Abstract
dMi-2 is a highly conserved ATP-dependent chromatin-remodeling factor that regulates transcription and cell fates by altering the structure or positioning of nucleosomes. Here we report an unanticipated role for dMi-2 in the regulation of higher-order chromatin structure in Drosophila. Loss of dMi-2 function causes salivary gland polytene chromosomes to lose their characteristic banding pattern and appear more condensed than normal. Conversely, increased expression of dMi-2 triggers decondensation of polytene chromosomes accompanied by a significant increase in nuclear volume; this effect is relatively rapid and is dependent on the ATPase activity of dMi-2. Live analysis revealed that dMi-2 disrupts interactions between the aligned chromatids of salivary gland polytene chromosomes. dMi-2 and the cohesin complex are enriched at sites of active transcription; fluorescence-recovery after photobleaching (FRAP) assays showed that dMi-2 decreases stable association of cohesin with polytene chromosomes. These findings demonstrate that dMi-2 is an important regulator of both chromosome condensation and cohesin binding in interphase cells.
Author Summary
The packaging of DNA into chromatin is critical for the organization and expression of eukaryotic genes. Nucleosomes repress transcription by blocking the access of transcription factors and other regulatory proteins to DNA. Levels of chromatin organization above the level of the nucleosome—including chromosome folding, pairing, and looping—can also have profound effects on gene expression. However, the mechanisms by which higher-order chromatin structure is regulated and used to control gene expression remain poorly understood. Using Drosophila as a model organism, we have discovered an unanticipated role for dMi-2, a well-characterized ATP-dependent chromatin- remodeling factor, in the regulation of higher-order chromatin structure and cohesin dynamics in vivo. The inhibition of dMi-2 function causes chromosomes to condense, while elevated expression of dMi-2 triggers the decondensation of polytene and mitotic chromosomes and also destabilizes cohesin binding. Our findings suggest that dMi-2 may regulate transcription and cellular differentiation in other organisms, including humans, by altering higher-order chromatin structure or cohesin dynamics.
Introduction
The packaging of DNA into chromatin is critical for the organization and expression of eukaryotic genes [1], [2], [3]. The basic unit of chromatin structure, the nucleosome, contains the core histones H2A, H2B, H3 and H4. The association of nucleosomes with histone H1 and other linker histones facilitates their packaging into 30 nm fibers, which in turn are packaged into increasingly compact higher-order structures. Nucleosomes and other components of chromatin can repress transcription by blocking the access of regulatory proteins and the basal transcriptional machinery to DNA. There is growing evidence that levels of chromosome organization above the level of the nucleosome – including chromosome folding, pairing and looping – also play important roles in the regulation of gene expression. For example, condensin and cohesin, which were initially identified by their roles in mitosis and meiosis, modulate transcription by promoting long-range chromosomal interactions and DNA looping in interphase cells [4].
The repressive effects of nucleosomes on transcription are modulated by two general mechanisms: the covalent modification of nucleosomal histones and ATP-dependent chromatin remodeling [1], [5]. By altering the structure or positioning of nucleosomes, ATP-dependent chromatin-remodeling factors play critical roles in transcription and other nuclear processes. Dozens of chromatin-remodeling factors, including members of the SWI/SNF, ISWI, CHD and INO80 families, have been identified in organisms ranging from yeast to humans. By contrast, relatively little is known about how higher-order chromatin structure is regulated and exploited to control gene expression and other nuclear processes.
A major barrier to the identification of factors that regulate higher-order chromatin structure is the difficulty of visualizing the decondensed interphase chromosomes of diploid cells. This barrier can be overcome through the use of Drosophila melanogaster as a model organism. During Drosophila development, many tissues undergo multiple rounds of DNA replication in the absence of cytokinesis, leading to the formation of huge polytene chromosomes containing hundreds of aligned sister chromatids. These transcriptionally active chromosomes are indistinguishable from the interphase chromosomes of diploid cells in most respects. Genetic studies in Drosophila have identified numerous factors that regulate polytene chromosome structure, including ISWI, an ATP-dependent chromatin-remodeling factor. The loss of ISWI function leads to the decondensation of salivary gland polytene chromosomes, possibly due to failure to assemble chromatin containing the linker histone H1 [6], [7], [8]. This striking phenotype led us to investigate the potential involvement of another ATP-dependent chromatin-remodeling factor, Drosophila Mi-2 (dMi-2), in the regulation of higher-order chromatin structure.
dMi-2 functions as the ATPase subunit of multiple chromatin-remodeling complexes, including the NuRD (Nucleosome Remodeling and Deacetylase) complex and dMec (Drosophila MEP-1 containing complex) [9]. NuRD is highly conserved in metazoans and is thought to repress transcription via its chromatin-remodeling and histone deacetylase activities [10], [11], [12]. dMec is the most abundant dMi-2 complex in Drosophila and has been implicated in SUMO-dependent transcriptional repression [13], [14]. Mi-2 plays an important role in cell fate specification in organisms ranging from nematodes to vertebrates. For example, Mi-2 helps maintain the distinction between the germline and soma during C. elegans embryogenesis [15]; regulates the terminal differentiation of B lymphocytes into plasma cells in mammals [16]; and participates in the transcriptional repression of HOX genes by Hunchback and Polycomb in Drosophila [17]. dMi-2 is also required for the efficient expression of heat-shock genes in Drosophila, indicating that its function is not limited to transcriptional repression [18].
Here we report an unanticipated role for Mi-2 in the regulation of higher-order chromatin structure in Drosophila. The loss of dMi-2 function causes salivary gland polytene chromosomes to lose their characteristic banding pattern and appear more condensed than normal. Conversely, the increased expression of dMi-2 in the salivary gland disrupts interactions between sister chromatids and triggers the decondensation of polytene chromosomes. Consistent with these findings, dMi-2 disrupts the association of cohesin with polytene chromosomes. Our studies reveal that dMi-2 is an important regulator of both chromosome condensation and cohesin binding in interphase cells.
Results
Generation and analysis of GAL4-regulated transgenes encoding wild-type and dominant-negative dMi-2 proteins
As a first step toward investigating the role of dMi-2 in the regulation of higher-order chromatin structure, we investigated phenotypes resulting from either the loss or gain of dMi-2 function. We could not examine the polytene chromosomes of individuals homozygous for loss-of-function dMi-2 alleles since they die relatively early in larval development [17], [19]. We therefore used the GAL4 system [20] to examine the consequences of expressing a dominant-negative form of the dMi-2 protein in the salivary gland. For this study, we used a mutation that deletes amino acids 932-1158 of the dMi-2 protein, including its bipartite ATP-binding site. This deletion eliminates the ATPase activity of dMi-2 in vitro [21] without disrupting its interaction with other proteins (Figure S1). We therefore reasoned that the expression of dMi-2Δ932-1158 should have a strong, dominant-negative effect on dMi-2 function in vivo, as has been observed for comparable mutations in other chromatin-remodeling factors [7], [22], [23].
Transgenic fly strains bearing GAL4-inducible transgenes encoding wild-type (UAS-dMi-2+) or dominant-negative (UAS-dMi-2Δ932-1158) dMi-2 proteins were generated by P element transformation. Transformants were crossed to GAL4 driver lines to generate progeny that express the dMi-2 transgenes in stage or tissue-specific patterns. The GAL4 system is inherently temperature-sensitive; higher levels of transgene expression are observed at higher temperatures [24]. This allowed us to modulate the expression of dMi-2 transgenes by varying the temperature at which progeny were reared.
The expression of dMi-2Δ932-1158 transgenes under the control of a ubiquitously expressed GAL4 driver (da-GAL4) had a dramatic effect on viability. At 25°C, the majority of UAS-dMi-2Δ932-1158 6-5/+; da-GAL4/+ individuals completed larval development, but only 5% survived to adulthood (Table 1). The lethality resulting from the expression of dMi-2Δ932-1158 was moderately enhanced by a hypomorphic allele of dMi-2 (dMi-2f08103) and strongly enhanced by the null allele dMi-24 (Table 1). At 29°C, no individuals expressing dMi-2Δ932-1158 under the control of the da-GAL4 driver survived beyond the first or second larval instar (data not shown). This lethal phase is identical to that resulting from the complete loss of zygotic dMi-2 function [17], [19]. These findings confirmed that the expression of the catalytically inactive dMi-2Δ932-1158 protein has strong, dominant-negative effects on dMi-2 function in vivo.
Table 1. Analysis of GAL4-regulated transgenes.
| Genotype | number of copies | % survival to | |||
| dMi-2+ | UAS-dMi-2Δ932-1158 | UAS-dMi-2+ | pupae | adults | |
| dMi-24/da-GAL4 (a) | 1 | 0 | 0 | 93.9 | 92.8 |
| dMi-24/dMi-24 da-GAL4 (b) | 0 | 0 | 0 | 0.0 | 0.0 |
| UAS-dMi-2 Δ932-1158 6-5/+; da-GAL4/+(c) | 2 | 1 | 0 | 74.0 | 5.0 |
| UAS-dMi-2 Δ932-1158 6-5/+; dMi-2f08103/da-GAL4(d) | 1 | 1 | 0 | 23.0 | 9.6 |
| UAS-dMi-2 Δ932-1158 6-5/+; dMi-24/da-GAL4(e) | 1 | 1 | 0 | 0.0 | 0.0 |
| UAS-dMi-2+ 3-3/+; dMi-24/dMi-24 da-GAL4 (f) | 0 | 0 | 1 | 76.0 | 73.9 |
| UAS-dMi-2+ 3-3/+; da-GAL4/+ (g) | 2 | 0 | 1 | 13.9 | 12.1 |
| UAS-dMi-2+ 3-3/+; dMi-24/da-GAL4 (h) | 1 | 0 | 1 | 88.2 | 82.6 |
Data are shown for progeny of the following crosses:
dMi-24/TM6B, Tb males X w; da-GAL4 females;
w; dMi-24/TM6B, Tb males X w; dMi-24 da-GAL4/TM6B, Tb females;
w; P[w+, UAS-dMi-2 Δ932-1158]6-5 males X w; da-GAL4 females;
w; P[w+, UAS-dMi-2 Δ932-1158]6-5; dMi-2f08103/TM6B, Tb males X w; da-GAL4 females;
w; P[w+, UAS-dMi-2 Δ932-1158]6-5; dMi-24/TM6B, Tb males X w; da-GAL4 females;
w; P[w+, UAS-dMi-2+]3-3; dMi-24/TM6B, Tb males X w; dMi-24 da-GAL4/TM6B, Tb females;
w; P[w+, UAS-dMi-2+]3-3 males X w; da-GAL4 females;
w; P[w+, UAS-dMi-2+]3-3; dMi-24/TM6B, Tb males X w; da-GAL4 females.
We also investigated the effect of expressing wild-type dMi-2 protein on Drosophila development using the da-GAL4 driver. As expected, the UAS-dMi-2+ transgene rescued the recessive lethality of dMi-24; 74% of UAS-dMi-2+ 3-3/+; dMi-24 da-GAL4/dMi-24 survived to adulthood (Table 1). Surprisingly, the over-expression of dMi-2+ in a wild-type dMi-2 background resulted in larval lethality; only 12% of UAS-dMi-2+ 3-3/+; da-GAL4/+ individuals survived beyond the third-larval instar (Table 1). The lethality resulting from the expression of the UAS-dMi-2+ transgene was suppressed by a twofold reduction in the level of endogenous dMi-2 function; over 80% of UAS-dMi-2+ 3-3/+; dMi-24/da-GAL4 individuals survived to adulthood (Table 1). Thus, even a modest increase in the level of dMi-2 can be lethal.
Analysis of chromosome defects resulting from dMi-2 over-expression
The above findings demonstrated that the GAL4 system can be used to analyze phenotypes resulting from either the gain or loss of dMi-2 function. We therefore proceeded to characterize chromosome defects resulting from the expression of wild-type or dominant-negative dMi-2 proteins. We initially used the da-GAL4 driver to induce the expression of UAS-dMi-2Δ932-1158 and UAS-dMi-2+ transgenes in salivary gland nuclei. The increased expression of wild-type dMi-2 protein dramatically altered the structure of salivary gland polytene chromosomes; fixed polytene chromosomes squashes appeared much larger than normal; their banding pattern was severely disrupted; and it was often difficult to distinguish individual chromosome arms (compare Figure 1A and 1B). This phenotype was never observed in wild-type larvae (data not shown) or UAS-LacZ/+; da-GAL4/+ control larvae (Figure 1A). Chromosome defects were even more pronounced in larvae bearing two copies of the UAS-dMi-2+ transgene in addition to the da-GAL4 driver (Figure 1C). Similar results were obtained with a Sgs3-GAL4 driver which is expressed only in the salivary gland of third-instar larvae (Figure S2).
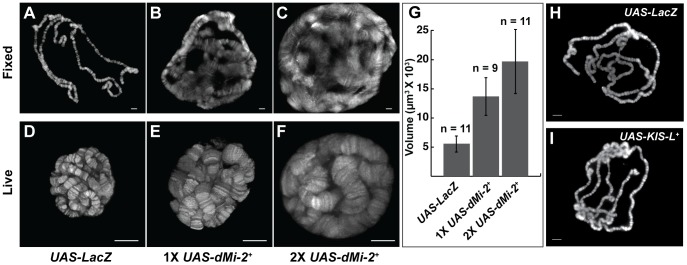
Figure 1. dMi-2 increases the size of polytene chromosomes in vivo.
(A–F) Salivary gland polytene chromosomes of late third-instar UAS-LacZ/+; da-GAL4/+ control larvae (A and D) or larvae bearing one (UAS-dMi-2+ 3-3/+; da-GAL4/+ [1X UAS-dMi-2+]) (B and E) or two (UAS-dMi-2+ 3-3/+; UAS-dMi-2+ 15-1/da-GAL4 (2X UAS-dMi-2+)] (C and F) copies of a UAS-dMi-2+ transgene. (A–C) Squashes of fixed polytene chromosomes stained with DAPI. (D–F) Live analysis of chromosomes of larvae expressing His2Av-GFP. The over-expression of dMi-2 increases the size of polytene chromosomes (compare B and C to A and E and F to D) and compromises their structural integrity, as evidenced by the disruption of the banding pattern (compare B and C to A) (G) Quantification of the volume of polytene chromosomes of control, 1X UAS-dMi-2+, and 2X UAS-dMi-2+ larvae imaged by live analysis. The over-expression of dMi-2 protein causes a two to fourfold increase in chromosome volume (1X UAS-dMi-2+: 1.4E+04±3.3E+03 µm3 and 2X UAS-dMi-2+: 1.97E+04±5.5E+03 µm3) compared to UAS-Lac-Z (5.6E+03±1.4E+03 µm3). (H–I) Over-expression of KIS-L has no obvious effect on the structure of polytene chromosomes. DAPI staining of polytene chromosomes squashes from UAS-LacZ/ey-GAL4 (H) and UAS-KIS-L+ 20-7/ey-GAL4 (I) individuals. A, B, C, D, E F, H and I scale bars are 10 µm. Larvae were reared at 29°C.
To gain a more accurate impression of the effect of increased dMi-2 expression on chromosome structure, we used live analysis to visualize the chromosomes of larvae expressing a fluorescently tagged core histone (His2Av-GFP). The salivary gland chromosomes of third-instar larvae expressing one or two UAS-dMi-2+ transgenes under the control of the da-GAL4 driver displayed a clear banding pattern but were much larger than normal (compare Figure 1D to 1E and 1F). The increased expression of dMi-2 led to a two to fourfold increase in chromosome volume (Figure 1G). The severe disruption of the chromosome banding pattern evident in fixed polytene chromosome squashes was not observed by live analysis (compare Figures 1B and 1C to 1E and 1F). This difference suggests the over-expression of dMi-2 makes chromosomes unusually sensitive to mechanical stress, leading to their disruption during the squashing procedure. Taken together, these observations suggest that the elevated expression of dMi-2 increases the size of polytene chromosomes while compromising their structural integrity.
The analysis of chromosome defects resulting from the loss of dMi-2 function
We next examined the consequences of expressing the dominant-negative dMi-2Δ932-1158 protein in salivary gland nuclei. A significant fraction of UAS-dMi-2Δ932-1158 6-5/+; da-GAL4/+ individuals survive to adulthood at 25°C; at 29°C, they do not survive beyond the first or second larval instar (Table 1 and data not shown). To reduce dMi-2 function without blocking larval development, individuals of this genotype were shifted from 25°C to 29°C before the end of the first larval instar (0–48 hours after egg laying). Under these conditions, the loss of dMi-2 function disrupted the banding pattern of salivary gland chromosomes and caused them to appear much thinner than normal (compare Figure 2B and 2D to 2A and 2C). Similar results were obtained using a GAL4-regulated transgene encoding a catalytically inactive form of the dMi-2 protein (dMi-2K761R) in which a conserved lysine in the ATP-binding site is replaced with an arginine (MM and AB, unpublished data). Live analysis revealed chromosome defects similar to those observed in fixed chromosome squashes (compare Figure 2F and 2H to 2E and 2G). The ability of dMi-2 to increase the size of salivary gland polytene chromosomes is therefore dependent on its ATPase activity.
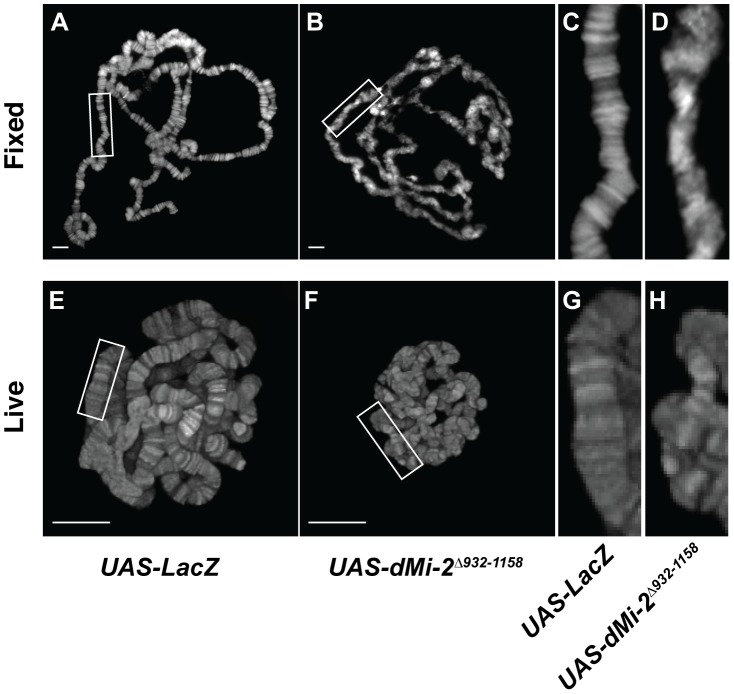
Figure 2. The loss of dMi-2 function alters chromosome structure.
Salivary gland polytene chromosomes of late third-instar UAS-LacZ/+; da-GAL4/+ control larvae (A and C) or larvae expressing the dominant-negative dMi-2Δ932-1158 protein (UAS-dMi-2 Δ932-1158 6-5/+; da-GAL4/+) (B and D). (A–D) Squashes of fixed polytene chromosomes stained with DAPI. (E–H) Live analysis of chromosomes of larvae expressing His2Av-GFP. The expression of dMi-2Δ932-1158 reduces the size of polytene chromosomes and disrupts their banding pattern. (Compare C and D to A and B, and G and H to E and F). A, B, E and F scale bars are 10 µm.
The CHD protein KIS-L does not regulate higher-order chromatin structure in vivo
Drosophila kismet (KIS) was identified in genetic screens for Polycomb antagonists and subsequently shown to encode a member of the CHD subfamily of chromatin-remodeling factors [25], [26], [27]. The chromosomal distributions of KIS-L and dMi-2 are highly overlapping; both proteins are enriched at less condensed, transcriptionally active regions of chromatin (Figure S5A) [28]. These similarities prompted us to investigate whether the over-expression of KIS-L also alters the structure of salivary gland polytene chromosomes. Transgenic fly strains bearing GAL4-inducible transgenes encoding wild-type KIS-L protein, UAS-KIS-L+, were generated by P element transformation; the da-GAL4 driver was used to drive the ubiquitous expression of these transgenes. As observed for dMi-2, the over-expression of the wild-type KIS-L protein is lethal; UAS-KIS-L+ 20-7/+; da-GAL4/+ individuals failed to survive beyond late larval or early pupal stages of development (data not shown). Unlike dMi-2, however, the increased expression of KIS-L did not cause obvious changes in the structure of salivary gland polytene chromosomes (Figure 1H and 1I). The ability of dMi-2 to alter chromosome structure therefore does not appear to be a property common to all CHD proteins.
The increased expression of wild-type dMi-2 triggers relatively rapid changes in chromosome structure
The chromosome defects described above resulted from the continuous expression of wild-type or dominant-negative dMi-2 proteins over a five to seven day period. To help distinguish between primary and secondary consequences of dMi-2 over-expression, we used a modification of the GAL4 system that permits the precise temporal regulation of GAL4-responsive transgenes through the use of a temperature-sensitive GAL80 repressor (GAL80ts) [29]. At the permissive temperature (18°C), GAL80ts binds to and inhibits the GAL4 activator. At the restrictive temperature (29°C), GAL4 function is restored, leading to the rapid activation of the GAL4-responsive target gene. The use of this system allowed us to monitor the time course of changes in chromosome structure resulting from increased dMi-2 expression.
UAS-dMi-2+ 3-3/+; UAS-dMi-2+ 15-1/da-GAL4 tubP-Gal80ts individuals were shifted from 18°C to 29°C at the middle of the third larval instar to activate UAS-dMi-2+ expression. RT-PCR was used to monitor the expression of the endogenous dMi-2+ gene, the UAS-dMi-2+ transgenes and the overall level of dMi-2 RNA during the subsequent 24 hours. The expression of the UAS-dMi-2+ transgenes, but not the endogenous dMi-2 gene, was rapidly activated after the shift to the restrictive temperature, leading to roughly a twofold increase in the total level of dMi-2 RNA within seven hours (Figure 3A). The level of dMi-2 RNA remained constant for the remainder of the experiment. Changes in chromosome structure were evident in squashed polytene chromosomes within ten to twelve hours after the shift to 29°C and became increasingly pronounced over the following twelve hours (Figure S3). The relatively short lag time between the increase in dMi-2 expression and alterations in chromosome morphology suggests that dMi-2 may directly regulate higher-order chromatin structure.
Figure 3. dMi-2 triggers rapid changes in chromosome structure without altering DNA replication.
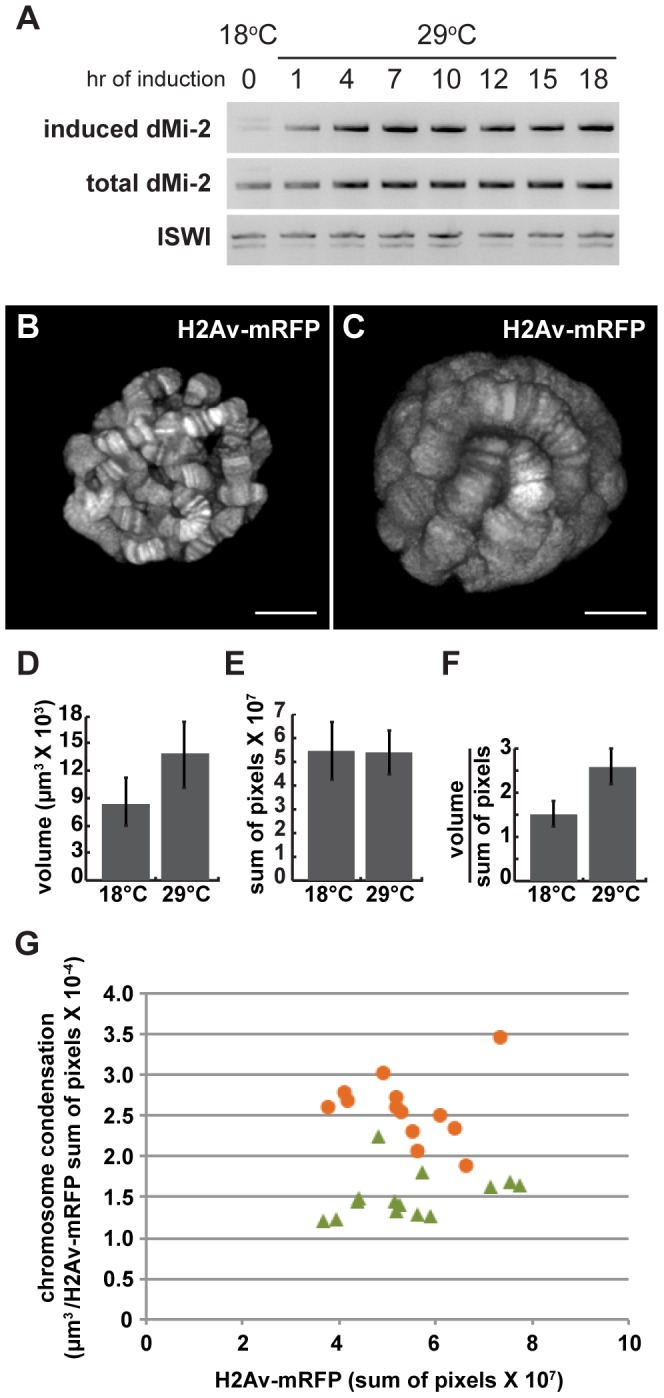
Third-instar UAS-dMi-2+ 3-3/+; UAS-dMi-2+ 15-1/da-GAL4, Gal80ts larvae were shifted from 18°C to 29°C to activate UAS-dMi-2+ expression. (A) Changes in dMi-2 RNA levels were monitored by RT-PCR following the shift from 18° to 29°C using primers that specifically amplify the RNA encoded by UAS-dMi-2+ (induced) or both the induced and endogenous dMi-2 RNAs (induced+endogenous). The level of ISWI RNA was assayed as an internal control. (B and C) Live analyses of salivary gland nuclei of UAS-dMi-2+ 3-3/+; UAS-dMi-2+ 15-1/da-GAL4 GAL80ts maintained at 18°C (B; n = 13) or shifted to 29°C for 27 hours (C; n = 14). (D) Quantification of the volume of salivary gland polytene chromosomes of larvae shown in B and C. (E) Quantification of His2Av-mRFP1 fluorescence in the same larvae shown in B and C. (F) Quantification of chromosome compaction, as assessed by the ratio of volume in µm3 to His2Av-mRFP1 fluorescence per nucleus for the larvae shown in B and C. (G) Scatter plot of the data used to generate D–F. Chromatin compaction vs. DNA content are compared for single nuclei of larvae maintained at 18°C (green triangles) or 29°C (orange circles) for 27 hours. B and C scale bars are 10 µm.
dMi-2 does not increase the size of polytene chromosomes by altering DNA replication
Salivary gland polytene chromosomes consist of more than a thousand closely aligned sister chromatids formed by repeated rounds of DNA replication in the absence of cytokinesis. Additional rounds of DNA replication could therefore account for the huge size of salivary gland polytene chromosomes in larvae that express increased levels of dMi-2. To address this possibility, the above experiments were repeated using larvae expressing a core histone tagged with RFP (His2Av-mRFP1); this allowed us to use live analysis to directly compare changes in chromosome volume, DNA content (as estimated by the level of His2Av-mRFP1 fluorescence) and chromosome compaction (the ratio of chromosome volume to DNA content) resulting from increased dMi-2 expression. UAS-dMi-2+ 3-3/His2Av-mRFP1; UAS-dMi-2+ 15-1/da-GAL4 tubP-Gal80ts individuals were shifted from 18°C to 29°C to activate UAS-dMi-2+ expression late in the third larval instar. 27 hours after the shift to 29°C, live analysis revealed a significant increase in polytene chromosome volume, but not His2Av-mRFP1 fluorescence, compared to larvae that had been maintained at 18°C, resulting in almost a twofold decrease in chromosome compaction (Figure 3B–G). Similar results were obtained when we compared the area of salivary gland polytene chromosome squashes to DNA content as estimated by DAPI fluorescence (data not shown). Increased dMi-2 expression thus triggers the decondensation of polytene chromosomes as opposed to additional rounds of replication.
We next examined the effect of the dominant-negative dMi-2 Δ932-1158 protein on chromosome compaction using the GAL4-GAL80ts system. UAS-dMi-2 Δ932-1158 6-5/His2Av-mRFP1; +/da-GAL4 tubP-Gal80ts individuals were shifted from 18°C to 29°C to activate dMi-2 Δ932-1158 expression late in the third larval instar. 48 hours after the shift to 29°C, live analysis revealed significant decreases in both polytene chromosome volume and His2Av-mRFP1 fluorescence relative to larvae maintained at 18°C (Figure 4A–D). A modest increase in chromosome compaction was also observed (Figure 4E); this effect was relatively striking when nuclei with similar DNA content were compared (Figure 4F). These findings provide additional evidence that dMi-2 is an important regulator of chromosome compaction in vivo.
Figure 4. The loss of dMi-2 function promotes chromosome compaction.
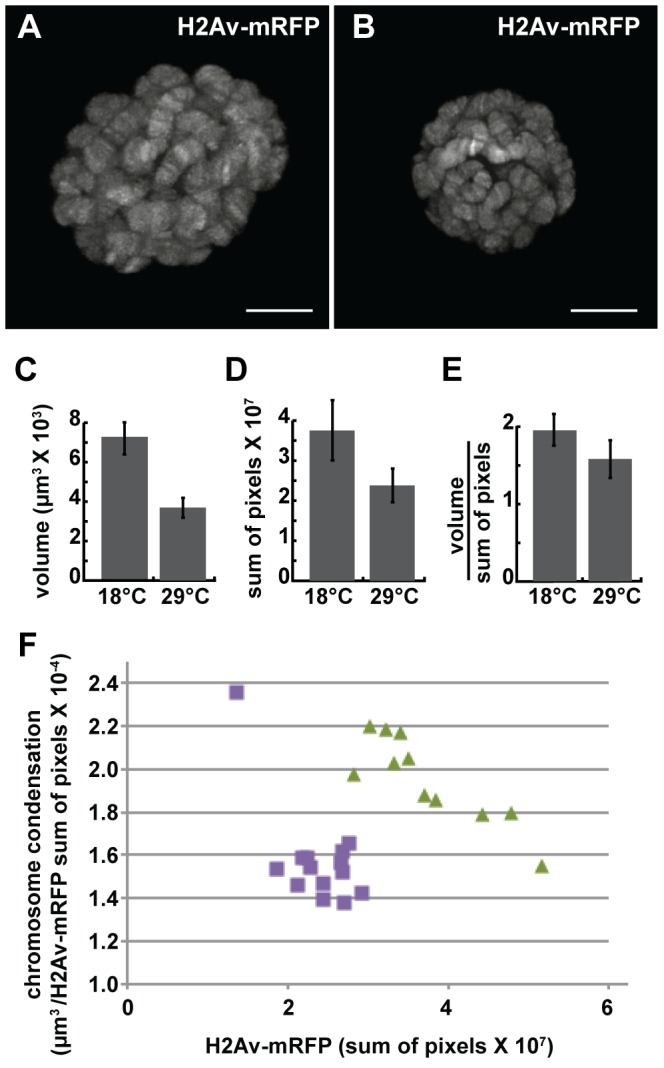
Third-instar UAS-dMi-2 Δ932-1158 6-5/His2Av-mRFP1; +/da-GAL4 tubP-Gal80ts larvae were shifted from 18°C to 29°C to activate dMi-2 Δ932-1158 expression. (A and B) Live analyses of salivary gland nuclei of UAS-dMi-2 Δ932-1158 6-5/His2Av-mRFP1; +/da-GAL4 tubP-Gal80ts larvae maintained at 18°C (A; n = 11) or shifted to 29°C for 48 hours (B; n = 14). (C–E) Quantification of the volume (C), His2Av-mRFP1 fluorescence (D), and compaction (E) of salivary gland chromosomes of larvae maintained at 18°C or 29°C for 48 hours after the temperature shift. (F) Scatter plot of the data used to generate C–E. Chromatin compaction vs. DNA content are compared for single nuclei of larvae maintained at 18°C (green triangles) or 29°C (purple circles) for 48 hours. A and B scale bars are 10 µm.
Loss of dMi-2 function reduces the size of heat-shock puffs
Activation of the hsp70 heat-shock (HS) genes is accompanied by the formation of transcriptionally active HS puffs at loci 87A and 87C, a process that is generally attributed to chromosome decondensation. dMi-2 is recruited to HS genes following their activation [18]. We asked if loss of dMi-2 function would interfere with HS-induced chromosome puffing. We subjected third-instar larvae to a 20 minute HS at 37°C and visualized puff formation at 87A and 87C by indirect immunofluorescence using a RNA Pol II antibody. Under these conditions, all the polytene chromosomes of control larvae exhibited strong (67%) to moderate (33%) decondensation at 87A and 87C as judged by a loss of DAPI signal and an accumulation of RNA polymerase II (Figure 5, upper panels). By contrast, the extent of puff formation and the amount of polymerase II signal was significantly reduced in transgenic larvae expressing catalytically inactive dMi-2 (dMi-2K761R, raised at 25°C prior to heat-shock; Figure 5, lower panels); no puffs or only moderate puffing was observed in 14 and 53% of the chromosomes of these larvae, respectively. This result suggests that dMi-2 function is important for a local and rapid chromosome decondensation event.
Figure 5. dMi-2 is required for full decondensation of heat-shock loci.
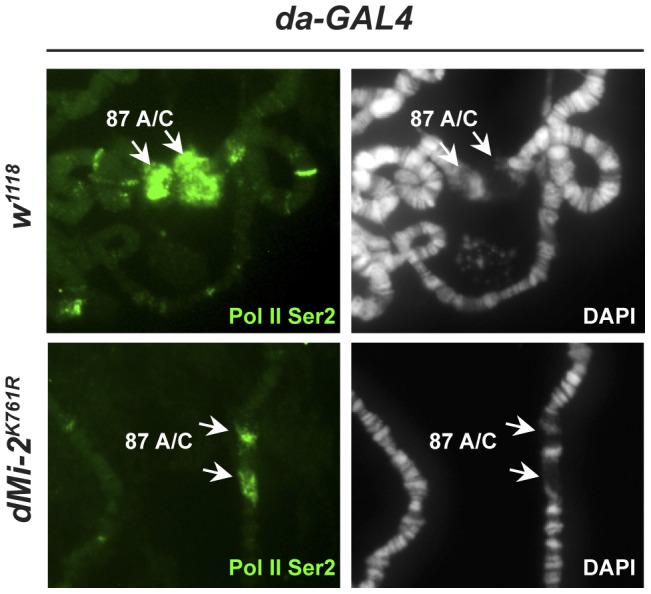
w1118 control and transgenic larvae expressing dominant-negative dMi-2K761R were subjected to a 20 min heat-shock at 37°C. Polytene chromosomes were visualized by staining with DAPI (right panels) or indirect immunofluorescence using an antibody against Pol II Ser2 (left panels). Arrows indicate the hsp70 gene containing loci 87A and 87C.
dMi-2 does not regulate higher-order chromatin structure by altering histone H1 assembly
The reduced expression of histone H1, a linker histone that promotes the formation of 30 nm fibers, causes polytene chromosome defects that are remarkably similar to those resulting from the increased expression of dMi-2 [8], [30]. This similarity suggested that dMi-2 might regulate chromosome structure by antagonizing the assembly of chromatin containing histone H1. To investigate this possibility, we examined whether the over-expression of dMi-2 decreases the association of histone H1 with chromatin. The live analysis of larvae expressing GFP-tagged histone H1 revealed that the elevated expression of dMi-2 increased the size of polytene chromosomes relative to control larvae without reducing the levels of bound histone H1 (Figure 6A–C). The over-expression of dMi-2 also did not reduce ISWI expression or the level of histone H1 in chromatin extracted from salivary glands as assayed by Western blotting (Figure 6D; Figure S4). The expression of dominant-negative dMi-2 in salivary gland nuclei caused a slight decrease in histone H1 levels (Figure S4). Thus, dMi-2 does not promote chromosome decondensation by repressing histone H1 expression or assembly.
Figure 6. dMi-2 does not promote chromatin decondensation by antagonizing histone H1 assembly.
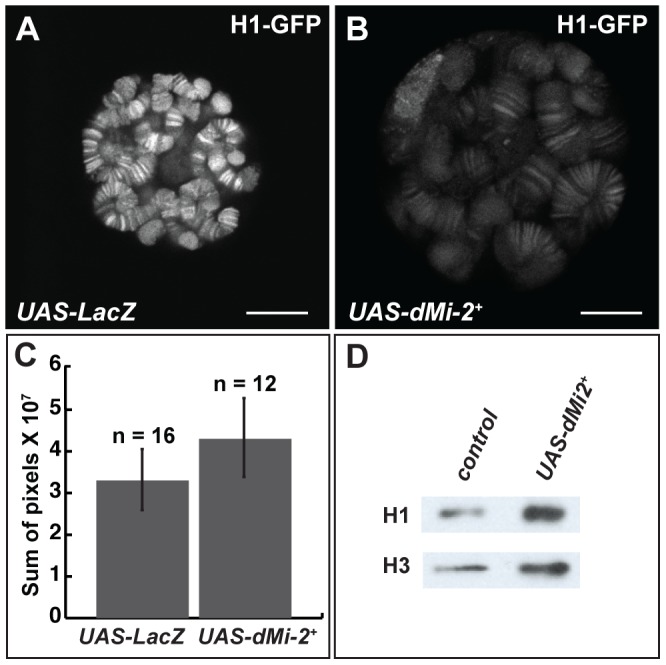
(A–B) Live analysis of salivary gland nuclei of late third-instar UAS-LacZ/+; ey-GAL4/+ (UAS-LacZ) control larvae (A) and UAS-dMi-2+ 3-3/+; UAS-dMi-2+ 15-1/ey-GAL4 (UAS-dMi-2+) larvae (B) expressing H1-GFP. Scale bars are 10 µm. (C) Quantification of H1-GFP fluorescence in larvae shown in A and B. The exposure times used to capture the images are identical; the number of glands analyzed is noted. (D) Protein blot showing the relative levels of histones H1 and H3 in chromatin extracted from late third-instar UAS-dMi-2+ 3-3/+; da-GAL4/+ (UAS-dMi-2+) and UAS-LacZ/+; da-GAL4/+ (UAS-LacZ) larvae raised at 29°C.
dMi-2 regulates interactions between sister chromatids in larval salivary gland polytene chromosomes
The disruption of interactions between aligned chromatids could also account for the changes in polytene chromosome structure caused by elevated dMi-2 expression. To test this hypothesis, we used the LacO/LacI-GFP system [31] to monitor the effect of dMi-2 over-expression on the structure of a specific locus by live analysis. In this system, a GFP-tagged Lac repressor (LacI-GFP) binds to tandem copies of Lac operator (LacO) sequences inserted at a single chromosomal location (Figure 7A). This assay has been successfully used to detect disruption of the normal parallel alignment of the chromatids in polytene chromosomes [32]. For this study, we used an insertion of a LacO transgene in an interband on the second chromosome and a transgene encoding LacI-GFP repressor under the control of the Hsp83 promoter. Control UAS-dMi-2+ 3-3/LacO LacI-GFP; UAS-dMi-2+ 15-1/da-GAL4 tubP-Gal80ts individuals were reared at 18°C to the end of the third larval instar (240 hours) and heat-shocked for 60 minutes at 37°C to activate LacI-GFP expression. To over-express dMi-2, individuals of the same genotype were reared at 18°C for (9 days); shifted to 29°C for 27 hours to activate the UAS-dMi-2+ transgenes; and heat-shocked for 60 minutes at 37°C to activate LacI-GFP expression. Polytene chromosomes of control larvae showed a compact band of GFP fluorescence signal perpendicular to the chromosome axis (Figure 7B and D). In larvae expressing elevated levels of dMi-2, however, the GFP signal was dispersed, with dots of LacI-GFP fluorescence scattered over a fivefold larger area (Figure 7C and E). This striking phenotype suggests that dMi-2 may play an unanticipated role in chromosome cohesion in vivo.
Figure 7. dMi-2 regulates chromosome cohesion in the larval salivary gland.
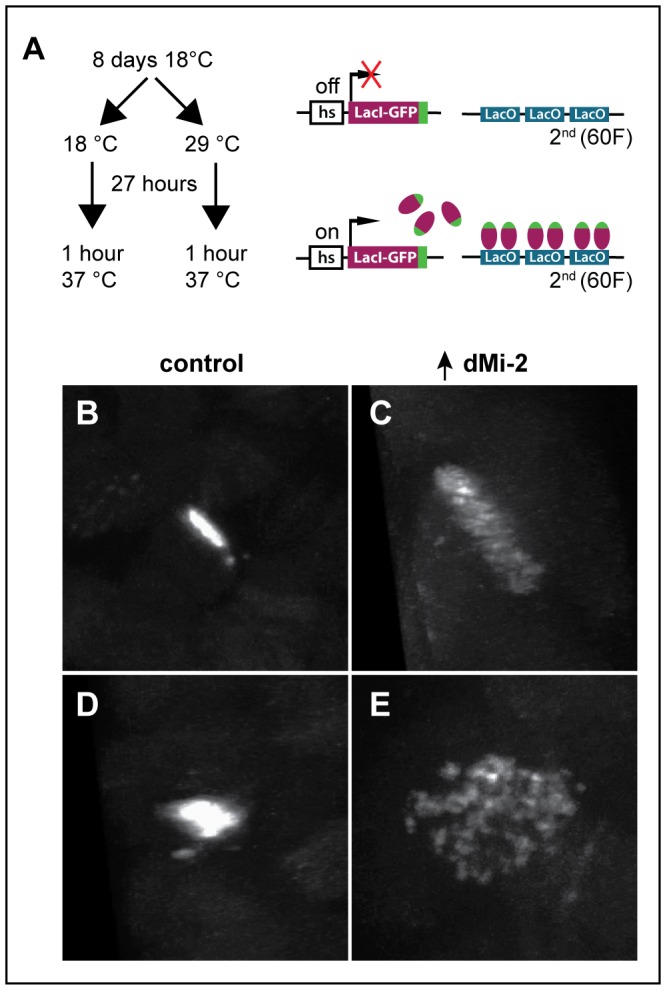
(A) Overview of LacI-GFP/LacO tethering assay. The GFP-tagged LacI fusion protein (LacI-GFP) binds 256 tandem LacO sequences inserted at position 60F on the second chromosome allowing the detection of the locus in living cells. The HS83 heat-shock promoter (hs) that drives LacI-GFP is activated at 37°C. (B-E) Live analysis of the LacI-GFP signal at 60F in the polytene chromosomes of third-instar larvae. UAS-dMi-2+ 3-3/HS83-LacI-GFP LacO; UAS-dMi-2+ 15-1/da-GAL4 GAL80ts individuals were cultured at 18°C until the middle of the third larval instar, maintained at 18°C (B and D) or shifted to 29°C (C and E) for 27 hours, and heat-shocked at 37°C for 1 hour to induce LacI-GFP expression. Both longitudinal (B and C) and transverse (D and E) views of the lacO array are shown.
dMi-2 destabilizes the association of cohesin with chromosomes
Cohesin is essential for sister chromatid cohesion in eukaryotic cells, and also plays important roles in gene expression, DNA repair, genome stability and chromatin organization [33], [34], [35], [36]. Four subunits of cohesin–Smc1, Smc3, Rad21 (Scc1/Mcd1) and stromalin (SA/Scc3/Stag2)–form a ring-like structure that encircles chromosomes from telophase until mitosis [33], [37]. Cohesin is loaded onto chromosomes by the kollerin complex containing the Nipped-B adherin protein, and is removed by the releasin complex and the separase protease to allow chromosome separation during anaphase [38].
As a first step toward investigating potential interactions between dMi-2 and cohesin, we compared the distributions of dMi-2; the SA cohesin subunit, and the Nipped-B adherin on salivary gland polytene chromosomes of wild-type larvae. Interestingly, the distributions of both SA and Nipped-B were nearly identical to that of dMi-2 (Figure 8B and C). All three proteins were preferentially associated with less condensed, transcriptionally active regions of polytene chromosomes, as evidenced by the co-localization of dMi-2 and elongating RNA polymerase II (Pol II Ser2) (Figure 8A and Figure S5 A–F) [28].
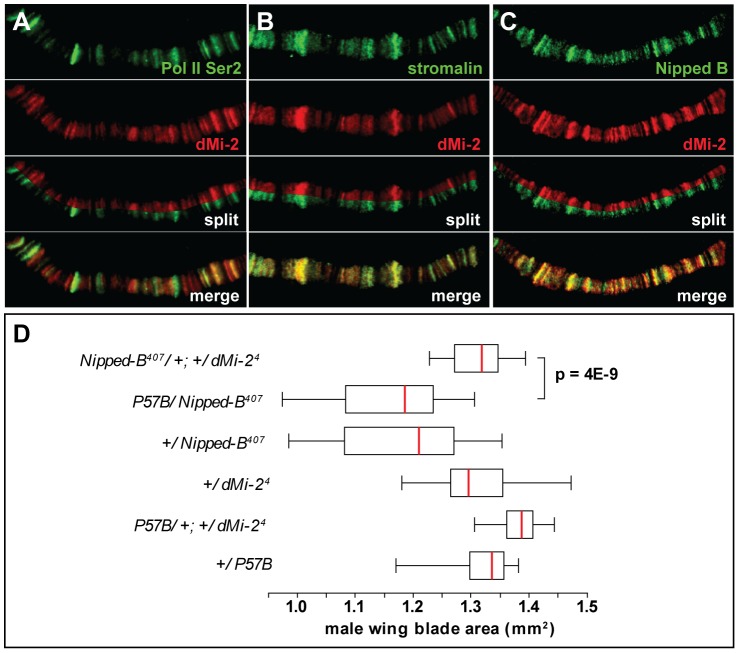
Figure 8. dMi-2 colocalizes with cohesin.
(A–C) Magnified images of salivary gland polytene chromosomes stained with antibodies against dMi-2 (red) and Pol II Ser2 (A, green), stromalin (B, green) and Nipped B (C, green). Note the extensive overlap between the chromosomal distributions of the four proteins. (D) Reducing dMi-2 gene dosage suppresses the small wing blade phenotype of individuals heterozygous for the Nipped-B407 null allele. A minimum of twenty adult male wing blade areas was measured for each of the indicated genotypes, and the distributions of blade areas are presented as box-plots. For each genotype, the chromosome to the left of the separator (/) came from the male parent, and the chromosome to the right came from the female parent. The P57B chromosome is the wild-type chromosome in which the Nipped-B407 mutation was induced by γ rays [39]. For the +/Nipped-B407, +/dMi-24 and +/P57B genotypes, the wild-type chromosomes came from an Oregon R male parent.
We used genetic experiments to test the possibility that Nipped-B and dMi-2 functionally interact, as suggested by their co-localization. Heterozygous null Nipped-B407 mutations reduce adult wing blade size relative to wild-type alleles, including the wild-type allele on the parental chromosome (P57B) on which Nipped-B407 was induced (Figure 8D) [39]. The reduced wing size mirrors overall reduced body size, similar to that seen in mouse and human Nipped-B (Nipbl, NIPBL) heterozygotes (MG, DD unpublished) [40], [41], [42]. The heterozygous dMi-24 mutation suppresses the reduced wing size caused by heterozygous Nipped-B407. This genetic interaction suggests that dMi-2 antagonizes the function of the Nipped-B cohesin loading factor.
We examined whether the over-expression of dMi-2 alters polytene chromosome structure by altering the expression of cohesin subunits. The over-expression of dMi-2 for twenty-four hours using the GAL4-GAL80ts system caused the dramatic decondensation of polytene chromosomes, but did not reduce the level of the Smc1 protein as assayed by western blotting (Figure 9A). The over-expression of dMi-2 slightly increased Smc1, SA, and Rad21 RNA in the salivary gland, as assayed by RT-PCR (Figure 9B–D). Although the cause of this increase is not known, it is clear that reduced cohesin expression is not responsible for dMi-2-dependent chromosome decondensation.
Figure 9. The over-expression of dMi-2 does not decrease cohesin levels.
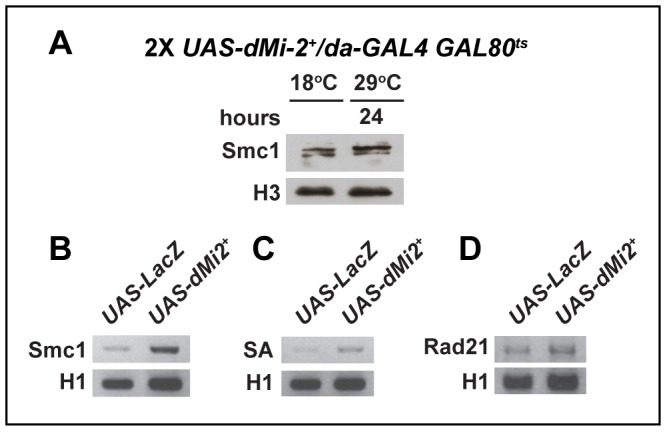
(A) Protein blot of salivary gland chromatin extracted from UAS-dMi-2+ 3-3/+; UAS-dMi-2+ 15-1/da-GAL4 GAL80ts individuals raised at 18°C until the late third-instar stage and then shifted to 29°C for 24 hours to induce UAS-Mi-2+ expression. The blot was probed with antibodies against Smc1 and histone H3 as a control. (B, C and D) RT-PCR analysis of Smc1, SA, and Rad21 RNA levels in the salivary glands of UAS-LacZ/+; da-GAL4/+ (UAS-LacZ) control larvae and UAS-dMi-2+ 3-3/+; UAS-dMi-2+ 15-1/da-GAL4 (UAS-Mi2+) larvae raised at 29°C. Histone H1 RNA levels are shown as a control.
We next investigated whether dMi-2 antagonizes binding of cohesin to polytene chromosomes. Fluorescence recovery after photobleaching (FRAP) assays were used to analyze interactions between Smc1 and polytene chromosomes in third-instar larvae that express modest levels of EGFP-tagged Smc1 in addition to normal levels of endogenous Smc1 [43]. A previous study defined the existence of three dynamic forms of the cohesin complex in salivary glands: unbound, weakly bound and stably bound to chromosomes, with short and long equilibration half-lives for the weak and stable forms, respectively [43]. Moderate over-expression of dMi-2 with two transgenes at 25°C had a dramatic impact on the association of cohesin with polytene chromosomes. In UAS-dMi-2+ 3-3/EGFP-Smc1; UAS-dMi-2+ 15-1/da-GAL4 larvae, there was a much faster equilibration between the bleached and the unbleached halves of the nucleus (Figure 10A), and the chromosomal half-life of the stable binding form decreased from 425 to 173 sec (Figure 10B). In addition, the fraction of stable EGFP-Smc1 binding decreased from 13% of the total to 7%, with a corresponding increase in the unbound fraction (Figure 10C). As expected, the nuclei with moderate dMi-2 overexpression were 1.6-fold larger in volume (9,784±627 µm3) on average than the control nuclei (5,986±337 µm3). Given the unusually long chromosomal residence time of stable cohesin, nuclear volume does not significantly affect the measured residence time through recapture of released cohesin [43]. A very large volume increase would be expected to increase recapture and cause an apparent increase in the measured chromosomal half-life instead of the observed decrease. These findings support the idea that dMi-2 promotes the dissociation of cohesin from polytene chromosomes. Reducing Nipped-B gene dosage similarly decreases the amount of stable cohesin, but in contrast to dMi-2 overexpression, does not alter its chromosomal residence time [43]. Thus it is unlikely that dMi-2 promotes cohesin dissociation by reducing Nipped-B activity.
Figure 10. dMi-2 decreases the stability and extent of cohesin chromosome binding.
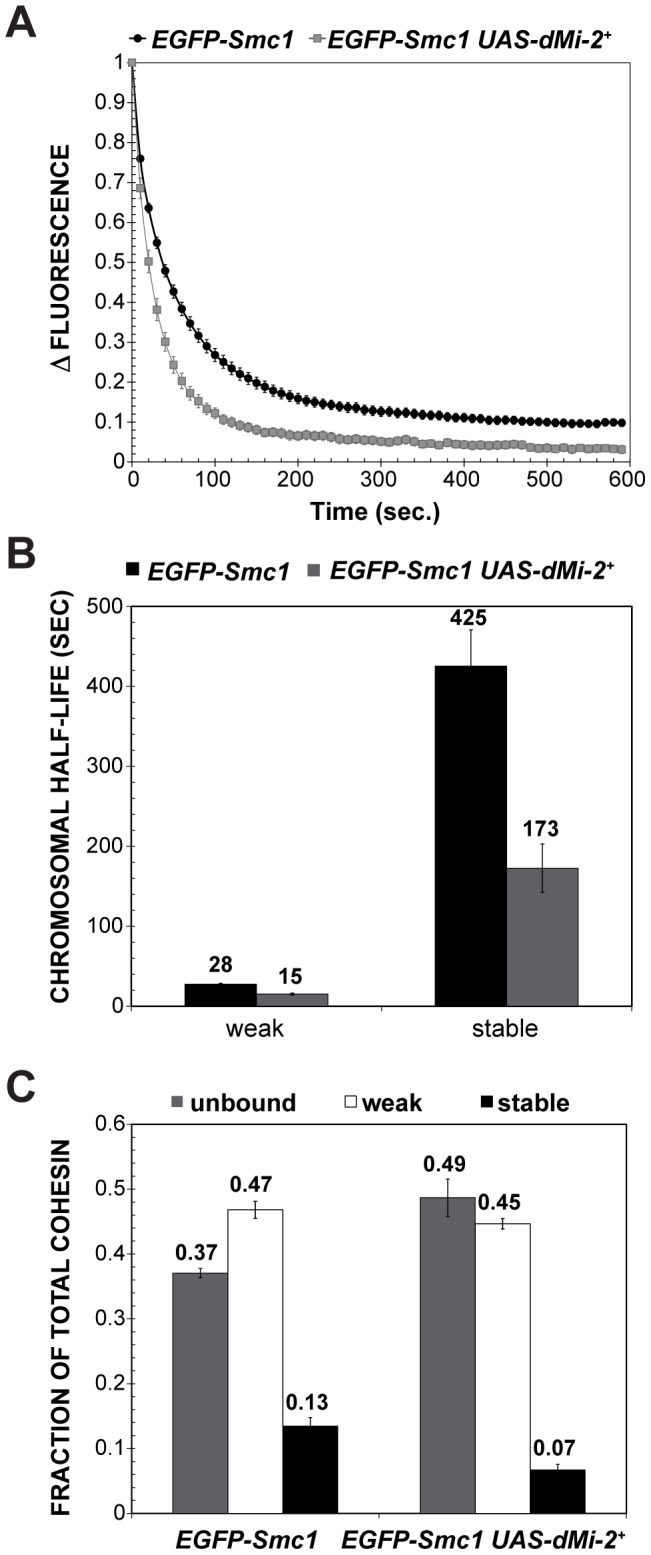
(A) FRAP recovery curves for EGFP-Smc1 in wild-type salivary gland nuclei, and salivary gland nuclei in which da-GAL4 drives the expression of two UAS-dMi-2+ transgenes (EGFP-Smc1 da-GAL4/UAS-dMi-2+ 3-3; UAS-dMi-2+ 15-1/+) at 25°C. This induced a 1.6-fold average increase in nuclear volume (see text). EGFP-Smc1 was bleached in half the nucleus, and the curves show the decreasing difference in fluorescence intensity (Δ fluorescence) between the bleached and unbleached halves over time as the unbleached EGFP-Smc1 re-equilibrates throughout the nucleus. The curves shown are the average of at least 29 nuclei, and the error bars show the standard error of the means. (B) Curve-fitting distinguishes three cohesin fractions, unbound (determined by the loss of fluorescence in the unbleached half during bleaching, not shown), a weak binding fraction, and a stable binding fraction, presumed to be bound topologically. (C) Both the half-life of the stable binding cohesin (upper right panel) and total fraction of cohesin that binds in the stable mode were decreased by dMi-2 overexpression.
dMi-2 regulates higher-order chromatin structure in diploid cells
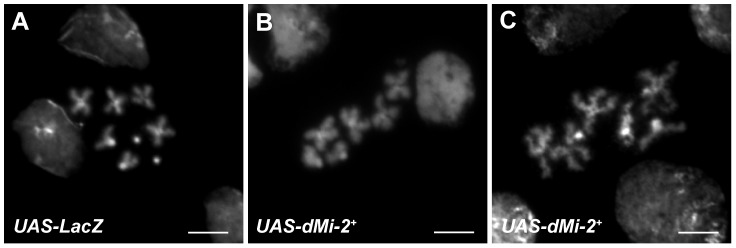
To investigate whether dMi-2 affects an aspect of chromatin organization unique to polytene chromosomes, we examined whether its increased expression alters the structure of mitotic chromosomes in wing imaginal discs, a diploid tissue that is particularly well suited for cytological studies. The elevated expression of dMi-2 in UAS-dMi-2+ 3-3/+; UAS-dMi-2+ 15-1/da-GAL4 larvae caused mitotic chromosomes to appear much less compact than normal (data not shown) without altering the mitotic index (1.4 vs. 1.3 in control larvae). To characterize this phenotype in more detail, we examined the metaphase chromosomes of imaginal discs treated with colchicine to induce mitotic arrest prior to fixation. In colchicine-treated wing discs of larvae over-expressing dMi-2, 20% of the metaphase chromosomes appeared disorganized and less compact than normal (compare figure 11B and C to A). Similar defects were not observed in colchicine-treated imaginal discs of control larvae. Roughly one-third of the abnormal metaphase chromosomes were also significantly longer than normal (third chromosome length = 3.3±0.4 µm (n = 43) vs. 5.4±1.0 µm (n = 63); compare figure 11C to A). These striking defects indicate that dMi-2 plays a general role in the regulation of higher-order chromatin structure in both diploid and polytene cells.
Figure 11. dMi-2 alters the structure of mitotic chromosomes.
DAPI staining of mitotic chromosomes from the wing imaginal discs of UAS-LacZ/+; da-GAL4/+ (UAS-LacZ) (A) and UAS-dMi-2+ 3-3/+; UAS-dMi-2+ 3-3/da-GAL4 (UAS-dMi-2+) (B and C) third-instar larvae raised at 29°C. A,B and C scale bars are 5 µm.
Discussion
dMi-2 promotes chromosome decondensation in vivo
Several lines of evidence suggest that the chromosome decondensation resulting from the increased expression of the dMi-2 protein reflects its normal function. The inducible transgene used in our studies rescued the recessive lethality of a dMi-2 null allele indicating that it encodes a fully functional dMi-2 protein. Even a modest (two to threefold) increase in dMi-2 levels triggered chromosome decondensation; this effect was relatively rapid and was dependent on dMi-2 ATPase activity. The expression of a dominant-negative dMi-2 protein had the opposite effect on chromosome structure; the loss of dMi-2 function made polytene chromosomes appear more condensed than normal and blurred the distinction between bands and interbands. Taken together, these observations provide strong evidence that dMi-2 promotes chromosome decondensation in vivo.
dMi-2 promotes local chromosome decondensation
HS puff formation exemplifies an unusually rapid and localized chromatin decondensation event. The precise mechanisms driving puff formation are not clear. HS gene activation is accompanied by nucleosome depletion, activation of PARP, extensive PARylation and the recruitment of topoisomerase, HSF, RNA polymerase II and several elongation factors as well as dMi-2 [18], [44], [45], [46]. Expression of dominant-negative dMi-2 protein reduces the transcriptional activation of HS genes but the underlying mechanisms have not been resolved [18]. Here, we have found that expression of a dominant negative dMi-2 protein leads to the formation of HS puffs with reduced size. This suggests that dMi-2 effects on chromosome structure are not always global. Instead, our results indicate that dMi-2 can be recruited to specific chromatin regions by environmental stimuli to contribute to the rapid local decondensation of chromatin. It is conceivable that the compromised decondensation of HS loci in dMi-2 mutants is one of the reasons for the observed loss in transcriptional output [18].
dMi-2 and ISWI regulate higher-order chromatin structure via distinct mechanisms
To date, only one other Drosophila chromatin-remodeling factor, ISWI, has been implicated in the regulation of higher-order chromosome structure [7], [47], [48]. ISWI promotes the association of the H1 linker histone with interphase chromosomes [6], [8]. The loss of ISWI function leads to chromosome decondensation accompanied by the loss of histone H1, and the elimination of histone H1 causes chromosome defects that are remarkably similar to those resulting from dMi-2 over-expression [6], [8]. Although this similarity suggested that dMi-2 and ISWI have antagonistic effects on higher-order chromatin structure and histone H1 assembly, we failed to detect decreases in ISWI and histone H1 expression or histone H1 assembly following dMi-2 over-expression. dMi-2 and ISWI therefore appear to regulate distinct aspects of higher-order chromatin structure.
dMi-2 regulates the dynamic association of cohesin with interphase chromosomes
Cohesin has been the topic of intensive study due to its critical role in sister chromatid cohesion during mitosis, and its roles in gene regulation and DNA repair. The complex forms a ring-like structure that encircles chromosomes beginning in telophase, and mediates sister chromatid cohesion upon DNA replication [33]. Cohesin binding is dynamic, but unusually stable compared to most DNA-binding proteins. Interphase cohesin is continuously loaded by the kollerin complex containing Nipped-B and released from chromosomes by the releasin complex containing Pds5 and Wapl [38], [43]. Our studies revealed an intriguing connection between dMi-2 and the cohesin complex, and argue that dMi-2 facilitates removal of cohesin from chromosomes during interphase. This activity is not restricted to situations in which dMi-2 is expressed at unusually high levels, since a twofold reduction in dMi-2 dosage counteracts the developmental consequences of reduced dosage of Nipped-B. Our findings add dMi-2 to the list of factors that regulate cohesin binding.
Cohesin regulates transcription by multiple mechanisms, including long-range interactions between insulators, enhancers and promoters via the formation of DNA loops, repression in collaboration with Polycomb proteins, and controlling transition of paused polymerase to elongation [34], [36], [49]. The observed suppression of a dominant Nipped-B mutant phenotype by reduced dMi-2 gene dosage suggests that regulation of cohesin chromosome binding may be one mechanism by which dMi-2 controls gene expression.
The live analysis of a LacO array tagged with GFP in living cells is consistent with a potential role for dMi-2 in chromosome cohesion. The array is organized in a compact disc due to cohesion between precisely aligned chromatids. The over-expression of dMi-2 caused the LacO array to disperse into hundreds of discrete foci, presumably due to the disruption of interactions between sister chromatids. The over-expression of dMi-2 also disrupted the organization of mitotic chromosomes along their longitudinal axes, possibly by interfering with chromosomal interactions in cis that contribute to the organization of chromosome shape [50], [51].
Our findings show that dMi-2 plays unanticipated roles in both the regulation of higher-order chromosome structure and cohesin dynamics. Is there a causal relationship between the two activities? The sudden removal of cohesin in late larval development by targeted proteolysis does not dramatically alter polytene structure [52] and thus cohesin may not be critical for maintenance of polytene structure once fully established. However, genetic studies of pds5 have revealed a role for both cohesin binding and sister chromatid cohesion in forming the normal structure of polytene chromosomes [53]. A pds5 null allele and an allele encoding an N-terminally truncated protein alter polytene chromosome structure in distinctive ways, but in both cases the size and normal banding pattern are disrupted [53]. Taken together, the above considerations prevent us from concluding that dMi-2 promotes chromosome decondensation by destabilizing cohesin binding. However, because dMi-2 over-expression causes a large reduction in both the amount of stable cohesin and its chromosomal residence time, we can conclude that cohesin binding has been reduced, and that it is also likely that cohesion is affected.
The internal diameter of cohesin is ∼35 by 50 nm; it can therefore encircle only one 30 nm or two 10 nm chromatin fibers [54]. Interactions between cohesin complexes are thought to contribute to chromatid cohesion and presumably anchor chromatin loops to form “hubs” of high transcriptional activity [55], [56], [57]. The destabilization of cohesin binding therefore may be a secondary consequence of changes in chromatin structure catalyzed by dMi-2. Further work will be necessary to test this possibility and clarify the causal relationship, if any, between changes in chromosome structure and cohesin binding catalyzed by dMi-2.
It is intriguing that dMi-2, an antagonist of cohesin binding and well-characterized transcriptional repressor, co-localizes with cohesin at sites of active transcription. Although cohesin subunits and Nipped-B were not identified as stable subunits of dMi-2 containing complexes in cultured cells [13], [58], the extensive overlap between their chromosomal distributions suggests that chromatin structure and gene activity may be dependent on a fine balance of opposing dMi-2 and cohesin activities. Cohesin selectively binds and regulates active genes that have paused RNA polymerase, and can both positively and negatively regulate these genes by multiple mechanisms, including controlling the transition of paused polymerase to elongation [49]. It is possible that dMi-2 may also influence this transition by regulating cohesin binding and the chromatin structure at the pause sites. Intriguingly, mouse Mi-2ß and the NuRD complex bind active and poised gene promoters in thymocytes, and have both negative and positive effects on expression of these genes [59]. The Mi-2/NuRD complex regulates the expression of genes involved in lymphocyte differentiation [16], [60] and is also involved in stem cell renewal and determination [61], [62]. As in Drosophila, mammalian cohesin also regulates many genes critical for growth and development [34]. Our findings raise the interesting possibility that Mi-2 may regulate cellular differentiation in vertebrates by modulating chromosome condensation and cohesin activity.
Materials and Methods
Drosophila stocks and crosses
Flies were raised on cornmeal, agar, yeast and molasses medium, supplemented with methyl paraben and propionic acid. Mutations, chromosome aberrations, strains and abbreviations used in this study are described in Table S1 or Flybase (www.flybase.org) unless otherwise indicated. The amorphic dMi-24 allele is a frameshift mutation that blocks the production of functional dMi-2 protein [17]. The hypomorphic dMi-2f08104 allele contains a PiggyBac transposon insertion in the first intron of the dMi-2 gene. The GAL4 and GAL4/GAL80ts system and sgs-GAL4, ey-GAL4 and da-GAL4 drivers were used to activate the expression of UAS transgenes (Table S1) [20], [29], [63].
Generation of Gal4-regulated transgenes
DNA fragments encoding C-terminal Flag-tagged dMi-2+ and dMi-2Δ932-1158 proteins were amplified from dMi-2 cDNA clones [21] by PCR using the primers described in Table S2, subcloned into the NotI and XbaI sites of pVL1392, and transferred into the P-element transformation vector pUAST. To generate a GAL4-regulated transgene encoding N-terminal Flag-tagged KIS-L, we first subcloned the DNA fragment generated by the hybridization of two oligonucleotides shown in Table S2 into the EcoR1 and Not1 sites of pUAST to generate the pUAST-NFLAG vector. A 2.3 kb DNA fragment containing a Not1 site immediately upstream of the KIS-L start codon was amplified from the kis2 cDNA [27] by PCR using the primers shown in Table S2. The resulting 2.3 kb Not1-EcoR1 fragment, together with the partially overlapping kis2, kis30 and kis40a cDNA clones [27], was used to generate an approximately 17 kb Not1-Kpn1 fragment containing the entire KIS-L coding region and subcloned into the Not1 and Kpn1 sites of pUAST-NFLAG. To generate a GAL4-regulated transgene encoding C-terminal GFP-tagged histone H1, a fragment was amplified from Drosophila genomic DNA using the primers shown in Table S2, subcloned into the EcoR1 and Xho1 sites of pENTR1A and transferred into the P-element transformation vector pTWG [20], [64], [65], [66]. Transformants were generated using by P-element-mediated transformation using the Df(1)w67c2 strain [67]. Homozygous viable transformants used in this study include UAS-dMi-2+ 3-3, UAS-dMi-2Δ932-1158 6-5, UAS-KIS-L+ 20-7 and UAS-H1-GFP 2-1 on the second chromosome and UAS-dMi-2+ 15-1 on the third chromosome (Table S1).
Viability studies
To determine the viability of individuals bearing dMi-2 mutations and transgenes, progeny of the crosses shown in Table 1 were scored. For each cross, ∼400 embryos were collected on grape juice/agar plates, transferred to vials and cultured at 25°C. Percent survival to each stage was calculated as the ratio between the number of individuals counted and the number of individuals expected for each genotype. The data for each of the crosses shown in Table 1 represent the average of at least two experiments.
Analysis of fixed polytene and mitotic chromosomes
To analyze polytene chromosomes, salivary glands of late third-instar larvae were dissected in 0.7% NaCl and fixed in 1.85% formaldehyde/45% acetic acid as previously described [6]. Mitotic chromosomes of wing imaginal discs were examined as described previously for Drosophila neuroblasts [68].
To analyze the effect of dMi-2Δ932-1158 expression on chromosome structure, the progeny of da-GAL4 females crossed to UAS-dMi-2 Δ932-1158 6-5 males were shifted from 25°C to 29°C 48 hours after egg deposition. To analyze the effect of dMi-2 over-expression on chromosome structure, da-GAL4 females, Sgs3-GAL4 females or ey-GAL4 females were crossed to UAS-dMi-2+ 3-3 males, UAS-dMi-2+ 15-1 males, or UAS-dMi-2+ 3-3 UAS-dMi-2+ 15-1 males; the resulting progeny were raised at 29°C. To analyze the effect of Kismet over-expression on chromosome structure, the progeny of ey-GAL4 females crossed to UAS-KIS-L+ 20-7 males were raised at 25°C. To analyze the effect of dMi-2K761R expression on puff formation, third-instar larvae from da-GAL4 females crossed to UAS-dMi-2K761R males were shifted from 25°C to 37°C to induce the heat shock. Heat shock puff formation was induced by incubating two to four third-instar larvae in a 0.2 ml PCR tube for 20 minutes at 37°C; salivary glands were immediately dissected from the larvae in 0.7% NaCl pre-warmed to 37°C. Polytene chromosomes were fixed, squashed and stained with DAPI or antibodies [6].
Fixed preparations of polytene and mitotic chromosomes were analyzed with a Zeiss Axioskop 2 plus fluorescent microscope equipped with an AxioCam HRm CCD camera and images were acquired with a HCX PL APO CS 63.0×1.40 OIL objective and Axiovision 4.6.3 software [Zeiss]. For DNA quantification, images of DAPI stained chromosomes were captured using identical exposure times. Volocity 5.4 software (Perkin Elmer) was used to calculate the sum of pixel intensities within chromosomes boundaries and to calculate the Pearson's correlation coefficient for the co-localization of dMi-2 with Stromalin and Nipped-B.
Live analysis of polytene chromosomes
Live analysis of polytene chromosomes was performed as previously described [8] in larvae bearing transgenes encoding GFP or mRFP1-tagged histone H2Av [69]. The level of histone H1 associated with chromosomes was analyzed using a transgene encoding GFP-tagged histone H1 (UAS-H1-GFP; Table S1) under the control of the ey-GAL4 driver. Volocity software (release 5.4, Perkin Elmer) was used for three-dimensional reconstructions. For volume calculation we imaged sections of polytene nuclei every 0.5 µm. The change in chromatin compaction was established by calculating the ratio of nuclear volume to DNA content. The ratio for control samples was normalized to one.
Antibodies and biochemical studies
Antibodies used in this study include rabbit polyclonal antibodies against dMi-2 (dMi-2C and dMi-2N) [70], Drosophila histone H1 (dH1 S72) [71], rabbit polyclonal Drosophila ISWI 196 [72], rabbit polyclonal Drosophila Rpd3 [70], Drosophila Smc1 [53], and histone H3 (Abcam, ab1791); a mouse monoclonal antibody against RNA PolIIoser2 (H5, Covance); and guinea pig antibodies against Stromalin and Nipped B [53], [73]. The preparation of salivary gland chromatin extracts, SDS-PAGE and protein blotting were performed as previously described [6]. Baculoviral expression of dMi-2, dMi-2Δ932-1158, CAF1p55, dp66 and dMBD2/3 in Sf9 cells, extract preparation and anti-FLAG co-immunoprecipitation was carried out as described [13].
GAL4 GAL80ts system
UAS-dMi-2+ 3-3/+; UAS-dMi-2+ 15-1/da-GAL4 Gal80ts and UAS-dMi-2 Δ932-1158 6-5/+; +/da-GAL4 Gal80ts individuals were shifted from 18°C to 29°C at the middle of the third larval instar to activate UAS-dMi-2+ and UAS-dMi-2 Δ932-1158 expression, respectively. The time course of changes in chromosome structure following the shift to 29°C was monitored via both live analysis and the analysis of polytene chromosome squashes as described above. RNA levels in the salivary gland of UAS-dMi-2+ 3-3/+; UAS-dMi-2+ 15-1/da-GAL4 Gal80ts individuals were monitored by RT-PCR using primers specific to the RNAs encoded by Smc1, SA, Rad21, the endogenous dMi-2+ gene or the UAS-dMi-2+ transgene. Primers from the dMi-2 coding region were used to measure the overall increase in total dMi-2 RNA levels (Table S2). RNA was isolated from salivary glands of ten UAS-dMi-2+ 3-3/+; UAS-dMi-2+ 15-1/da-GAL4 Gal80ts female larvae at specific times after the shift to 29°C and from ten control larvae maintained at 18°C using NucloSpin RNA XS (Clontech). cDNA was synthesized using the Superscript III First-Strand Synthesis System for RT-PCR kit (Invitrogen) and quantified with a nano-spectrophotometer using the NANODROP software. Identical amounts of cDNA were used for the PCR amplification of each sample.
LacI-GFP/LacO and confocal microscopy
The progeny of LacI-GFP LacO; da-GAL4 GAL80ts females crossed to UAS-dMi-2+ 3-3; UAS-dMi-2+ 15-1 males were allowed to develop to the third larval instar at 18°C (eight days). The expression of dMi-2 transgenes was activated by raising the temperature to 29°C for 27 hours, after which larvae were heat-shocked for 1–2 hours at 37°C to express LacI-GFP. After one hour of recovery at 29°C, salivary glands were dissected and polytene chromosomes were subjected to live analysis as described above. 0.2 µm sections of nuclei were imaged by confocal microscopy.
Effects of Nipped-B and dMi-2 mutations on wing size
Wings of adult progeny of crosses reared under uncrowded conditions at 25°C were mounted on microscope slides in Permount (Fisher Scientific) under coverslips. Digital images of the mounted wings were obtained using a 4X objective on a Nikon Microphot microscope (calibrated using a slide micrometer) and the blade areas measured using NIH ImageJ software (rsbweb.nih.gov/ij/). Statistical calculations and boxplots were generated using SSP software (economics files.pomona.edu/StatSite/ssp.html).
Fluorescence recovery after photobleaching (FRAP)
The EGFP-Smc1 fusion protein and its functional characteristics were described previously [43]. Fluorescence recovery after photobleaching (FRAP) and data analysis was performed as previously described using salivary glands from late third-instar UAS-dMi-2+ 3-3/Chip-EGFP-Smc1; UAS-dMi-2+15-1/da-GAL4 larvae raised at 25°C [43].
Supporting Information
dMi-2Δ932-1158 interacts with dNuRD subunits in vitro . Subunits of the dNuRD complex were expressed in Sf9 cells either alone (left panels), with flag-tagged dMi-2+ (middle panels) or with flag-tagged dMi-2Δ932-1158 (right panels) using recombinant baculoviruses. Whole cell extracts were immunoprecipitated with flag-beads. Immunoprecipitates were analysed by protein blotting using the antibodies indicated on the left. IN = input (extract); IP = immunoprecipitate.
(TIF)
The over-expression of dMi-2+ alters chromosome structure. (A–B) Salivary gland polytene chromosome squashes of UAS-LacZ/+; Sgs-GAL4/+ (UAS-LacZ) control larvae (A) and UAS-dMi-2+ 3-3/+; UAS-dMi-2+15-1/Sgs-GAL4 (UAS-dMi-2+) larvae (B) stained with DAPI. The over-expression of dMi-2 in late third-instar larvae under the control of the Sgs3-GAL4 driver increases the size of polytene chromosomes and disrupts their banding pattern (compare B to A). A and B scale bars are 10 µm.
(TIF)
The over-expression of dMi-2+ rapidly induces changes in chromosome structure. (A–C) DAPI stained salivary gland polytene chromosome squashes of UAS-dMi-2+ 3-3/+; UAS-dMi-2+ 15-1/da-GAL4 GAL80ts (UAS-dMi-2+) two (A) and ten (B) hours following the shift from 18 to 29°C to induce UAS-dMi-2+ expression. (C) UAS-LacZ/+; da-GAL4 GAL80ts/+ (UAS-LacZ) used as a control. Chromosome decondensation and disruption of the banding pattern was evident within 10 hours. A, B and C scale bars are 10 µm.
(TIF)
Effect of dMi-2 on histone H1 and ISWI expression in the salivary glands of third-instar larvae. (A) A Western blot of salivary gland proteins extracted from the salivary glands of larvae over-expressing wild-type dMi-2 for 24 hours was probed with antibodies against ISWI and histone H3 as a control. (B) A Western blot of proteins extracted from the salivary glands of larvae expressing dominant-negative dMi-2 for 24 hours was probed with antibodies against histone H1 and histone H3 as a control. The ratio of the histone H1 and H3 signals are indicated. Note that the over-expression of dMi-2 does not alter ISWI levels. The expression of dominant-negative dMi-2 leads to a slight decrease in histone H1 levels.
(TIF)
Cohesin colocalizes with dMi-2 and RNA Pol II. (A) Upper panel, magnified image of a portion of a salivary gland polytene chromosome stained with an antibody against dMi-2 (red) and DAPI (blue). Lower panel, linear plot profile showing that dMi-2 is associated primarily with less condensed regions.. The dashed lines indicate the corresponding location of dMi-2 bands in the plot. (B–D) Merged images of wild-type polytene chromosomes showing the colocalization of dMi-2 (red) with Pol II Ser2 (B, green), with stromalin (C, green) and with Nipped B (D, green). (E–G) Pairwise scatter plot of the intensities of overlapping pixels of dMi-2 and Pol II Ser2 (E), stromalin (F) and Nipped B (G) staining in the images shown in panels B, C and D, respectively. dMi-2 is represented by red dots while Pol II Ser2, Nipped B and stromalin are shown as green dots. The color of the dots indicates the level of colocalization, with yellow indicating perfect overlap. B, C and D scale bar is 10 µm.
(TIF)
The full genotypes of the stocks used in this study, their corresponding abbreviations and their sources are indicated.
(DOC)
List of forward and reverse oligonucleotide primers used for the construction of vectors and transgenes and the quantification of RNA levels by RT-PCR. The templates used, primer names, and corresponding sequences are shown.
(DOC)
Acknowledgments
We thank the Bloomington Drosophila Stock Center, Juerg Müller, and Giovanni Bosco for providing stocks used in this work. The H1 S72 antibody was kindly provided by Jim Kadonaga. We also thank the members of our laboratories for numerous helpful discussions during the course of this work and Ben Abrams and the UCSC Life Sciences Microscopy Center for technical support.
Funding Statement
This work was supported by National Institutes of Health grant GM49883 to JWT, and grants GM055683 and HD052860 to DD, DFG grant TRR81 “Chromatin changes in differentiation and malignancies” to AB, and California Institute for Regenerative Medicine training grant TG2-01157 to KMD. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Li B, Carey M, Workman JL (2007) The role of chromatin during transcription. Cell 128: 707–719. [DOI] [PubMed] [Google Scholar]
- 2. Sanyal A, Bau D, Marti-Renom MA, Dekker J (2011) Chromatin globules: a common motif of higher order chromosome structure? Curr Opin Cell Biol 23: 325–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bassett A, Cooper S, Wu C, Travers A (2009) The folding and unfolding of eukaryotic chromatin. Curr Opin Genet Dev 19: 159–165. [DOI] [PubMed] [Google Scholar]
- 4. Wood AJ, Severson AF, Meyer BJ (2010) Condensin and cohesin complexity: the expanding repertoire of functions. Nat Rev Genet 11: 391–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kouzarides T (2007) Chromatin modifications and their function. Cell 128: 693–705. [DOI] [PubMed] [Google Scholar]
- 6. Corona DF, Siriaco G, Armstrong JA, Snarskaya N, McClymont SA, et al. (2007) ISWI regulates higher-order chromatin structure and histone H1 assembly in vivo. PLoS Biol 5: e232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Deuring R, Fanti L, Armstrong JA, Sarte M, Papoulas O, et al. (2000) The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol Cell 5: 355–365. [DOI] [PubMed] [Google Scholar]
- 8. Siriaco G, Deuring R, Chioda M, Becker PB, Tamkun JW (2009) Drosophila ISWI regulates the association of histone H1 with interphase chromosomes in vivo. Genetics 182: 661–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kunert N, Brehm A (2009) Novel Mi-2 related ATP-dependent chromatin remodelers. Epigenetics 4: 209–211. [DOI] [PubMed] [Google Scholar]
- 10. Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL (1998) Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature 395: 917–921. [DOI] [PubMed] [Google Scholar]
- 11. Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D (1998) The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95: 279–289. [DOI] [PubMed] [Google Scholar]
- 12. Bouazoune K, Brehm A (2006) ATP-dependent chromatin remodeling complexes in Drosophila . Chromosome Res 14: 433–449. [DOI] [PubMed] [Google Scholar]
- 13. Kunert N, Wagner E, Murawska M, Klinker H, Kremmer E, et al. (2009) dMec: a novel Mi-2 chromatin remodelling complex involved in transcriptional repression. EMBO J 28: 533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stielow B, Sapetschnig A, Krüger I, Kunert N, Brehm A, et al. (2008) Identification of SUMO-dependent chromatin-associated transcriptional repression components by a genome-wide RNAi screen. Mol Cell 29: 742–754. [DOI] [PubMed] [Google Scholar]
- 15. Passannante M, Marti C, Pfefferli C, Moroni P (2010) Different Mi-2 complexes for various developmental functions in Caenorhabditis elegans . PLoS ONE [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fujita N, Jaye D, Geigerman C, Akyildiz A, Mooney M, et al. (2004) MTA3 and the Mi-2/NuRD complex regulate cell fate during B lymphocyte differentiation. Cell 119: 75–86. [DOI] [PubMed] [Google Scholar]
- 17. Kehle J, Beuchle D, Treuheit S, Christen B, Kennison JA, et al. (1998) dMi-2, a hunchback-interacting protein that functions in Polycomb repression. Science 282: 1897–1900. [DOI] [PubMed] [Google Scholar]
- 18. Murawska M, Hassler M, Renkawitz-Pohl R, Ladurner A, Brehm A (2011) Stress-induced PARP activation mediates recruitment of Drosophila Mi-2 to promote heat shock gene expression. PLoS Genet 7: e1002206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Khattak S, Lee BR, Cho SH, Ahnn J, Spoerel NA (2002) Genetic characterization of Drosophila Mi-2 ATPase. Gene 293: 107–114. [DOI] [PubMed] [Google Scholar]
- 20. Brand AH, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- 21. Bouazoune K (2005) dMi-2 chromatin binding and remodeling activities are regulated by dCK2 phosphorylation. J Biol Chem 280: 41912–41920. [DOI] [PubMed] [Google Scholar]
- 22. Elfring LK, Daniel C, Papoulas O, Deuring R, Sarte M, et al. (1998) Genetic analysis of brahma: the Drosophila homolog of the yeast chromatin remodeling factor SWI2/SNF2. Genetics 148: 251–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Armstrong JA, Papoulas O, Daubresse G, Sperling AS, Lis JT, et al. (2002) The Drosophila BRM complex facilitates global transcription by RNA polymerase II. EMBO J 21: 5245–5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Duffy JB (2002) GAL4 system in Drosophila: a fly geneticist's Swiss army knife. Genesis 34: 1–15. [DOI] [PubMed] [Google Scholar]
- 25. Daubresse G, Deuring R, Moore L, Papoulas O, Zakrajsek I, et al. (1999) The Drosophila kismet gene is related to chromatin-remodeling factors and is required for both segmentation and segment identity. Development 126: 1175–1187. [DOI] [PubMed] [Google Scholar]
- 26. Kennison JA, Tamkun JW (1988) Dosage-dependent modifiers of Polycomb and Antennapedia mutations in Drosophila. Proc Natl Acad Sci USA 85: 8136–8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Therrien M, Morrison DK, Wong AM, Rubin GM (2000) A genetic screen for modifiers of a kinase suppressor of Ras-dependent rough eye phenotype in Drosophila . Genetics 156: 1231–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Srinivasan S, Armstrong JA, Deuring R, Dahlsveen IK, McNeill H, et al. (2005) The Drosophila trithorax group protein Kismet facilitates an early step in transcriptional elongation by RNA Polymerase II. Development 132: 1623–1635. [DOI] [PubMed] [Google Scholar]
- 29. McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL (2003) Spatiotemporal rescue of memory dysfunction in Drosophila . Science 302: 1765–1768. [DOI] [PubMed] [Google Scholar]
- 30. Lu X, Wontakal SN, Emelyanov AV, Morcillo P, Konev AY, et al. (2009) Linker histone H1 is essential for Drosophila development, the establishment of pericentric heterochromatin, and a normal polytene chromosome structure. Genes Dev 23: 452–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Robinett CC, Straight A, Li G, Willhelm C, Sudlow G, et al. (1996) In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J Cell Biol 135: 1685–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hartl TA, Smith HF, Bosco G (2008) Chromosome alignment and transvection are antagonized by condensin II. Science 322: 1384–1387. [DOI] [PubMed] [Google Scholar]
- 33. Nasmyth K, Haering CH (2009) Cohesin: its roles and mechanisms. Annu Rev Genet 43: 525–558. [DOI] [PubMed] [Google Scholar]
- 34. Dorsett D (2011) Cohesin: genomic insights into controlling gene transcription and development. Curr Opin Genet Dev 21: 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mannini L, Liu J, Krantz ID, Musio A (2010) Spectrum and consequences of SMC1A mutations: the unexpected involvement of a core component of cohesin in human disease. Hum Mutat 31: 5–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Seitan VC, Merkenschlager M (2012) Cohesin and chromatin organisation. Curr Opin Genet Dev 22: 93–100. [DOI] [PubMed] [Google Scholar]
- 37. Haering CH, Farcas AM, Arumugam P, Metson J, Nasmyth K (2008) The cohesin ring concatenates sister DNA molecules. Nature 454: 297–301. [DOI] [PubMed] [Google Scholar]
- 38. Nasmyth K (2011) Cohesin: a catenase with separate entry and exit gates? Nat Cell Biol 13: 1170–1177. [DOI] [PubMed] [Google Scholar]
- 39. Rollins RA, Morcillo P, Dorsett D (1999) Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics 152: 577–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Krantz ID, McCallum J, DeScipio C, Kaur M, Gillis LA, et al. (2004) Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat Genet 36: 631–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tonkin ET, Wang TJ, Lisgo S, Bamshad MJ, Strachan T (2004) NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat Genet 36: 636–641. [DOI] [PubMed] [Google Scholar]
- 42. Kawauchi S, Calof AL, Santos R, Lopez-Burks ME, Young CM, et al. (2009) Multiple organ system defects and transcriptional dysregulation in the Nipbl(+/−) mouse, a model of Cornelia de Lange Syndrome. PLoS Genet 5: e1000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gause M, Misulovin Z, Bilyeu A, Dorsett D (2010) Dosage-sensitive regulation of cohesin chromosome binding and dynamics by Nipped-B, Pds5, and Wapl. Mol Cell Biol 30: 4940–4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Petesch SJ, Lis JT (2008) Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell 134: 74–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zobeck KL, Buckley MS, Zipfel WR, Lis JT (2010) Recruitment timing and dynamics of transcription factors at the Hsp70 loci in living cells. Mol Cell 40: 965–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Petesch SJ, Lis JT (2012) Activator-induced spread of Poly(ADP-Ribose) polymerase promotes nucleosome loss at Hsp70. Mol Cell 45: 64–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Corona DF, Clapier CR, Becker PB, Tamkun JW (2002) Modulation of ISWI function by site-specific histone acetylation. EMBO Rep 3: 242–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Badenhorst P, Voas M, Rebay I, Wu C (2002) Biological functions of the ISWI chromatin remodeling complex NURF. Genes Dev 16: 3186–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fay A, Misulovin Z, Li J, Schaaf CA, Gause M, et al. (2011) Cohesin selectively binds and regulates genes with paused RNA polymerase. Curr Biol 21: 1624–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Guacci V, Koshland D, Strunnikov A (1997) A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae . Cell 91: 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lavoie BD, Hogan E, Koshland D (2002) In vivo dissection of the chromosome condensation machinery: reversibility of condensation distinguishes contributions of condensin and cohesin. J Cell Biol 156: 805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pauli A, van Bemmel JG, Oliveira RA, Itoh T, Shirahige K, et al. (2010) A direct role for cohesin in gene regulation and ecdysone response in Drosophila salivary glands. Curr Biol 20: 1787–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dorsett D, Eissenberg JC, Misulovin Z, Martens A, Redding B, et al. (2005) Effects of sister chromatid cohesion proteins on cut gene expression during wing development in Drosophila . Development 132: 4743–4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Haering CH, Lowe J, Hochwagen A, Nasmyth K (2002) Molecular architecture of SMC proteins and the yeast cohesin complex. Mol Cell 9: 773–788. [DOI] [PubMed] [Google Scholar]
- 55. Sofueva S, Hadjur S (2012) Cohesin-mediated chromatin interactions–into the third dimension of gene regulation. Briefings in functional genomics 11: 205–216. [DOI] [PubMed] [Google Scholar]
- 56. Hirano T (2006) At the heart of the chromosome: SMC proteins in action. Nat Rev Mol Cell Biol 7: 311–322. [DOI] [PubMed] [Google Scholar]
- 57. Misteli T (2007) Beyond the sequence: cellular organization of genome function. Cell 128: 787–800. [DOI] [PubMed] [Google Scholar]
- 58. Reddy BA, Bajpe PK, Bassett A, Moshkin YM, Kozhevnikova E, et al. (2010) Drosophila transcription factor Tramtrack69 binds MEP1 to recruit the chromatin remodeler NuRD. Mol Cell Biol 30: 5234–5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang J, Jackson AF, Naito T, Dose M, Seavitt J, et al. (2011) Harnessing of the nucleosome-remodeling-deacetylase complex controls lymphocyte development and prevents leukemogenesis. Nat Immunol 13: 86–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kim J, Sif S, Jones B, Jackson A, Koipally J, et al. (1999) Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity 10: 345–355. [DOI] [PubMed] [Google Scholar]
- 61. Yoshida T, Hazan I, Zhang J, Ng SY, Naito T, et al. (2008) The role of the chromatin remodeler Mi-2 in hematopoietic stem cell self-renewal and multilineage differentiation. Genes Dev 22: 1174–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Reynolds N, Latos P, Hynes-Allen A, Loos R, Leaford D, et al. (2012) NuRD suppresses pluripotency gene expression to promote transcriptional heterogeneity and lineage commitment. Cell Stem Cell 10: 583–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gerber M, Eissenberg JC, Kong S, Tenney K, Conaway JW, et al. (2004) In vivo requirement of the RNA polymerase II elongation factor elongin A for proper gene expression and development. Mol Cell Biol 24: 9911–9919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, et al. (2002) A monomeric red fluorescent protein. Proc Natl Acad Sci USA 99: 7877–7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, et al. (2002) A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol 20: 87–90. [DOI] [PubMed] [Google Scholar]
- 66. Rorth P (1998) Gal4 in the Drosophila female germline. Mech Dev 78: 113–118. [DOI] [PubMed] [Google Scholar]
- 67.Spradling AC (1986) P-element mediated transformation. In: Drosophila: a practical approach. Roberts DB, editor. Oxford: IRL Press. 175–197.
- 68. Cenci G, Siriaco G, Raffa GD, Kellum R, Gatti M (2003) The Drosophila HOAP protein is required for telomere capping. Nat Cell Biol 5: 82–84. [DOI] [PubMed] [Google Scholar]
- 69. Clarkson M, Saint R (1999) A His2AvDGFP fusion gene complements a lethal His2AvD mutant allele and provides an in vivo marker for Drosophila chromosome behavior. DNA Cell Biol 18: 457–462. [DOI] [PubMed] [Google Scholar]
- 70. Brehm A, Langst G, Kehle J, Clapier CR, Imhof A, et al. (2000) dMi-2 and ISWI chromatin remodelling factors have distinct nucleosome binding and mobilization properties. EMBO J 19: 4332–4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ner SS, Travers AA (1994) HMG-D, the Drosophila melanogaster homologue of HMG 1 protein, is associated with early embryonic chromatin in the absence of histone H1. EMBO J 13: 1817–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tsukiyama T, Daniel C, Tamkun J, Wu C (1995) ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140 kDa subunit of the nucleosome remodeling factor. Cell 83: 1021–1026. [DOI] [PubMed] [Google Scholar]
- 73. Gause M, Webber HA, Misulovin Z, Haller G, Rollins RA, et al. (2008) Functional links between Drosophila Nipped-B and cohesin in somatic and meiotic cells. Chromosoma 117: 51–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
dMi-2Δ932-1158 interacts with dNuRD subunits in vitro . Subunits of the dNuRD complex were expressed in Sf9 cells either alone (left panels), with flag-tagged dMi-2+ (middle panels) or with flag-tagged dMi-2Δ932-1158 (right panels) using recombinant baculoviruses. Whole cell extracts were immunoprecipitated with flag-beads. Immunoprecipitates were analysed by protein blotting using the antibodies indicated on the left. IN = input (extract); IP = immunoprecipitate.
(TIF)
The over-expression of dMi-2+ alters chromosome structure. (A–B) Salivary gland polytene chromosome squashes of UAS-LacZ/+; Sgs-GAL4/+ (UAS-LacZ) control larvae (A) and UAS-dMi-2+ 3-3/+; UAS-dMi-2+15-1/Sgs-GAL4 (UAS-dMi-2+) larvae (B) stained with DAPI. The over-expression of dMi-2 in late third-instar larvae under the control of the Sgs3-GAL4 driver increases the size of polytene chromosomes and disrupts their banding pattern (compare B to A). A and B scale bars are 10 µm.
(TIF)
The over-expression of dMi-2+ rapidly induces changes in chromosome structure. (A–C) DAPI stained salivary gland polytene chromosome squashes of UAS-dMi-2+ 3-3/+; UAS-dMi-2+ 15-1/da-GAL4 GAL80ts (UAS-dMi-2+) two (A) and ten (B) hours following the shift from 18 to 29°C to induce UAS-dMi-2+ expression. (C) UAS-LacZ/+; da-GAL4 GAL80ts/+ (UAS-LacZ) used as a control. Chromosome decondensation and disruption of the banding pattern was evident within 10 hours. A, B and C scale bars are 10 µm.
(TIF)
Effect of dMi-2 on histone H1 and ISWI expression in the salivary glands of third-instar larvae. (A) A Western blot of salivary gland proteins extracted from the salivary glands of larvae over-expressing wild-type dMi-2 for 24 hours was probed with antibodies against ISWI and histone H3 as a control. (B) A Western blot of proteins extracted from the salivary glands of larvae expressing dominant-negative dMi-2 for 24 hours was probed with antibodies against histone H1 and histone H3 as a control. The ratio of the histone H1 and H3 signals are indicated. Note that the over-expression of dMi-2 does not alter ISWI levels. The expression of dominant-negative dMi-2 leads to a slight decrease in histone H1 levels.
(TIF)
Cohesin colocalizes with dMi-2 and RNA Pol II. (A) Upper panel, magnified image of a portion of a salivary gland polytene chromosome stained with an antibody against dMi-2 (red) and DAPI (blue). Lower panel, linear plot profile showing that dMi-2 is associated primarily with less condensed regions.. The dashed lines indicate the corresponding location of dMi-2 bands in the plot. (B–D) Merged images of wild-type polytene chromosomes showing the colocalization of dMi-2 (red) with Pol II Ser2 (B, green), with stromalin (C, green) and with Nipped B (D, green). (E–G) Pairwise scatter plot of the intensities of overlapping pixels of dMi-2 and Pol II Ser2 (E), stromalin (F) and Nipped B (G) staining in the images shown in panels B, C and D, respectively. dMi-2 is represented by red dots while Pol II Ser2, Nipped B and stromalin are shown as green dots. The color of the dots indicates the level of colocalization, with yellow indicating perfect overlap. B, C and D scale bar is 10 µm.
(TIF)
The full genotypes of the stocks used in this study, their corresponding abbreviations and their sources are indicated.
(DOC)
List of forward and reverse oligonucleotide primers used for the construction of vectors and transgenes and the quantification of RNA levels by RT-PCR. The templates used, primer names, and corresponding sequences are shown.
(DOC)