivery of nanoparticles into cells. Typically, a drug carrier must be in a narrow size range from approximately 10 nm to 100 nm.26 Nanoparticles with zeta potentials between −30 mV and +30 mV typically tend to aggregate.27 The cytotoxicity of dextran-spermine and CXCR4 siRNAs/dextran-spermine nanoparticles was previously reported in vitro and in vivo, and it was identified as an effective carrier for siRNA systemic delivery.28–30 With respect to the lower weight-mixing ratio of CXCR4 siRNA I, II and dextran-spermine (1:5) and low doses of siRNAs used in the preparation of the nanoparticles, small sizes of nanoparticles (57.62 ± 2.51 nm) with suitable zeta potential (39.7 ± 0.2 mV) were obtained. Thus, it was expected that nanoparticles could be efficiently internalized into cells with minimal toxicity. In the field of cancer therapy, nanotechnology has provided researchers with expertise to explore new avenues for diagnosis and treatment of the disease.31 Utilization of nanotechnology has enabled the development of devices in nanometer sizes that could be designed to encapsulate agents but otherwise are generally toxic due to the doses intended for more applications.32 In the present study, dextran-spermine was used to evaluate the ability of CXCR4 siRNAs to knock down CXCR4 mRNA. The results revealed that CXCR4 siRNAs encapsulated in dextran-spermine could downregulate the expression of CXCR4 mRNA than naked CXCR4 siRNAs.
Since much research has been done on chemokine receptors, CXCR4 remains an attractive candidate for cancer metastatic therapy. The involvement of CXCR4 expression in colorectal cancer progression and metastases was first shown by Zeelenberg and Ruuls-Van Stalle33 and Rossi and Zlotnik.34 In our recent studies, we investigated the effect of silencing CXCR4 mRNA by CXCR4 siRNAs/dextran-spermine nanoparticles on chemotactic response of mouse colon carcinoma cells (CT.26WT) in vivo and the influence of this chemokine on tumor growth and serum ALP level.
Various combination strategies of siRNAsI, II of CXCR4 with and without dextran-spermine were compared in experimental metastatic animal models. Animals from groups A, B, C, D, E, and F were compared to determine whether lowering CXCR4 levels could affect serum ALP and block colorectal cancer metastasis to the liver, and to evaluate the efficacy of the pretreatment or post-treatment of siRNAs, as well as the efficacy of naked siRNAI, II and CXCR4 siRNAI, II/dextran-spermine nanoparticles. Animal study demonstrated that self-assembled siRNAs I, II/dextran-spermine nanoparticles used in twice-weekly treatment achieved better inhibition of CXCR4 expression than naked siRNAs I, II.
The average body weight of animals in groups A and E was 24.13 ± 0.51 g and 17.61 ± 0.69 g, respectively. There were significant differences among groups A, B, C, D, and E (P < 0.05), but no significant differences were observed within group F (P = 0.724). The significant difference was related to tumor growth. In group E, animals did not gain weight and showed significant differences with other groups (P = 0.001) (photos not shown).
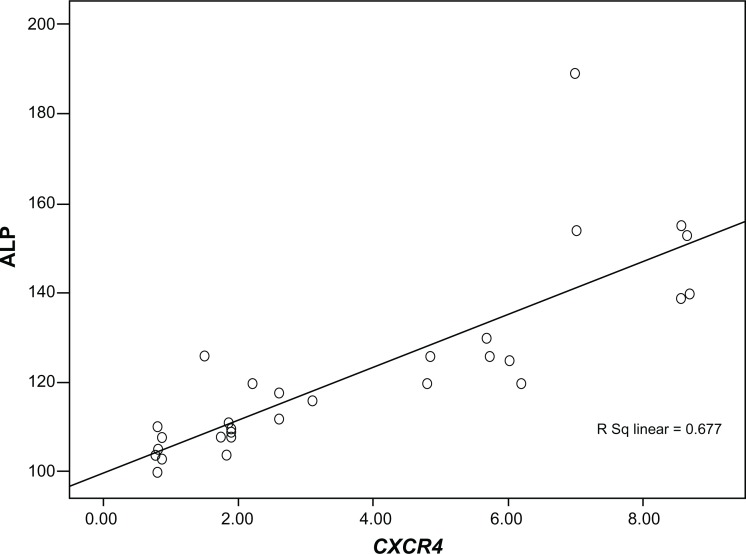
Among groups A to E, the highest levels of CXCR4 expression (8.09 ± 0.8-fold) and serum ALP enzyme (155 ± 18 IU/L) were detected in group A. On the contrary, the lowest levels of CXCR4 expression (2.49 ± 0.04-fold) and serum ALP enzyme (105 ± 4 IU/L) were found in group E. There was a significant correlation between CXCR4 expression and ALP among groups A, B, C, D, and E (Pearson correlation coefficient R = 0.823 and P = 0. 000) (Figure 6).
Figure 6.
Correlation coefficient between CXCR4 expression and serum alkaline phosphatase (ALP) level among groups A, B, C, D, and E.
Note: Pearson correlation coefficient R was 0.823 and P was 0.000.
Our results revealed that animals in group A developed liver metastasis. In contrast, animals in groups B, D, C, and E developed metastases less than those in groups A and C. Metastasis was barely visible in group E. The morphology of liver in group E was normal, whereas those from groups A, B, C, and D showed infiltrative lymphocytes between hepatic parenchyma and portal vein following metastasis of tumor cells to the liver (data not shown).35
Immunohistochemistry staining showed that after injection of CT26.WT cells, high CXCR4 protein expression was detected in the colon and distal ileum in group A, while moderate expression was detected in group D and faint expression was detected in group E. In group F, faint to negative expression was detected. These results clearly indicate that dextran-spermine nanoparticles efficiently encapsulated siRNA and downregulated CXCR4 mRNA more than naked CXCR4 siRNAs. The fluctuation in CXCR4 expression led to changes in serum ALP level in groups A, B, C, D, and E, which may be associated with colorectal cancer metastasis to the liver. Measurement of serum ALP can be applied economically to screen for liver metastases and to determine which patients should undergo a liver scan. More experiments should be carried out to find out the effect of silencing CXCR4 on pathways involved in serum ALP enzyme.
Conclusion
This study clearly showed that dextran-spermine nanoparticles significantly downregulated CXCR4 expression through CXCR4 silencing as well as good correlation between CXCR4 expression and serum ALP. The functional design of the hematopoietic chemokine receptor CXCR4 may facilitate metastasis to the liver. If validated by prospective studies, CXCR4 expression could be a potential predictive factor for recurrence of liver metastasis. This could improve the current staging of colorectal cancer by defining additional criteria for administration of systemic therapy in patients without overt signs of advanced disease. Perhaps most important is the identification of the CXCR4 receptor as a novel target for clinical therapy. The effect of CXCR4 silencing on serum ALP level may be a useful marker to predict liver metastasis in colorectal cancer and is very promising for future therapeutic strategies that may allow control of tumor spreading by blocking the CXCR4 receptor.
Acknowledgments
This study was performed through the National Science Council (NSC) of Taiwan, and was supported by the Research Grant of NSC 99-2314-B-011-001-MY3.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Geoghegan JG, Scheele J. Treatment of colorectal liver metastases. Br J Surg. 1999;86:158–169. doi: 10.1046/j.1365-2168.1999.01013.x. [DOI] [PubMed] [Google Scholar]
- 2.Muller A, Homey B, Soto H, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 3.Alenius H, Muller A, Soto H, et al. Involvement of chemokine receptors in breast cancer metastasis. J Nat Med. 2002;8:157–165. [Google Scholar]
- 4.Tachibana K, Hirota S, Iizasa H, et al. The chemokine receptor CXCR4 is essential for vascularization of the gastrointestinal tract. Nature. 1998;393:591–594. doi: 10.1038/31261. [DOI] [PubMed] [Google Scholar]
- 5.Ottaiano A, Napolitano M, Tatangelo F, Acquaviva AM, Ascierto PA, Scala S. Inhibitory effects of anti-CXCR4 antibodies on human colon cancer cells. Cancer Immunol Immunother. 2005;54:781–791. doi: 10.1007/s00262-004-0636-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liang Z, Yoon Y, Votaw J, et al. Silencing of CXCR4 blocks breast cancer metastasis. Cancer Res. 2005;65:967–971. [PMC free article] [PubMed] [Google Scholar]
- 7.Friedman LS, Martin P, Munoz SJ. Liver function tests and the objective evaluation of the patient with liver disease In: Hepatology: a textbook of liver disease. In: Zakim D, Boyer TD, editors. Philadelphia, PA: WB Saunders; 1996. pp. 791–833. [Google Scholar]
- 8.Reichling JJ, Kaplan MM. Clinical use of serum enzymes in liver diseases. Dig Dis Sci. 1988;33:1601–1614. doi: 10.1007/BF01535953. [DOI] [PubMed] [Google Scholar]
- 9.Saif W, Alexander D, Charles M. Serum alkaline phosphatase level as a prognostic tool in colorectal cancer: a study of 105 patients. J Appl Res. 2005;5(1):88–95. [PMC free article] [PubMed] [Google Scholar]
- 10.Kohne CH, Cunningham D, Di CF, et al. Clinical determinants of survival in patients with 5-fluorouracil-based treatment for metastatic colorectal cancer: results of a multivariate analysis of 3825 patients. Ann Oncol. 2002;13:308–317. doi: 10.1093/annonc/mdf034. [DOI] [PubMed] [Google Scholar]
- 11.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2002;411(6836):494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 12.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by doublestranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 13.Liang Z, Wu T, Lou H. Inhibition of breast cancer metastasis by selective synthetic polypeptide against CXCR4. Cancer Res. 2004;64:4302–4308. doi: 10.1158/0008-5472.CAN-03-3958. [DOI] [PubMed] [Google Scholar]
- 14.Banan M, Puri N. The ins and outs of RNAi in mammalian cells. Curr Pharm Biotechnol. 2004;5:441–450. doi: 10.2174/1389201043376643. [DOI] [PubMed] [Google Scholar]
- 15.Dorsett Y, Tuschl T. siRNAs: applications in functional genomics and potential as therapeutics. Nature reviews. Drug Discovery. 2004;3:318–329. doi: 10.1038/nrd1345. [DOI] [PubMed] [Google Scholar]
- 16.Bartlett DW, Davis ME. Insights into the kinetics of siRNA-mediated gene silencing from live-cell and live-animal bioluminescent imaging. Nucleic Acids Res. 2006;34:322–333. doi: 10.1093/nar/gkj439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Azzam T, Raskin A, Makovitzki A, Brem H, Vierling P, Lineal M. Cationic polysaccharides for gene delivery. Macromolecules. 2002;35:9947–9953. [Google Scholar]
- 18.Azzam T, Eliyahu H, Makovitzki A, Domb AJ. Dextran-spermine conjugate: an efficient vector for gene delivery. Macromol Symp. 2003;195:247–261. [Google Scholar]
- 19.Hosseinkhani H, Azzam T, Tabata Y, Domb AJ. Dextran-spermine polycation: an efficient non-viral vector for in vitro and in vivo gene transfection. Gene Therapy. 2004;11:194–203. doi: 10.1038/sj.gt.3302159. [DOI] [PubMed] [Google Scholar]
- 20.Eliyahu H, Makovitzki A, Azzam T, Zlotkin A, Joseph A, Gazit D. Novel dextran-spermine conjugates as transfecting agents: comparing water-soluble and micellar polymers. Gene Therapy. 2005;12:494–503. doi: 10.1038/sj.gt.3302395. [DOI] [PubMed] [Google Scholar]
- 21.Hosseinkhani H, Hosseinkhani M, Khademhosseini A, Gabrielson NP, Pack DW, Kobayashi H. DNA nanoparticles encapsulated in 3-D tissue engineered scaffold enhance osteogenic differentiation of mesenchymal stem cells. J Biomed Mater Res A. 2008;85:47–60. doi: 10.1002/jbm.a.31327. [DOI] [PubMed] [Google Scholar]
- 22.Hosseinkhani H, Inatsugu Y, Hiraoka Y, Inoue S, Shimokawa H, Tabata Y. Impregnation of plasmid DNA into three-dimensional scaffolds and medium perfusion enhance in vitro DNA expression of mesenchymal stem cells. J Tissue Eng. 2005;11:1459–1475. doi: 10.1089/ten.2005.11.1459. [DOI] [PubMed] [Google Scholar]
- 23.Hosseinkhani H, Tabata Y. Self assembly of DNA complexes with polycations for the delivery of genetic materials into cells. J Nanosci Nanotechnol. 2006;6:1–9. doi: 10.1166/jnn.2006.507. [DOI] [PubMed] [Google Scholar]
- 24.Nagane K, Jo J, Tabata Y. Promoted adipogenesis of rat mesenchymal stem cells by transfection of small interfering RNA complexed with a cationized dextran. J Tissue Eng Part A. 2010;16(1):21–31. doi: 10.1089/ten.TEA.2009.0170. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Robby A, Joseph M. Strategies in the design of nanoparticles for therapeutic applications. Nat Rev Drug Discov. 2010;9:615–627. doi: 10.1038/nrd2591. [DOI] [PubMed] [Google Scholar]
- 27.Mohanraj VJ, Chen Y. Nanoparticles, a review. TJPR. 2006;5:561–573. [Google Scholar]
- 28.Eliyahu H, Joseph A, Schillemans JP, Azzam T, Domb AJ, Barenholz Y. Characterization and in vivo performance of dextran-spermine polyplexes and DOTAP/cholesterol lipoplexes administered locally and systemically. Biomaterials. 2007;28:2339–2349. doi: 10.1016/j.biomaterials.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 29.Abedini F, Hosseinkhani H, Ismail M, et al. In vitro intracellular trafficking of biodegradable nanoparticles dextran-spermine in cancer cell lines. Int J Nanotechnol. 2011;8:712–723. [Google Scholar]
- 30.Abedini F, Ismail M, Hosseinkhani H, et al. Toxicity evaluation of dextran-spermine polycation as a tool for genetherapy in vitro. J Cell Animal Biol. 2010;4:170–176. [Google Scholar]
- 31.Nie S, Xing Y, Kim GJ, Simons JW. Nanotechnology applications in cancer. Annu Rev Biomed Eng. 2007;9:257–288. doi: 10.1146/annurev.bioeng.9.060906.152025. [DOI] [PubMed] [Google Scholar]
- 32.Siddiqui IA, Adhami VM, Chamcheu JC, Mukhtar H. Impact of nanotechnology in cancer: emphasis on nanochemoprevention. Int J Nanomedicine. 2012;7:591–605. doi: 10.2147/IJN.S26026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeelenberg S, Ruuls-Van Stalle L. The chemokine receptor CXCR4 is required for outgrowth of colon carcinoma micrometastases. Cancer Res. 2003;63:3833–3839. [PubMed] [Google Scholar]
- 34.Rossi D, Zlotnik A. The biology of chemokines and their receptors. Annu Rev Immunol. 2000;18:217–242. doi: 10.1146/annurev.immunol.18.1.217. [DOI] [PubMed] [Google Scholar]
- 35.Abedini F, Ismail M, Hosseinkhani H, et al. Effects of CXCR4 siRNA/dextran-spermine nanoparticles on CXCR4 expression and serum LDH levels in a mouse model of colorectal cancer Metastasis to liver. Cancer Manag Res. 2011;3:301–309. doi: 10.2147/CMR.S11678. [DOI] [PMC free article] [PubMed] [Google Scholar]