Abstract
The production of antimicrobial peptides (AMPs) is a major defense mechanism against pathogen infestation and of particular importance for insects relying exclusively on an innate immune system. Here, we report on the characterization of three AMPs from the carpenter ant Camponotus floridanus. Due to sequence similarities and amino acid composition these peptides can be classified into the cysteine-rich (e.g. defensin) and glycine-rich (e.g. hymenoptaecin) AMP groups, respectively. The gene and cDNA sequences of these AMPs were established and their expression was shown to be induced by microbial challenge. We characterized two different defensin genes. The defensin-2 gene has a single intron, whereas the defensin-1 gene has two introns. The deduced amino acid sequence of the C. floridanus defensins is very similar to other known ant defensins with the exception of a short C-terminal extension of defensin-1. The hymenoptaecin gene has a single intron and a very peculiar domain structure. The corresponding precursor protein consists of a signal- and a pro-sequence followed by a hymenoptaecin-like domain and six directly repeated hymenoptaecin domains. Each of the hymenoptaecin domains is flanked by an EAEP-spacer sequence and a RR-site known to be a proteolytic processing site. Thus, proteolytic processing of the multipeptide precursor may generate several mature AMPs leading to an amplification of the immune response. Bioinformatical analyses revealed the presence of hymenoptaecin genes with similar multipeptide precursor structure in genomes of other ant species suggesting an evolutionary conserved important role of this gene in ant immunity.
Introduction
Insects have evolved multiple innate defense mechanisms to respond to microbial invasion [1], [2], [3], [4]. Early protective measure involves the “constitutive” immediate-acting defenses including phagocytes and reactive oxygen species. At later time points, an inducible immune response is mounted which mainly involves the production of antimicrobial peptides (AMPs) [5], [6]. It is believed that this late-acting humoral response is required to kill those bacteria that have survived the immediate host's constitutive defenses [5]. In 1981, the first AMPs were described from the cecropia moth [7]. In the past two decades a multitude of AMPs have been identified produced by many different organisms ranging from animals to plants. Most AMPs have a low molecular weight (<10 kDa), are membrane-active and display hydrophobic and/or cationic properties. Based on structural characteristics, insect AMPs can be divided into several groups, mainly α-helical peptides (e.g. cecropin), cysteine-rich peptides (e.g. defensin), proline-rich peptides (e.g. drosocin), and glycine-rich peptides (e.g. hymenoptaecin) [8].
Several AMPs from Hymenopteran species have been reported so far [9], [10], [11], [12], [13]. The few defensins known from Hymenopterans are short cationic peptides characterized by three stabilizing disulfide bridges. These peptides appear to act primarily against gram-positive bacteria by interference with acidic phospholipids of the cytoplasmic membrane and the formation of voltage-dependent channels [14], [15]. They are synthesized as an inactive precursor peptide with a signal- and a pro-sequence. Processing of the precursors leads to the active peptides. Apis mellifera has two structurally different defensin genes (defensin-1 and defensin-2) [16]. Defensin-1 is characterized by the presence of two introns and three exons, whereat the last exon encodes a short C-terminal extension known from bee defensins only. In contrast, defensin-2 possesses a single intron [16]. Several defensin-2 genes of a variety of ant species, including Formica, Lasius and Myrmica species, have been described previously [12]. The comparison and determination of codon substitution frequencies revealed positive selection in the mature region of the ant defensins, while the signal- and pro-regions of the AMPs appear to have evolved neutrally [12].
Hymenoptaecins are glycine-rich AMPs with activity against gram-negative and gram-positive bacteria and have been reported so far from Hymenopterans only [17]. The reported antibacterial effects of the A. mellifera hymenoptaecin suggest that its lethal consequences against E. coli are secondary to sequential permeabilization of the outer and inner membranes of these gram-negative bacteria [17]. The bee hymenoptaecins are endowed with a signal- and a pro-sequence which after processing give rise to the mature and active AMP [9], [10], [18]. In contrast, the two similar hymenoptaecins from the wasp Nasonia vitripennis encode multipeptide precursors with an AMP-like region at the position corresponding to the propeptide of bee hymenoptaecins [11], [19].
Here, we report the identification and molecular characterization of three AMP genes from the carpenter ant Camponotus floridanus. All ants are eusocial. Colonies of C. floridanus may contain, up to several thousand individuals. Such huge and dense colonies of genetically very similar organisms may pose specific problems to hygiene issues and pathogen defense [20]. Most interestingly, ants of this genus are exceptional in that they generally lack the so-called metapleural gland, which is known from other ants to be a major depository of antimicrobial compounds [21], [22]. Moreover, Camponotus and closely related genera harbor an obligate intracellular endosymbiont in specialized cells, the bacteriocytes, in their midgut, which need to be tolerated by the host's defense mechanisms [23], [24]. The recently published genome sequence of C. floridanus and subsequent bioinformatical analyses revealed the presence of two AMP genes encoding defensins [25], [26], which have significant similarities with defensins known from other ant species [12], [26], [27]. In addition to these defensin genes, a suppression subtractive hybridization approach also detected the presence of a hymenoptaecin gene in C. floridanus [28], which has not been annotated in the genome. In order to gain a better insight into the antimicrobial repertoire of C. floridanus, the present study aimed at the characterization of these antimicrobial peptides on the molecular level and comparison to AMPs encoded by other ant species. Most importantly, we show that in comparison to bee hymenoptaecins the C. floridanus hymenoptaecin gene is much longer and encodes a multipeptide precursor with structural similarities to apidaecin precursors from A. mellifera [17], the proteolytic processing of which possibly leads to a massive amplification of the antimicrobial response [13], [29].
Materials and Methods
Insect rearing and bacterial challenge
Founding queens of C. floridanus were collected in Florida near Orchid Island in August 2001 and were then kept in a climate chamber at Würzburg University as described before [30]. For pathogen challenge, C. floridanus major workers were injected with heat-killed Serratia marcescens (2×105 cells/individual). At 24 h after injection, midgut and fat body were collected and kept in RNAlater (Ambion/Applied Biosystems, USA) until RNA preparation.
DNA extraction and total RNA isolation
Total RNA from midgut and fat body of injected and naive major workers was extracted using TRIzol® Reagent (Invitrogen, Darmstadt, Germany) and purified through RNeasy mini kit columns (Qiagen, Hilden, Germany) with on-column DNase digestion (RNase-Free DNase Set, Qiagen) as described in the manufacturer's procedures. After purification, the RNA concentration of each sample was measured by the Nanodrop® spectrophotometer. RNA used in Northern blot analysis was additionally checked by PCR for gDNA contamination. C. floridanus genomic DNA was extracted from six larvae as described before [31].
Sequencing of full-length cDNAs and genes
The complete sequences of the transcripts of interest were obtained by 3′ and 5′ RACE, performed with the SMART RACE cDNA Amplification Kit including the Advantage II PCR kit (Clontech, Heidelberg, Germany). For defensin-1 and hymenoptaecin, the nucleotide sequences of the 3′- and 5′- primers (GSP1 and GSP2) were designed on the corresponding EST (GenBank Acc. No. for EST from hymenoptaecin: HS410972 and for EST from defensin: HS410966) obtained from a suppression subtractive hybridization (SSH) experiment [28]. For defensin-2 RACE primers were designed according to the C. floridanus genome sequence. Primers used for RACE were Cfl_def-1-GSP1 and Cfl_def-1-GSP2 for defensin-1, Cfl_def-2-GSP1 and Cfl_def-2-GSP2 for defensin-2 and Cfl_hym-GSP1 and Cfl_hym-GSP2 for hymenoptaecin (see Table S1). The first-strand cDNA used for 5′ and 3′-RACE were produced by using 1 μg of total RNA from Serratia-injected workers prepared for the SSH method, and using the primers provided in the kit. Amplification of the RACE products was carried out according to the manufacturer's instructions.
Oligonucleotide primers were then designed from the obtained RACE cDNA sequences and used for PCR amplification of the full length cDNAs und genes. Primers used were Cfl_def-1_flsF and Cfl_def-1_flsR for defensin-1, Cfl_def-2_flsF and Cfl_def-2_flsR for defensin-2 and Cfl_hym_flsF and Cfl_hym_flsR for hymenoptaecin (see Table S1).
In each case resulting PCR-products were purified with the PCR Purification Kit (Qiagen), inserted into the plasmid vector pGEM (Promega) and transformed into E. coli DH5α cells (Invitrogen). The plasmids from several different clones were then extracted for sequencing with the UltraPrep Kit (Molzym, Bremen, Germany) according to the manufacturer's instructions. The sequences were generated by Seqlab (Sequence Laboratories Göttingen) with the vector primers M13 forward (5′-GTTTTCCCAGTCACGAC-3′) and M13 reverse (5′-CAGGAAACAGCTATGAC-3′).
Hymenoptaecin probes for Northern and Southern blot analysis
In order to obtain specific C. floridanus hymenoptaecin probes for hybridization experiments, the inserts from two subtracted hymenoptaecin cDNA clones ([28], GenBank Acc. No. HS410972 and HS410975)) were amplified using PCR cycler and the Mol Taq PCR system (Molzym). Primers used were as follows: for the 270 bp hymenoptaecin 5′-probe: Cfl_hym_5′F and Cfl_hym_5′R; for the 270 bp hymenoptaecin repeat-probe Cfl_hym_repF and Cfl_hym_repR (see Table S1). PCR conditions were as follows: denaturation at 95°C for 3 min followed by 32 cycles of denaturation at 95°C for 15 s, annealing at 56°C for 15 s, and extension at 72°C for 1 min. PCR products were purified with the PCR Purification Kit (Qiagen).
Northern blot analysis
For verification of C. floridanus hymenoptaecin full length mRNA, Northern blot analysis was performed. Total RNA from immune-challenged and naive workers (25 µg per lane) was separated on a 1.0% formaldehyde agarose gel and transferred onto a nylon blotting membrane (Amersham Hybond N+, GE Healthcare, UK). Membranes were prehybridized in roller bottles with 10 ml of Amersham Rapid-hyp Buffer (GE Healthcare, UK) for 30 min at 65°C. The hymenoptaecin 5′-fragment (see above) was radioactively labelled with 60 mCi of [α-32P]-dATP with the DecaLabel Kit (Fermentas, Germany) according to the manufacturer's specifications. The labelled cDNA probe was purified with Illustra Microspin S-200 HR Columns (GE Healthcare, UK). Membranes were rinsed and then washed two times for 15 min at 65°C in 50 ml of wash buffer (2× SSC, 0.1% SDS), sealed in saran wrap and exposed to a storage phosphor screen (GE Healthcare, UK) for 2 days. Screens were scanned on a Typhoon 9200 Variable Mode Imager (GE Healthcare, UK) with a resolution of 50 microns.
Southern blot analysis
In order to characterize the C. floridanus hymenoptaecin gene locus, Southern blot analysis was performed. Digested DNA from immune-challenged and healthy workers (about 30 µg per lane) was separated on a 1.0% agarose gel and transferred onto a nylon blotting membrane (Amersham Hybond N+, GE Healthcare, UK). Membranes were prehybridized in roller bottles with 10 ml of hybridization buffer (0.5 M NaHPO4 (pH 7∶4), 1 mM EDTA, 0.7% SDS) for 30 min at 65°C. The hymenoptaecin repeat-fragment (see above) was labeled with Rediprime II DNA Labeling System (Amersham) according to the manufacturer's specifications. After addition of the heat-denatured probe (5 min at 95°C, cooled down on ice) hybridization was continued for 20 h at 62°C in a rotatory oven. Membranes were rinsed and then washed two times for 30 min at 65°C in 20 ml of wash buffer (0.04 M NaHPO4 (pH 7∶4), 1 mM EDTA, 0.5% SDS), sealed in saran wrap and exposed to a storage phosphor screen (GE Healthcare, UK) for 2 days. Screens were scanned on a Typhoon 9200 Variable Mode Imager (GE Healthcare, UK) with a resolution of 50 microns.
Expression and purification of recombinant Camponotus hymenoptaecin
One of the putative mature Camponotus hymenoptaecins (Cfl-hym) was amplified by PCR with oligonucleotides Cfl_hymNdeI_F and Cfl_hymBamHI_R (see Table S1). The PCR product was inserted into pET-15b vector (Novagen, Merck KGaA, Darmstadt, Germany) at NdeI and BamHI sites with a thrombin cleavage site (LVPRGS) for subsequent removal of the N-terminal 6xHis-Tag. The resulting pET-15b-Cfl-hym was transformed into E. coli Rosetta 2(DE3)pRARE2 (Novagen, Merck KGaA) for protein expression. Expression of the recombinant Cfl-hym peptide was induced with 0.1 mM IPTG at an optical density of 0.5 at 600 nm. Cells were harvested after 5 h at 37°C and pelleted by centrifugation. Cell pellets were stored at −80°C until further purification.
Recombinant Cfl-hym peptide was purified from insoluble inclusion bodies under denaturing conditions. All purification steps were monitored by SDS-PAGE analysis using Tris-glycine gels. Thawed cell pellets were resuspended in resuspension buffer (100 mM NaH2PO4, 10 mM Tris-HCl, 8 M Urea, pH 8.0) and lysed by gently vortexing. Cellular debris was removed by centrifugation and the cleared lysate was mixed with 1 ml PerfectPro Ni-NTA Agarose (5 Prime GmbH, Hamburg, Germany). The lysate-resin mixture was then loaded on a column and washed twice with wash buffer (the resuspension buffer adjusted to pH 6.3). Finally, the fusion proteins were eluted by elution buffers (the resuspension buffer adjusted to pH 5.9 and subsequently pH 4.5). Suitable elution fractions were combined and carefully dialyzed to a final dialysis buffer concentration of 400 mM NaCl, 1 M Urea, 20 mM Tris-HCl, 20% glycerol at pH 7.4. In order to remove the N-terminal 6xHis-Tag, recombinant peptides were digested with thrombin using the Thrombin Cleavage Capture Kit (Novagen, Merck KGaA) according to the manufacturer's instructions. The cleaved peptide was concentrated by ultrafiltration using Amicon Ultra-4 centrifugal filter devices (Millipore, Schwalbach, Germany). For buffer exchange the concentrated protein solution was diluted in dialysis buffer (400 mM NaCl, 1 M Urea, 20 mM Tris-HCl, 20% glycerol at pH 7.4) to a volume of 4 ml and subsequently concentrated again as described above. This procedure was repeated four times. Protein concentrations were measured using the method of Bradford with bovine serum albumin as standard.
Inhibition zone assays
Inhibition zone assays were performed in order to determine antibacterial activity of the purified Cfl-hym peptide. Microorganisms used in the inhibition zone assays were E. coli D31 [32] as a gram-negative bacterium and Bacillus subtilis as a gram-positive bacterium. In each case 3–4×105 bacteria were diluted in 3 ml preheated Luria broth (LB) containing 0.75% agarose. The mixture was spread out evenly on preheated LB agar plates. After settling, blanc discs (Oxoid, Thermo Fisher Scientific, Schwerte, Germany) were put on the agar plates and 4.5 nmol Cfl-hym peptide (in dialysis buffer) were applied on top. Furthermore dialysis buffer alone was applied as a negative control and 4 µg kanamycin as a positive control. The plates were incubated at 37°C overnight. On the next day the clear zone of inhibition was documented by photography.
Bioinformatical prediction of proteins in ant genomes
Gene prediction was performed for the published genomes of the ant species C. floridanus, Atta cephalotes, Harpegnathos saltator, Pogonomyrmex barbatus, Solenopsis invicta, Linepithema humile, and Acromyrmex echinatior using the gene prediction pipeline maker (version 2.11-beta) [33]. Therefore, the genomic contigs were prefiltered by BLASTX (version 2.2.24). All contigs having hits against published hymenoptaecin or defensin proteins were used for the gene prediction pipeline. The identified C. floridanus cDNA sequences and all available protein sequences from A. mellifera or other ants were used as EST and protein evidence for the gene predictions. Augustus with its Nasonia model was used for de novo gene predictor [34]. The obtained gene predictions were manually curated.
Phylogenetic tree reconstruction for mature hymenoptaecin peptides
The sequences for the cDNAs and proteins resulting from the gene prediction and the real sequences obtained from C. floridanus were analyzed by the ProP server (version 1.0) [35]. All cDNAs were fragmented according to the cleavage sites predicted by ProP. All obtained single domain cDNA fragments were aligned by translator [36] with default settings and the resulting alignment was cleaned by Gblocks [37] with the default settings from the translatorX website. The phylogenetic tree was reconstructed by PhyML (version 3.0.1) [38] under the GTR+I+G+F model with 100 bootstrap replicates as implemented in seaview (version 4.3.0) [39]. Branches with a bootstrap support below 40 were combined using iTOL [40] and the tree was drawn with the software FigTree (version 1.3.1).
Phylogenetic tree reconstruction and tree reconciliation for defensin peptides
For C. floridanus and S. invicta two defensin peptides were predicted. Therefore, we added Ixodes scapularis to the sequence set as outgroup (GenBank Acc. No.: XP_002436104.1) and the two A. mellifera defensins defensin-1 (GenBank Acc. No.: NM_001011616.2) and defensin-2 (GenBank Acc. No.: NM_001011638.1). The whole sequence set was aligned by MUSCLE (version 3.8.31) [41]. The tree was reconstructed by BioNJ [42]. Therefore, the observed amino acid frequencies were used. Branches with a bootstrap support below 40 were combined using iTOL [40]. The species tree for tree reconciliation was derived from Brady et al. 2006 and Gadau et al. 2012 [43], [44]. For tree reconciliation the software Notung (version 2.6) [45], [46] was used. The gene tree and the reconciled tree were drawn using FigTree (version 1.3.1).
Results
Cloning and sequence analysis of a hymenoptaecin encoding cDNA of C. floridanus
The search for immune inducible genes in C. floridanus by a SSH approach revealed the presence of a cDNA encoding a homologue of hymenoptaecin, an AMP known from other hymenopteran species [28]. The subsequent attempts to define the 5′- and 3′ends of the cDNA resulted in complex patterns. As expected the 5′RACE of the hymenoptaecin cDNA revealed one product. However, the 3′RACE resulted in several products of different length. Further investigation of these products revealed that all were hymenoptaecin derived 3′RACE products with an identical 3′UTR and a poly-A tail. The various molecules with different length resulted from a 309-nucleotide sequence which was repeated several times but in different copy number in the different amplification products. To confirm the existence of these deduced cDNAs the full length hymenoptaecin cDNAs were amplified with primers binding near the 5′and 3′-ends. Several clones with insert size varying from 681 bp to 2536 bp were identified and analyzed (Fig. 1). All of the examined cDNA sequences contained a constant 5′- and 3′end, embracing a region of variable length containing one to six copies of the 309-nucleotide repeat sequence. All putative precursor proteins deduced from the different amplification products are composed of a signal peptide of 19 amino acids (aa), a propeptide of 26 aa and a mature peptide region of differing length in dependence of the repeat number. The latter seems to be further processed into multiple mature AMPs in accordance with the presence of proprotein convertase cleavage sites Arg (R)/Lys (K), as predicted by ProP 1.0 [35]. Several different hymenoptaecin peptide variants could be deduced from the analyzed ESTs. Considering all obtained cDNA sequences and the size of the major product, we deduced a major hymenoptaecin mRNA (GenBank Acc. No.: HQ315784) of 2536 bp containing an ORF of 2373 bp (781 aa) corresponding to six repeats of the putative mature peptide sequence. Each of the repeated units consists of a coding sequence for the mature hymenoptaecin peptide, preceded by coding regions for a spacer sequence (EAEP) and a putative proprotein cleavage site (RR or KR) (Fig. 2). Further analysis of the deduced mature peptides showed that all putative hymenoptaecin peptides are 97 aa long and start with a glutamine (Q) at their N-terminus. Only the first peptide of each precursor, the so called hymenoptaecin-like peptide, displayed an exception consisting of 108 aa due to an N-terminal insertion (Fig. 2) and starting with a glycine (G). In sum, the domain composition of the Camponotus hymenoptaecin resembled the multipeptide precursor structure of bee apidaecins [29].
Figure 1. PCR amplification of full length hymenoptaecin gene and cDNA.
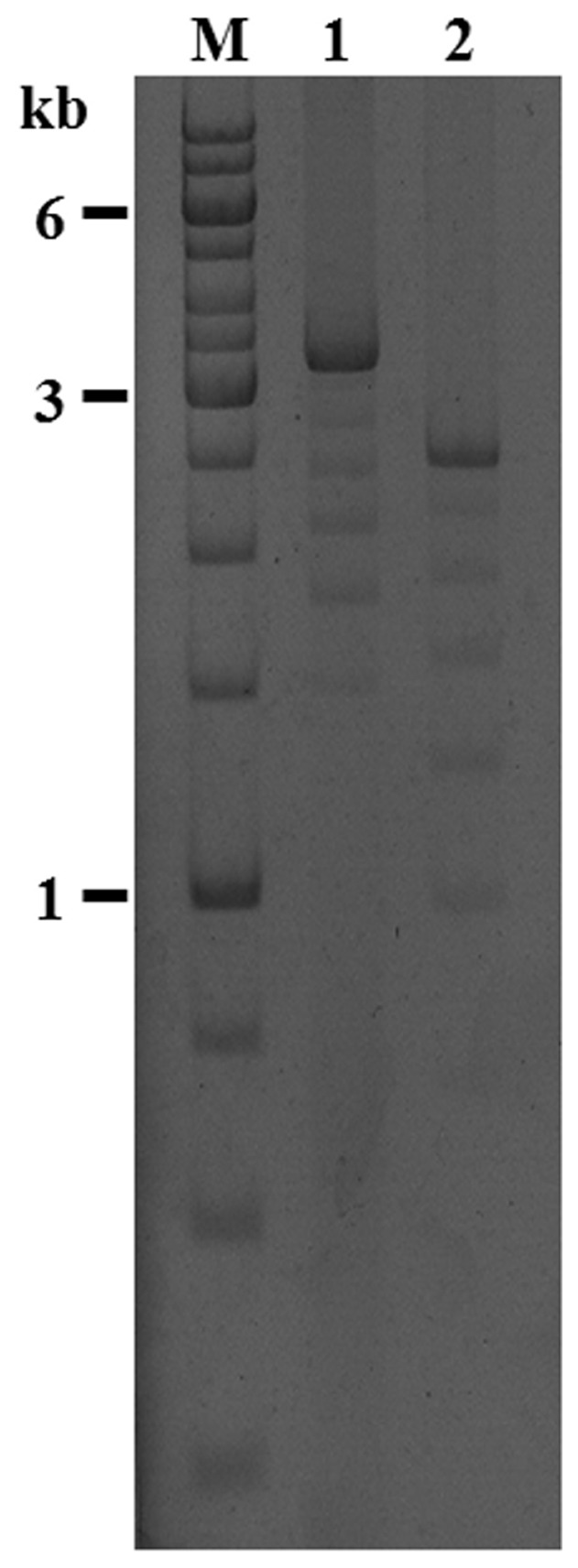
The PCR-products from gDNA (lane 1) and cDNA (lane 2) were separated on a 1.2% agarose gel alongside molecular size markers (lane M, GeneRuler 1 kb DNA Ladder, Fermentas) and analyzed with EtBr staining. The major bands correspond to the full length hymenoptaecin gene- (3356 bp lane 1) and cDNA-product (2536 bp, lane 2). The minor bands are technical artefacts with variable repeat numbers caused by the tandem repeats.
Figure 2. Alignment of HLD (hymenoptaecin-like domain) and all HDs (hymenoptaecin domains) from the same C. floridanus hymenoptaecin multipeptide precursor protein.
Grey boxes indicate conserved residues. The insertion in the hymenoptaecin-like domain (top) is clearly visible.
Genomic organization of hymenoptaecin
As described above, by comparison of all possible repeat versions from the sequenced hymenoptaecin cDNAs, we found a high number of different deduced hymenoptaecin peptide variants. Therefore we addressed the question whether this diversity was caused by the existence of multiple hymenoptaecin genes in the C. floridanus genome or by alternative splicing of large transcripts from a single gene. To solve this question, we amplified the hymenoptaecin gene(s) by PCR using conditions for the amplification of large fragments using the Cfl_hym_fls forward and reverse primers (see Table S1). Similar to the amplification of the hymenoptaecin cDNAs the PCR with gDNA yielded a ladder of amplified products ranging from 1500 bp to 3356 bp in length (Fig. 1). Comparing gDNA to mRNAs, we located one phase 0 intron of 820 bp in size, which is present after the codon coding for histidine number 39 (Fig. 3). As no other introns were found, the observed variation in repeat numbers cannot be explained by alternative splicing of exons coding for the repeats. Therefore, we investigated the question, whether hymenoptaecin variants may be encoded by a multigene family. In Southern Blot analysis of Bsu15I-digested DNA from multiple insects a fragment of about 2.9 kb in size hybridized with a cDNA probe derived from the repeat sequences (Fig. 4). The size of this fragment was consistent with that expected from digestion of the main PCR-product containing six repeats. Since no other signal was found, this result suggests that hymenoptaecin is encoded by a single gene which harbours six repeated sequence motifs (Fig. 3). This is also confirmed by Northern Blot analysis which resulted in a major band of the expected size of the mature transcript and a larger minor band which probably represents the unspliced primary transcript, since its size perfectly matches the predicted size (Fig. 4). In sum, our data suggest the existence of a single C. floridanus hymenoptaecin gene (GenBank Acc. No.: HQ315784) and we suppose that the above described variable repeat numbers after PCR amplification were a technical artefact caused by the tandem repeats.
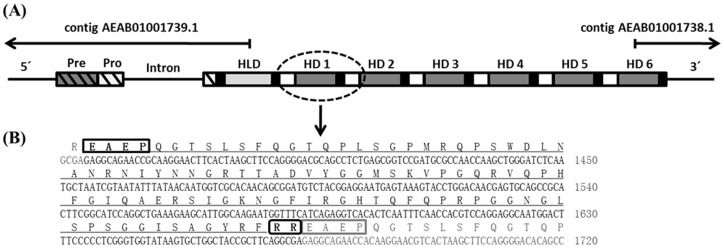
Figure 3. Structure of the C. floridanus hymenoptaecin gene locus.
(A) Schematic structure of the hymenoptaecin gene containing a single intron within the region coding for the hymenoptaecin propeptide. The deduced multipeptide precursor peptide consists of a signal-sequence (Pre, grey hatched box) and a pro-sequence (Pro, white hatched box), followed by a hymenoptaecin-like domain (HLD, light grey box) and six repeated hymenoptaecin domains (HD 1–6, dark grey boxes). The hymenoptaecin domains are flanked by the two putative processing sites EAEP (white boxes) and RR (black boxes). (B) The nucleotide and deduced amino acid sequence of a hymenoptaecin repeat unit are shown and the putative processing sites are boxed.
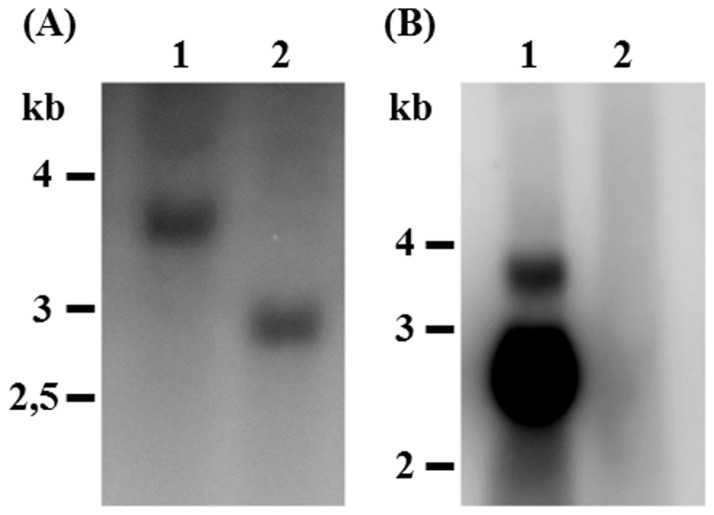
Figure 4. Southern blot (A) with C. floridanus genomic DNA using a 32P-labelled hymenoptaecin fragment corresponding to one of the repeats as a probe.
Genomic DNA (35 µg per lane) was digested with EcoRI (lane 1) and with Bsu15I (lane 2), separated by gel electrophoresis and hybridized with the above mentioned DNA fragment. Northern blot (B) with total RNA of C. floridanus using a 32P-labelled cDNA fragment corresponding to the 5′end of the hymenoptaecin gene as a probe. Total RNA (25 µg per lane) was isolated from midgut and fat body of major workers injected with heat-killed Serratia marcescens (2×105 bacteria/ant) in the haemocoel (lane 1) or untreated animals (lane 2). The major band corresponds to the spliced mature transcript, while the minor band very likely is the unspliced precursor. The position of molecular size markers is indicated on the left side of each figure. All hybridizing bands have the expected molecular size.
Antibacterial activity of recombinant C. floridanus hymenoptaecin
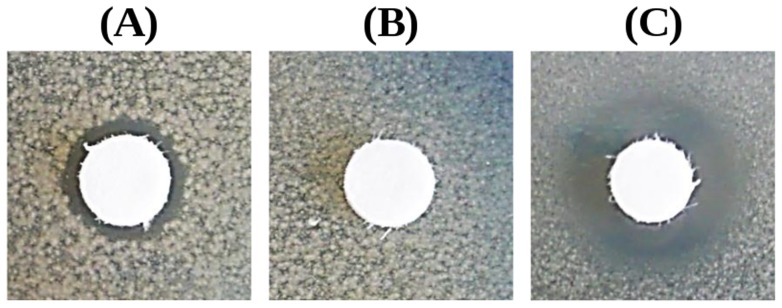
The 6xHis-tagged hymenoptaecin fusion protein (Cfl-hym) of 12.7 kDa was expressed in an insoluble form in E. coli Rosetta 2(DE3)pRARE2 (Novagen, Merck KGaA) and thus had to be purified under denaturing conditions. In order to keep the peptide soluble after digestion and refolding steps, dialysis buffer (containing 1 M Urea and pH 7.4) was most suitable. The antibacterial activity of the purified Cfl-hym peptide was tested against gram-negative E. coli D31 and gram-positive Bacillus subtilis with inhibition zone assays. Dialysis buffer alone was included as a negative control. Under the experimental conditions Cfl-hym was only active against E. coli D31 (Fig. 5).
Figure 5. Antibacterial activity of 4.5 nmol recombinant Cfl-hym peptide (A) against E. coli D31.
Dialysis buffer alone (B) was applied as a negative control and 4 µg kanamycin (C) as a positive control.
Phylogenetic analysis of hymenoptaecin peptides and comparison of the different hymenoptaecin multipeptide precursors
The recently established genome sequences of six different ant species revealed the presence of at least one AMP belonging to the hymenoptaecin family in each ant species [25], [47], [48], [49], [50]. For some of the ant species hymenoptaecin proteins were annotated. First we used these predicted proteins and found unusual domain compositions. Additionally, some of the proteins show a lack of crucial elements like recognition sequences for signal- and propeptides. Therefore, we investigated the predicted proteins on genome level using the published genome drafts. On genome level we could identify the problems which lead to the wrong prediction results. The sequence region, which apparently codes for the hymenoptaecin peptide(s) in the species Atta cephalotes, Linepithema humile, Pogonomyrmex barbatus and Solenopsis invicta seems to span contig boundaries, which were filled with N's during the scaffolding. This is based on the general problem to assemble short read sequences from next generation sequencing methods through regions with repetitive sequence elements. Three different hymenoptaecin precursor proteins were predicted for the genome of the ant species Acromyrmex echinatior (GenBank Acc. No.: EGI65977, EGI65978 and EGI65979) [48]. However, from the analysis of the A. echinatior genome we could deduce one putative hymenoptaecin multipeptide precursor, which combines the three predicted ones due to missing stop codons between the predicted proteins. The obtained hymenoptaecin peptide region of this precursor is extremely long and codes for 23 putative mature hymenoptaecin peptides (Fig. 6F). The two annotated hymenoptaecin precursor proteins from Harpegnathos saltator seemed to be plausible and contain either four (GenBank Acc. No.: EFN79831) or six (GenBank Acc. No.: EFN79832) mature AMPs, respectively.
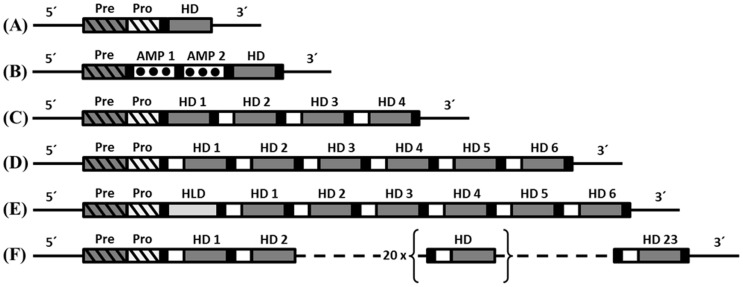
Figure 6. Schematic structure of the hymenoptaecin precursors from different hymenopteran species:
A) Apis mellifera (GenBank Acc. No.: NP_001011615) or Bombus ignitus (GenBank Acc. No.: ACA04900); B) Nasonia vitripennis: (GenBank Acc. No.: NP_001165829 XP_001607881); C) Harpegnathos saltator 1 (GenBank Acc. No.: EFN79831); D) Harpegnathos saltator 2 (GenBank Acc. No.: EFN79832); E) Camponotus floridanus (GenBank Acc. No.: HQ315784); F) Acromyrmex echinatior (hymenoptaecin multipeptide precursor deduced from genome draft). The various domains are marked as follows: signal-sequence (grey hatched box), pro-sequence (white hatched box), hymenoptaecin-like domain (HLD, light grey box), hymenoptaecin domains (HD 1–6, dark grey boxes), proline-rich AMP-like peptide (AMP 1–2, white dotted boxes). The hymenoptaecin domains are flanked by the putative processing sites EAEP (EANP for Harpegnathos) (white box) and RR (or RxxR) (black box).
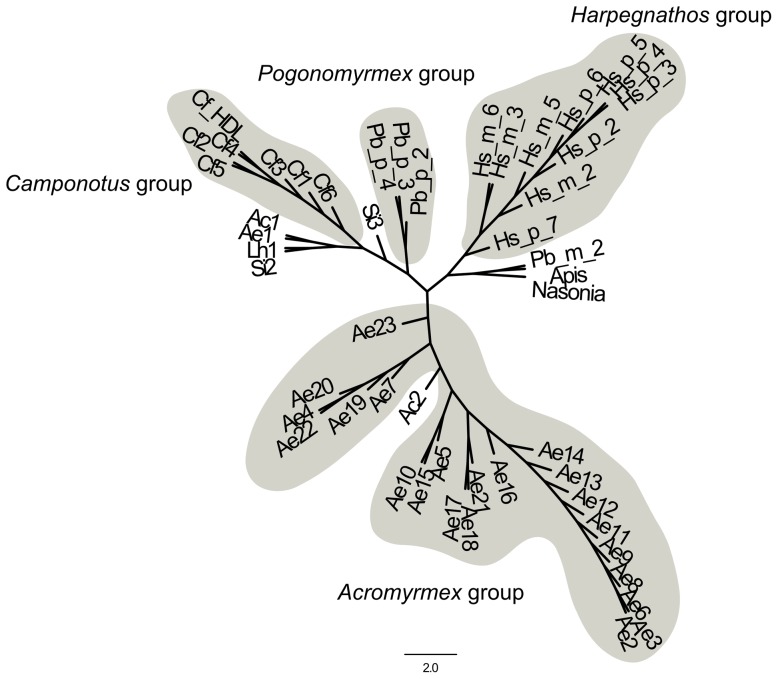
Fig. 6 shows a schematic comparison of the domain structures of hymenoptaecins from different hymenopteran species. Interestingly, the deduced hymenoptaecin precursor proteins from ant species are all multidomain proteins with remarkable similarities to the multipeptide precursor of C. floridanus hymenoptaecin. They have varying numbers of hymenoptaecin domains (HDs), which are all flanked by the putative spacer region EAEP (EANP for Harpegnathos) and processing sites RR (or RxxR, as predicted by ProP 1.0). Our phylogenetic analysis of the hymenoptaecins based on the existing set of proteins suggests an intra-species accumulation of the single domains within the proteins (Fig. 7, see also Fig. S1).
Figure 7. Phylogenetic analysis of hymenoptaecin domains from different ant species.
Shown is the unrooted tree of the single domains of the hymenoptaecins of the ant species, N. vitripennis, and A. mellifera. The proteins were cleaved at the sites predicted by ProP, followed by the alignment by translatorX. The tree was reconstructred by PhyML with a GTR+I+G+F model with 100 bootstrap replicates. The domains of the species with a complete hymenoptaecin protein form clades and are named as groups according there genus name and are indicated by their grey background. The domains which are outside these groups result from missing data from the predicted genes. The gene models are incomplete due to long N-stretches in the genomic sequences based on the scaffolding process. Nevertheless, the distinct groups formed by the complete proteins suggest an intra-species mechanism for the accumulation of the single domains.
Cloning and sequence analysis of C. floridanus defensins
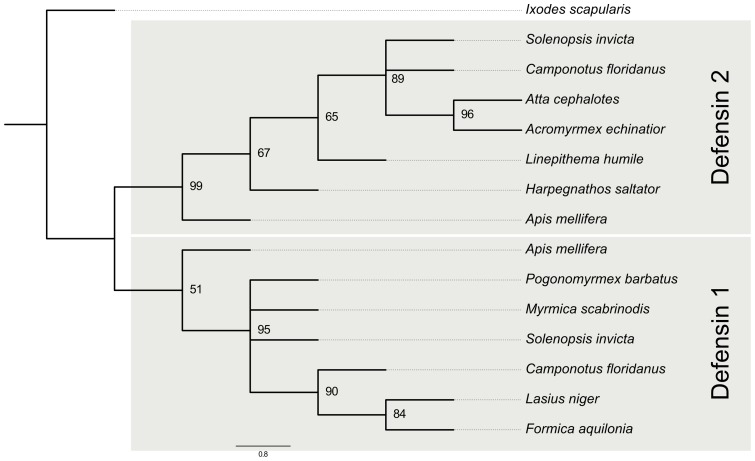
The SSH approach performed to identify immune inducible genes as well as the screening of the genome sequence resulted in the identification of two different sequences coding for defensin-like AMPs in C. floridanus. Phylogenetic analysis allocated these sequences as homologues to defensin-1 and defensin-2 from A. mellifera (Fig. 8).
Figure 8. Phylogenetic analysis of defensins from different ant species.
All defensin sequences were aligned by MUSCLE and a BioNJ-tree with 100 bootstrap replicates was calculated. Branches with a bootstrap support below 40 were removed. Other bootstrap values are indicated. The genes for the gene tree were generated using the gene prediction pipeline maker and hand curated. The gene tree was rooted at the defensin from Ixodes scapularis (GenBank Acc. No.: XP_002436104.1). The A. mellifera defensin-1 forms a clade with proteins formerly described as defensin-2, which gives a first indication that they could be renamed accordingly. However, some of the proteins form a clade with the A. mellifera defensin-2. Moreover, two species S. invicta and C. floridanus, own both defensins. Therefore, we suggest a duplication event at the LCA of A. mellifera and the ant species.
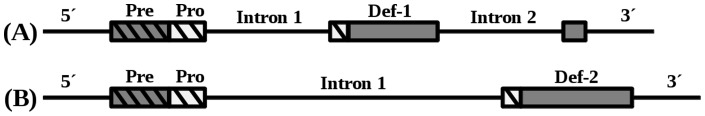
5′RACE and 3′RACE of the defensin-1 cDNA suggested a full length mRNA sequence of 535 bp (GenBank Acc. No.: JN989495), which was verified by amplification with primers near the 5′- and 3′-ends. The deduced C. floridanus defensin prepropeptide is 102 amino acids (aa) long, including a signal peptide of 17 aa and a propeptide of 40 aa, followed by a mature peptide of 44 aa, as predicted by ProP 1.0 [35]. The subsequent cloning of the corresponding defensin gene revealed the presence of three exons (64, 229 and 13 bp) and two introns (399 and 360 bp). The first intron is a phase 1 intron, which is located within the codon of glutamic acid number 22. The second intron is a phase 2 intron at the alanine residue number 98 following a so-called CXC motif characteristic for defensins (Fig. 9A).
Figure 9. Schematic structure of the defensin genes from C. floridanus.
(A) The gene encoding defensin-1 (GenBank Acc. No.: JN989495) is composed of three exons and two introns. The first intron is located within the region coding for the propeptide and the second is located within the region coding for the mature defensin peptide. (B) The gene encoding defensin-2 (GenBank Acc. No.: JQ693412) contains only one intron, which is also located within the propeptide coding region. Both deduced precursor peptides consist of a signal-sequence (Pre, grey hatched box) and a pro-sequence (Pro, white hatched box), followed by the mature defensin peptide (Def, grey box).
In contrast, the defensin-2 gene contains only two exons (97 and 194 bp), which are separated through one phase 1 intron (1043 bp) located within the codon of threonine number 33 (Fig. 9B). The full length mRNA sequence of defensin-2 (GenBank Acc. No.: JQ693412) is 1238 bp long and encodes a prepropeptide of 97 aa consisting of a signal peptide of 18 aa, a propeptide of 36 aa and a mature defensin peptide of 43 aa, as predicted by ProP 1.0 [35].
Phylogenetic analysis of defensin peptides
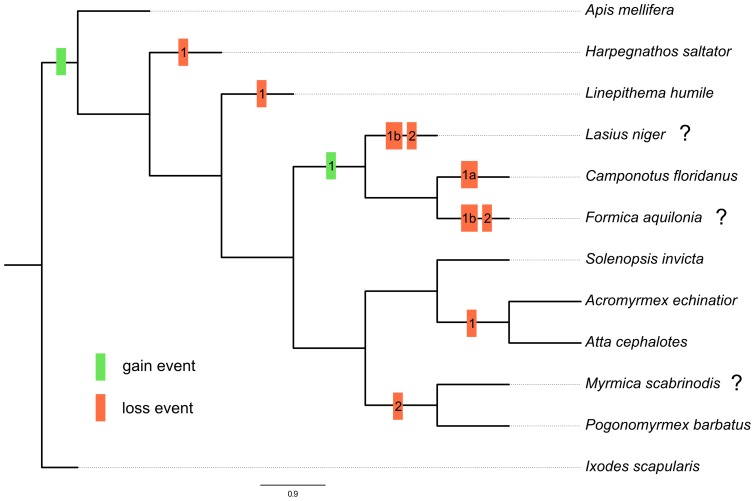
The comparison of the deduced amino acid sequences of the C. floridanus defensins with other ant defensins suggests a duplication event in the last common ancestor (LCA) of the bee A. mellifera and the ant species (Fig. 10). Nevertheless, almost all ant species have lost one of their defensin proteins. Only C. floridanus and S. invicta possess two defensins. Moreover, most of the proteins forming a clade with the A. mellifera defensin-1 were formerly described as defensin-2 homologues. Therefore, these proteins should be renamed to defensin-1 (Fig. 8, see also Fig. S2). Furthermore, an additional duplication event of defensin-1 peptide seems to have taken place in the LCA of Lasius niger, C. floridanus and Formica aquilonia, due to the grouping of the defensin-1 proteins in the phylogenetic tree (Fig. 8), which does not represent the species tree (Fig. 10). Nevertheless, the species lost either their defensin-1a (C. floridanus) or their defensin-1b (L. niger and F. aquilonia; Fig. 10).
Figure 10. Reconciled species tree of ant defensins.
This tree was generated by Notung. The ant species L. niger (GenBank Acc. No.: ACB46517.1), Myrmica scabrinodis (GenBank Acc. No.: ACB46524.1), and F. aquilonia (GenBank Acc. No.: Q5BU36.1) are examples for the sequences generated by [12], [27]. For these species no whole genome is available, which is indicated by the question mark behind the species. The gain and loss events are indicated by green and red boxes. The gain event at the LCA of A. mellifera and the ant species generated the defensin-1/2 peptide. The LCA of L. niger, C. floridanus and F. aquilonia had an additional duplication event of its defensin-1 peptide, but the species lost either their defensin-1a or their defensin-1b.
Discussion
AMPs are essential components of the insect immune system. Here we describe the identification and initial characterization of AMP genes of the ant C. floridanus encoding two defensins and a hymenoptaecin. Our results, taken together with the genome sequence of this social insect, indicate that these are the only three genes in C. floridanus encoding AMPs. Thus, similar to A. mellifera, a tendency to reduce the immune gene repertoire was suggested for ant species possibly due to hygiene measures on the colony level [13], [20].
Characterization of C. floridanus hymenoptaecin
In the recently published genome sequence of C. floridanus the hymenoptaecin gene escaped detection possibly due to sequencing problems of this gene carrying multiple direct repeats [25]. However, two contigs (AEAB01001738.1 and AEAB01001739.1) were found which harbour the 5′- and 3′- ends of the gene (Fig. 3). Since no other contigs encoding DNA sequences resembling the hymenoptaecin gene were discovered, the genomic data also supports the existence of a single hymenoptaecin gene, thus confirming that the above described variable repeat numbers after PCR amplification were a technical artefact caused by the tandem repeats. The characterization of the hymenoptaecin revealed a very peculiar modular composition of the deduced peptide(s) as compared to hymenoptaecins of other Hymenoptera. The hymenoptaecins known from other hymenopterans such as A. mellifera [9] and B. ignitus [10] show significant sequence homology to the hymenoptaecin domains repeated several times in the multipeptide precursor of the C. floridanus hymenoptaecin, suggesting structural and functional similarities. The A. mellifera hymenoptaecin is 93 aa long, including a 2-pyrrolidone-5-caboxylic acid at the N-terminus, which is derived from glutamine [17]. With the exception of the first so-called hymenoptaecin-like domain, the six deduced mature C. floridanus hymenoptaecins are 97 aa long and all start with a glutamine residue. Therefore, an amino-terminal blocking by forming 2-pyrrolidone-5-carboxylic acid is very likely also for the C. floridanus hymenoptaecin peptides. One of the putative mature hymenoptaecin peptides was overexpressed in E. coli and shown to exhibit moderate antibacterial activity. The possibility of an amino-terminal blocking might further increase the antibacterial potency of Cfl-hym peptides.
In contrast to the A. mellifera hymenoptaecin the C. floridanus hymenoptaecin has a complex precursor organization. A comparable precursor organization is known for the N. vitripennis hymenoptaecin, which encodes three AMP-like peptides, including one with similarity to the hymenoptaecin domains of C. floridanus [19]. Overall, the C. floridanus hymenoptaecin precursor structure is more similar to the multipeptide precursor structure of apidaecins, consisting of several repeated units [29]. As for apidaecin precursors we assume that the mature hymenoptaecin peptides are released by a three step mechanism, which is similar to maturation procedure of the yeast alpha-mating factor, since the repeats are flanked by repeating -X-A- (or -X-P-) sequences [51]. The initial processing is probably mediated by the KEX2-encoded endoprotease, which cuts at the C-terminus of the basic dipeptides Arg/Lys (RK) or Arg/Arg (RR) [52]. The next step is the C-terminal maturation via the KEX1-encoded carboxypeptidase, which removes both basic residues [53]. The last step is the N-terminal maturation of the spacer-mature peptides by a dipeptidyl aminopeptidase that removes E/D-A/P dipeptides [51]. Homologues of the respective enzymes are present in C. floridanus (GenBank Acc. No. EFN61704 to CAA96915 (E-value: 3E-35), EFN64345 to CAA96143 (E-value: 7E-94) and EFN67964 to NP_014862 (E-value: 5E-55)).
Despite the similarities in the multipeptide precursors, the C. floridanus hymenoptaecin differs from the A. mellifera apidaecin with regard to the gene structure. The latter one consists of several exons, each encoding a functional and distinct apidaecin peptide [13]. In contrast the hymenoptaecin mature peptide regions are encoded by a single exon only. This intronless gene structure prohibits the possibility of generating different transcripts by splice variation. Nevertheless, the multipeptide precursor structure of the Camponotus hymenoptaecin gene would allow the amplification of the antibacterial response despite the presence of only a single gene, as it was already suggested for apidaecins [29]. Furthermore, we also find surprisingly high sequence variability in our pooled samples. This high level of individual sequence variation has also been described for apidaecin exons from different bees [13], [29].
Phylogenetic analysis of hymenoptaecin peptides
The genome sequences of other ant species [25], [47], [48], [49], [50] revealed the presence of at least one gene locus encoding a hymenoptaecin precursor with similar domain structure as the C. floridanus hymenoptaecin. The presence of such multidomain hymenoptaecins in all ant species indicates an ancient origin of this gene structure early in the evolution of ants. However, the genome sequences indicate problems during the assembly and the gene prediction step of the genome projects and only a few sequences seem to be complete. All investigated complete hymenoptaecin peptide regions are encoded by a single exon only, which is evidence for exon duplication [54]. Our preliminary phylogenetic analysis of mature hymenoptaecin peptides suggests that the duplication event occurred independently in each species after separation (Fig. 7). Ongoing studies will reveal the full length hymenoptaecin precursor sequences from other ant species by direct sequencing and will deliver insights into the evolutionary history of the hymenoptaecin protein family in ants.
Phylogenetic analysis of ant defensin peptides
Bioinformatical prediction of defensins revealed the presence of at least one defensin gene in all investigated ant genomes with homology to defensin-1 or defensin-2 from A. mellifera (Fig. 8). We show that C. floridanus and S. invicta encode both defensin genes. Therefore, we suggest that the LCA of the ants and A. mellifera encoded both defensins. Based on the assumption that the genomes of L. niger, F. aquilonia, and M. scabrinodis contain only the published defensin genes, the reconciled gene tree (Fig. 10) exhibits many gain and loss events. Multiple duplication and loss events indicate a high adaptive potential and evolutionary plasticity of the antimicrobial peptides in ants. The C. floridanus mature defensin-1 and defensin-2 peptide sequences are well conserved with other ant defensins. However, the defensin-1 gene comprises three exons and two introns, in contrast to other characterized ant defensin genes [12], [27], which have two exons and one intron. Interestingly, a similar intron-exon composition is also known for other hymenopteran defensin-1 genes, e.g. from A. mellifera [16], B. ignitus [10] and N. vitripennis [11], while the Drosophila defensin gene does not carry any intron at all [55]. In contrast to other insect defensins, the bee defensin-1 has an extra stretch of 11 amino acids at its C-terminus, which encodes an additional C-terminal α-helical domain [9]. The C. floridanus defensin-1 has a short C-terminal extension of three amino acids in length. The precursors of the bee defensins have an extra amino acid, a glycine (G), at their C-termini, which seem to be amidated as suggested in the mature A. mellifera defensin [9]. As the deduced C. floridanus defensin-1 also ends with a G, it mayas well be amidated. According to this the mature C. floridanus defensin-1 is 3 amino acids longer than all other known ant defensins and it is so far the only known ant defensin which has an additional exon that is lacking from most other insects [56]. Further investigations will reveal, if this C-terminal extension can also be found in defensins from other ant species or if it is a special feature of C. floridanus.
Conclusions
The data reported here in combination with the recently published ant genome sequences indicate that the hypothesis of a reduced immune gene repertoire in social insects cannot easily be adopted for ant species. The genome drafts of C. floridanus and H. saltator [25] suggest indeed a comparable low number of genes encoding AMPs. However, this low number may to a certain extent be counteracted by the amplification of hymenoptaecin domains which are encoded as large precursor proteins with multiple bioactive domains. Sequence variations in the mature peptides may also lead to diversification of the immune response. Furthermore, P. barbatus even has more AMP genes than A. mellifera [50]. Detailed analyses of the complete antimicrobial repertoire from different ant species will deliver a better classification of the individual defense capabilities of these social insects.
Supporting Information
Alignment of hymenoptaecin domains from different ant species. The cDNAs of the ants were fragmented according to the cleavage sites predicted by ProP. Afterwards, all cDNA fragments were aligned by translatorX with default settings by muscle and the resulting alignment was cleaned by Gblocks with the default settings from the translatorX website.
(TIFF)
Alignment of defensin peptides from different ant species. The predicted peptide sequences for the seven ant species, the defensin-1 and defensin-2 of Apis mellifera (NM_001011616.2, NM_001011638.1), and the defensin of Ixodes scapularis (XP_002436104.1) were aligned by muscle with default settings.
(TIFF)
Primers used for characterization of C. floridanus hymenoptaecin and defensins.
(PDF)
Acknowledgments
We thank the Department of Zoology II, University of Würzburg, for the generous help with the rearing of C. floridanus ants.
Funding Statement
This work was funded by the priority program SFB567/C2 and the grant GR1243/8-1 of the Deutsche Forschungsgemeinschaft (DFG), and by the EU COST (European Cooperation in Science and Technology) action FA0701 “Arthropod symbioses: from fundamental studies to pest management”. C. Ratzka was kindly supported by the program “Chancengleichheit für Frauen in Forschung und Lehre”. The publication of this manuscript was funded by the “Open Access Publishing” program of the DFG and the University of Würzburg. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Lemaitre B, Hoffmann J (2007) The host defense of Drosophila melanogaster . Annu Rev Immunol 25: 697–743. [DOI] [PubMed] [Google Scholar]
- 2. Strand MR (2008) The insect cellular immune response. Insect Sci 15: 1–14. [Google Scholar]
- 3. Ganesan S, Aggarwal K, Paquette N, Silverman N (2011) NF-kappaB/Rel proteins and the humoral immune responses of Drosophila melanogaster . Curr Top Microbiol 349: 25–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Feldhaar H, Gross R (2008) Immune reactions of insects on bacterial pathogens and mutualists. Microbes Infect 10: 1082–1088. [DOI] [PubMed] [Google Scholar]
- 5. Haine ER, Moret Y, Siva-Jothy MT, Rolff J (2008) Antimicrobial defense and persistent infection in insects. Science 322: 1257–1259. [DOI] [PubMed] [Google Scholar]
- 6. Bulet P, Stocklin R, Menin L (2004) Anti-microbial peptides: from invertebrates to vertebrates. Immunol Rev 198: 169–184. [DOI] [PubMed] [Google Scholar]
- 7. Steiner H, Hultmark D, Engstrom A, Bennich H, Boman HG (1981) Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature 292: 246–248. [DOI] [PubMed] [Google Scholar]
- 8. Reddy KV, Yedery RD, Aranha C (2004) Antimicrobial peptides: premises and promises. Int J Antimicrob Agents 24: 536–547. [DOI] [PubMed] [Google Scholar]
- 9. Casteels-Josson K, Zhang W, Capaci T, Casteels P, Tempst P (1994) Acute transcriptional response of the honeybee peptide-antibiotics gene repertoire and required post-translational conversion of the precursor structures. J Biol Chem 269: 28569–28575. [PubMed] [Google Scholar]
- 10. Choi YS, Choo YM, Lee KS, Yoon HJ, Kim I, et al. (2008) Cloning and expression profiling of four antibacterial peptide genes from the bumblebee Bombus ignitus . Comp Biochem Physiol B Biochem Mol Biol 150: 141–146. [DOI] [PubMed] [Google Scholar]
- 11. Tian C, Gao B, Fang Q, Ye G, Zhu S (2010) Antimicrobial peptide-like genes in Nasonia vitripennis: a genomic perspective. BMC Genomics 11: 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Viljakainen L, Pamilo P (2008) Selection on an antimicrobial peptide defensin in ants. J Mol Evol 67: 643–652. [DOI] [PubMed] [Google Scholar]
- 13. Evans JD, Aronstein K, Chen YP, Hetru C, Imler JL, et al. (2006) Immune pathways and defence mechanisms in honey bees Apis mellifera . Insect Mol Biol 15: 645–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cornet B, Bonmatin JM, Hetru C, Hoffmann JA, Ptak M, et al. (1995) Refined three-dimensional solution structure of insect defensin A. Structure. 3: 435–448. [DOI] [PubMed] [Google Scholar]
- 15. Maget-Dana R, Ptak M (1997) Penetration of the insect defensin A into phospholipid monolayers and formation of defensin A-lipid complexes. Biophys J 73: 2527–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Klaudiny J, Albert S, Bachanova K, Kopernicky J, Simuth J (2005) Two structurally different defensin genes, one of them encoding a novel defensin isoform, are expressed in honeybee Apis mellifera . Insect Biochem Mol Biol 35: 11–22. [DOI] [PubMed] [Google Scholar]
- 17. Casteels P, Ampe C, Jacobs F, Tempst P (1993) Functional and chemical characterization of Hymenoptaecin, an antibacterial polypeptide that is infection-inducible in the honeybee (Apis mellifera). J Biol Chem 268: 7044–7054. [PubMed] [Google Scholar]
- 18. Xu P, Shi M, Chen XX (2009) Antimicrobial peptide evolution in the Asiatic honey bee Apis cerana . PLoS ONE 4: e4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gao B, Zhu S (2010) Characterization of a hymenoptaecin-like antimicrobial peptide in the parasitic wasp Nasonia vitripennis . Process Biochem 45: 139–146. [Google Scholar]
- 20. Cremer S, Armitage SA, Schmid-Hempel P (2007) Social immunity. Curr Biol 17: R693–702. [DOI] [PubMed] [Google Scholar]
- 21. Maschwitz U, Koob K, Schildknecht H (1970) Ein Beitrag zur Funktion der Metathoracaldrüse der Ameisen. J Insect Physiol 16: 387–404. [Google Scholar]
- 22. Schlüns H, Crozier RH (2009) Molecular and chemical immune defenses in ants (Hymenoptera: Formicidae). Myrmecological News 12: 237–249. [Google Scholar]
- 23. Sauer C, Dudaczek D, Hölldobler B, Gross R (2002) Tissue localization of the endosymbiotic bacterium “Candidatus Blochmannia floridanus” in adults and larvae of the carpenter ant Camponotus floridanus . Appl Environ Microbiol 68: 4187–4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wernegreen JJ, Kauppinen SN, Brady SG, Ward PS (2009) One nutritional symbiosis begat another: phylogenetic evidence that the ant tribe Camponotini acquired Blochmannia by tending sap-feeding insects. BMC Evol Biol 9: 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bonasio R, Zhang G, Ye C, Mutti NS, Fang X, et al. (2010) Genomic comparison of the ants Camponotus floridanus and Harpegnathos saltator . Science 329: 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gruber CW, Muttenthaler M (2012) Discovery of defense- and neuropeptides in social ants by genome-mining. PLoS ONE 7: e32559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Viljakainen L, Pamilo P (2005) Identification and molecular characterization of defensin gene from the ant Formica aquilonia . Insect Mol Biol 14: 335–338. [DOI] [PubMed] [Google Scholar]
- 28. Ratzka C, Liang C, Dandekar T, Gross R, Feldhaar H (2011) Immune response of the ant Camponotus floridanus against pathogens and its obligate mutualistic endosymbiont. Insect Biochem Mol Biol 41: 529–536. [DOI] [PubMed] [Google Scholar]
- 29. Casteels-Josson K, Capaci T, Casteels P, Tempst P (1993) Apidaecin multipeptide precursor structure: a putative mechanism for amplification of the insect antibacterial response. EMBO J 12: 1569–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Feldhaar H, Straka J, Krischke M, Berthold K, Stoll S, et al. (2007) Nutritional upgrading for omnivorous carpenter ants by the endosymbiont Blochmannia . BMC Biol 5: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Heinze J, Gadau J, Hölldobler B, Nanda I, Schmid M, et al. (1994) Genetic-variability in the ant Camponotus floridanus detected by multilocus DNA-fingerprinting. Naturwissenschaften 81: 34–36. [Google Scholar]
- 32. Monner DA, Jonsson S, Boman HG (1971) Ampicillin-resistant mutants of Escherichia coli K-12 with lipopolysaccharide alterations affecting mating ability and susceptibility to sex-specific bacteriophages. J Bacteriol 107: 420–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cantarel BL, Korf I, Robb SM, Parra G, Ross E, et al. (2008) MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res 18: 188–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stanke M, Diekhans M, Baertsch R, Haussler D (2008) Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24: 637–644. [DOI] [PubMed] [Google Scholar]
- 35. Duckert P, Brunak S, Blom N (2004) Prediction of proprotein convertase cleavage sites. Protein Eng Des Sel 17: 107–112. [DOI] [PubMed] [Google Scholar]
- 36. Abascal F, Zardoya R, Telford MJ (2010) TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res 38: W7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Talavera G, Castresana J (2007) Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 56: 564–577. [DOI] [PubMed] [Google Scholar]
- 38. Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, et al. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321. [DOI] [PubMed] [Google Scholar]
- 39. Gouy M, Guindon S, Gascuel O (2010) SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27: 221–224. [DOI] [PubMed] [Google Scholar]
- 40. Letunic I, Bork P (2007) Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23: 127–128. [DOI] [PubMed] [Google Scholar]
- 41. Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gascuel O (1997) BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol Biol Evol 14: 685–695. [DOI] [PubMed] [Google Scholar]
- 43. Brady SG, Schultz TR, Fisher BL, Ward PS (2006) Evaluating alternative hypotheses for the early evolution and diversification of ants. Proc Natl Acad Sci U S A 103: 18172–18177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gadau J, Helmkampf M, Nygaard S, Roux J, Simola DF, et al. (2012) The genomic impact of 100 million years of social evolution in seven ant species. Trends Genet 28: 14–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Durand D, Halldorsson BV, Vernot B (2006) A hybrid micro-macroevolutionary approach to gene tree reconstruction. J Comput Biol 13: 320–335. [DOI] [PubMed] [Google Scholar]
- 46. Vernot B, Stolzer M, Goldman A, Durand D (2008) Reconciliation with non-binary species trees. J Comput Biol 15: 981–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Suen G, Teiling C, Li L, Holt C, Abouheif E, et al. (2011) The genome sequence of the leaf-cutter ant Atta cephalotes reveals insights into its obligate symbiotic lifestyle. PLoS Genet 7: e1002007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nygaard S, Zhang G, Schiott M, Li C, Wurm Y, et al. (2011) The genome of the leaf-cutting ant Acromyrmex echinatior suggests key adaptations to advanced social life and fungus farming. Genome Res 21: 1339–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Smith CD, Zimin A, Holt C, Abouheif E, Benton R, et al. (2011) Draft genome of the globally widespread and invasive Argentine ant (Linepithema humile). Proc Natl Acad Sci USA 108: 5673–5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Smith CR, Smith CD, Robertson HM, Helmkampf M, Zimin A, et al. (2011) Draft genome of the red harvester ant Pogonomyrmex barbatus . Proc Natl Acad Sci USA 108: 5667–5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Julius D, Blair L, Brake A, Sprague G, Thorner J (1983) Yeast alpha factor is processed from a larger precursor polypeptide: the essential role of a membrane-bound dipeptidyl aminopeptidase. Cell 32: 839–852. [DOI] [PubMed] [Google Scholar]
- 52. Fuller RS, Brake A, Thorner J (1989) Yeast prohormone processing enzyme (KEX2 gene product) is a Ca2+-dependent serine protease. Proc Natl Acad Sci USA 86: 1434–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dmochowska A, Dignard D, Henning D, Thomas DY, Bussey H (1987) Yeast KEX1 gene encodes a putative protease with a carboxypeptidase B-like function involved in killer toxin and alpha-factor precursor processing. Cell 50: 573–584. [DOI] [PubMed] [Google Scholar]
- 54. Street TO, Rose GD, Barrick D (2006) The role of introns in repeat protein gene formation. J Mol Biol 360: 258–266. [DOI] [PubMed] [Google Scholar]
- 55. Dimarcq JL, Hoffmann D, Meister M, Bulet P, Lanot R, et al. (1994) Characterization and transcriptional profiles of a Drosophila gene encoding an insect defensin. A study in insect immunity. Eur J Biochem 221: 201–209. [DOI] [PubMed] [Google Scholar]
- 56. Froy O, Gurevitz M (2003) Arthropod and mollusk defensins–evolution by exon-shuffling. Trends Genet 19: 684–687. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of hymenoptaecin domains from different ant species. The cDNAs of the ants were fragmented according to the cleavage sites predicted by ProP. Afterwards, all cDNA fragments were aligned by translatorX with default settings by muscle and the resulting alignment was cleaned by Gblocks with the default settings from the translatorX website.
(TIFF)
Alignment of defensin peptides from different ant species. The predicted peptide sequences for the seven ant species, the defensin-1 and defensin-2 of Apis mellifera (NM_001011616.2, NM_001011638.1), and the defensin of Ixodes scapularis (XP_002436104.1) were aligned by muscle with default settings.
(TIFF)
Primers used for characterization of C. floridanus hymenoptaecin and defensins.
(PDF)