Summary
In vivo site-directed mutagenesis by ssDNA recombineering is a facile method to change the color of fluorescent proteins without cloning. Two different starting alleles of GFP were targeted for mutagenesis: gfpmut3* residing in the E. coli genome and egfp carried by a bacterial/mammalian dual expression lentiviral plasmid vector. Fluorescent protein spectra were shifted by subtle modification of the chromophore region and residues interacting with the chromophore of the fluorescent protein. Eight different fluorescent proteins (Violeta, Azure, Aqua, Mar, Celeste, Amarillo, Mostaza and Bronze) were isolated and shown to be useful in multicolor imaging and flow cytometry of bacteria and transgenic human stem cells. To make in vivo site-directed mutagenesis more efficient, the recombineering method was optimized using the fluorescence change as a sensitive quantitative assay for recombination. A set of rules to simplify mutant isolation by recombineering is provided.
Keywords: Recombineering, fluorescent proteins, gene targeting, mutagenesis, recombination, oligonucleotides
INTRODUCTION
The ability to easily change the spectra of fluorescent proteins expands the palette for routine molecular cell biology. While in vitro mutagenesis has been used to modify the spectra of green fluorescent protein, and many spectral variants are commercially available, we have developed a simple method for producing spectrally distinct variants on demand by making modest changes to fluorescent protein (FP) genes without subcloning. Based on previous studies (1–3) and our analysis of sequences encoding green, cyan, blue and yellow FP variants (Supporting Fig. 1), amino acids at positions 64 (F|L), 65 (G|T), 66 (Y|W|H) and 203 (S|T|Y) appear to be primarily responsible for the “color” of each protein, while other residues are less likely to be important for spectral shifts. We surmised that it might be possible to produce several spectrally distinct fluorescent proteins by oligonucleotide-directed mutagenesis in vivo by recombineering.
Recombineering uses viral recombination proteins (e.g. Beta protein expressed from a defective lambda prophage (4, 5)) to anneal a mutagenic oligonucleotide (oligo) to its homologous target sequence inside the cell (6). Unlike in vitro site-directed mutagenesis, recombineering can directly modify any size DNA molecule, including chromosomes and large BACs (7–9). As oligo-mediated recombineering is most efficient in bacteria, we manipulated gfp gene variants stably integrated into the E. coli genome (10) or in lentiviral plasmid vectors (11) in E. coli.
EXPERIMENTAL PROCEDURES
Cell culture
293T cells (obtained from Dr. Prya Rai) were grown in DMEM-high glucose medium with 10 % fetal bovine serum (FBS) and antibiotics-antimycotics (50 units/ml Penicillin G, 0.25 μg/ml Amphotericin B and 10 μg/ml Streptomycin) at 37°C in 5 % atmospheric CO2. MIAMI 3515 adult human stem cells (12) (isolated from whole bone marrow from a 20 year old living male donor) were grown in DMEM-low glucose, 3% FBS, 20 mM ascorbic acid12.9 nM arachidonic acid, 1.12 μM cholesterol, 290 nM DL-alpha tocopherol-acetate, 85.9 nM myristic acid, 69.4 nM oleic acid, 76.5 nM palmitic acid, 77.1 nM palmitoleic acid, 68.9 nM stearic acid and 100 U/mL penicillin and 0.1 mg/mL streptomycin under low oxygen conditions (3% O2, 5% CO2 and 92% N2) at 37°C. Media were changed every 2–3 days and the cells were detached using trypsin upon reaching ~60% confluency and pelleted. Pelleted cells were resuspended in medium and plated in 10 ng/ml fibronectin-treated flasks at 100 cells/cm2.
Plasmids
pDual-eGFP was created by ligating a PCR product (produced using 5′ phosphorylated primers (oligos 61 and 62) to amplify the T7 promoter region from pET28a) to SmaI digested pNL-eGFP/CEF (11) and screening transformed Rosetta-gami™2(DE3) colonies for gain of green fluorescence. The inserted sequence contains the T7 promoter, the LacI binding site (Lac operator), a Shine-Dalgarno sequence, a His6 tag and a T7 tag. The resulting His6 - T7 tag -eGFP fusion protein is strongly expressed from pDual-egfp in the Rosetta-gami™2(DE3) and the construct was verified by sequencing using oligo 44 as a primer. pDual-eGFP(W66) and pDual-eGFP(Stop66) were created from pDual-eGFP by recombineering with oligos 59 and 60 respectively in strain RIK473. pDual-eGFP(H66) and pDual-eGFP(Y66) were created from pDual-eGFP using the QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies) with oligos 68–71. Mutagenesis was confirmed by allele-specific PCR (Y66 with oligos 49 and 45 or oligos 72 and 46, W66 with oligos 50 and 45, Stop66 with oligos 53 and 45, H66 with oligos 73 and 46, T203 with oligos 45 and 74, Y203 with oligos 45 and 75) and sequencing using oligos 45 and 46. pCMV-VSV-G and pCMV-dR8.2dvpr used to create lentivirus transducing particles were gifts from Dr. Prya Rai. All oligo sequences are listed in Supporting Table 2 online.
Recombineering
Recombineering was performed essentially as in (4). Oligos were designed with the desired change centrally located with ≥35 nt of targeting homology at each side. Oligos were obtained from Sigma Genosys, PAGE purified, resuspended in TE buffer (10mM Tris 1 mM EDTA pH 7.55) to 100 ng/μl and stored at −20°C until used. All oligo sequences are listed in Supporting Table 2 online. Recombineering host strains were grown overnight in LB medium at 32°C, diluted 1/100 into fresh LB and grown at 32°C to mid-log phase. Recombinase expression was induced at 42°C for 15 minutes. Cells were chilled on ice for 10 minutes to stop induction and to prepare cells for electroporation. Cells were then washed in ice-cold water and concentrated 200-fold. 40 μl of induced competent cells were mixed with 1 μl of oligo (100 ng); for plasmid recombineering, 10 ng of the plasmid containing the target gene (13) was also added. Cells were electroporated in chilled 1 mm gap electroporation cuvettes using a Bio-Rad Gene Pulser at 1.75 KV, 200 ohms and 25 microfarads. Cells were allowed to recover in 1 ml LB for 30 min at 32°C before plating dilutions on LB agar medium and incubation at 32°C. For FP engineering in pDual-egfp by recombineering, cultures were grown overnight in LB broth and then plasmid DNA was isolated and used to transform the E. coli Rosetta-gami™2 strain to permit expression of FP genes from the T7 promoter. In all cases, fluorescent recombinant colonies were scored using a plate imager (DR46B Dark Reader transilluminator, Clare Chemical Research). Occasionally, recombinants were screened by allele-specific PCR. Recombination efficiency was calculated as the recombinant titer divided by the total titer. Transformation efficiency was determined by parallel electroporation of the recombineering strain with pUC19. Mutagenesis was validated by allele-specific PCR followed by sequencing. The detailed recombineering protocol is available in the online Supporting Methods.
Confocal Microscopy
E. coli grown to OD600 = 0.4 in LB at 32°C were pelleted and resuspended in 1.05 % K2HPO4, 0. 45 % KH2PO4, 0.005 % MgSO4.7H2O, 0.1 % (NH4)2SO4, 0.05 % sodium citrate and 0.2% glucose to 10% of the initial volume. 1 μl of the concentrated cells were added to slides with mounting medium (P7481 Prolong Antifade, Molecular Probes). Slides were covered and left to sit overnight before imaging. Images were collected with a Zeiss LSM710 confocal microscope. PerkinElmer/Improvision Velocity 64-bit software was used to create a maximum projection and Adobe Photoshop CS3 was used to merge the colors. MIAMI adult human stem cells were grown on Fibronectin-coated cover slips. Image acquisition was performed with a Leica SP5 confocal microscope using a 63x oil-immersion 1.3 numerical aperture objective. Confocal acquisition parameters were determined at the beginning of the study and the same parameters (e.g., gains, slit aperture, laser intensity) were used for all the images. Confocal optical sections were 0.6 μm thick. Field selection was performed using the DAPI channel to identify MIAMI cells. Adobe Photoshop was used to merge the colors.
Fluorometry
Protein extracts were prepared from overnight E. coli cultures that were subsequently pelleted, incubated with BugBuster and Benzonase, and clarified by centrifugation as recommended by Novagen. Excitation and emission spectra weredetermined using a PTI QuantumMaster spectrofluorometer with 2 nm slits. The average integrated one second samples detected ~104 –106 emitted photons. The data were collected using Felix (PTI), and analyzed using Excel (Microsoft) and PRISM (GraphPad). Each spectrum was corrected for background, normalized to its peak value and plotted.
Lentiviral transduction
2 × 106 293T cells were plated on 10 cm2 dishes. The next day, 4 μg of pDual-eGFP (or variants), 4 μg of pHR’8Δ2 R and 0.4 μg of pCMV-VSV-G were mixed with 24 μl of Fugene 6 and 400 μl DMEM, incubated for 15–30 min, and then added to the 293T cells. Cells were incubated overnight in a BL2+ incubator. The next day, the medium was changed to MIAMI cell medium. The following day, media containing transducing particles were collected, filtered with a 0.45 μm syringe filter, and added to growing MIAMI cells. MIAMI cells were incubated with transducing particles overnight and then the media was changed to MIAMI cells media.
Flow cytometry analysis
About 107 cells were harvested, washed with PBS and resuspended in DMEMgfp (Evrogen, MC101). Just before flow analysis, cells were vortexed and filtered. Samples were analyzed with the Accuri cytometer using a blue laser and narrow bandpass filters. Data were analyzed using C-flow (Accuri).
RESULTS AND DISCUSSION
GFPmut3* variants created in the E. coli genome
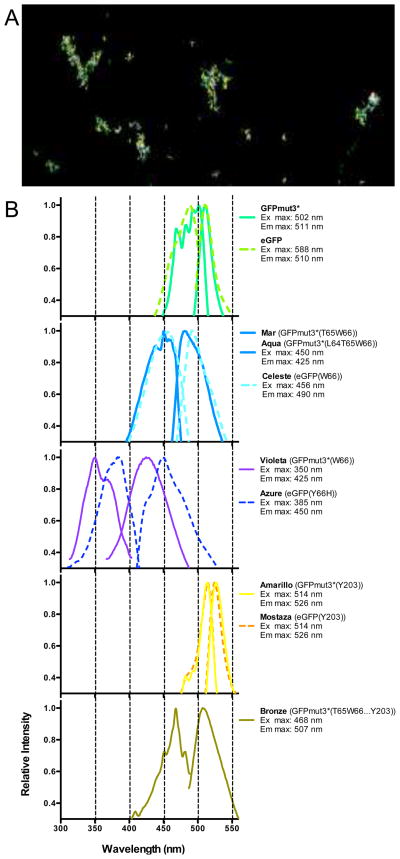
To change the color of GFPmut3*, bacteria transiently expressing Beta were electroporated with mutagenic oligos encoding the desired substitution of one or more nucleotides near the chromophore sequences of the gfpmut3* gene. Recombinants were identified by a change in colony fluorescence or by allele-specific PCR. Protein extracts prepared from isolates of each recombinant class were analyzed by fluorescence spectroscopy (Fig. 1). We corroborated that changes in the chromophore region of GFPmut3*(F64G65Y66…T203) produced spectral changes from green fluorescence. GFPmut3*(F64G65W66…T203) produced a blue shifted variant (Violeta). GFPmut3*(F64T65W66…T203) and GFPmut3*(L64T65W66…T203) produced cyan variants (Aqua and Mar, respectively). GFPmut3*(F64G65Y66…Y203) produced a yellow variant (Amarillo) while GFPmut3*(L64T65W66…Y203) produced a greener cyan variant (Bronze). We determined these variants could be distinguished by microscopy (Fig. 1a) and flow cytometry (e.g. Supporting Fig. 2). Even the modestly shifted Aqua variant is easily resolved from GFPmut3*, demonstrating the utility of oligo recombineering for producing useful variants for multi-color studies.
Figure 1. Fluorescent proteins created by in vitro (conventional) and in vivo (recombineering) site-directed mutagenesis.
Mostaza and Azure FP were created by in vitro site-directed mutagenesis while all others were created by recombineering. (A) Confocal image of Aqua, GFPmut3* and Amarillo E. coli. (B) Excitation and emission spectra of fluorescent protein extracts.
Recombineering optimization
Initially, while creating the mutants using standard recombineering methods, the frequency of mutant recombinants was low (<0.1%). Therefore, we evaluated how to make the protocol more efficient. In our studies, we learned that the efficiency of mutagenesis was heavily influenced by the outgrowth time following electroporation, the oligo sequence, the structure of the heteroduplex recombination intermediate and the transformation efficiency. The study of each parameter is shown in Figure 2 and discussed below.
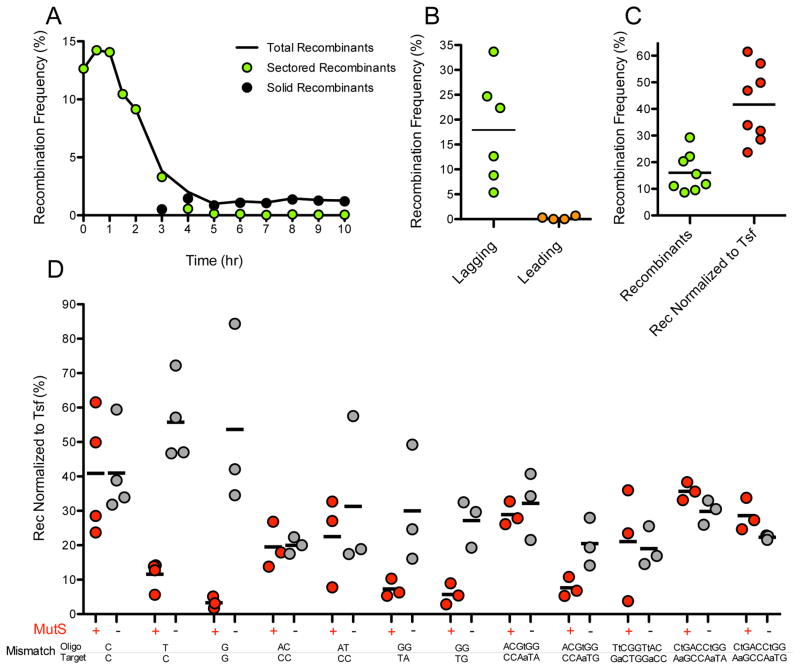
Figure 2. ssDNA Recombineering can be very efficient.
Unless specified, strain RIK423 was recombined with the lagging strand 99-base oligo 11. Recombination frequency is reported in each panel as the % of green colonies among the total number of colonies. (A) Segregation of recombinants during outgrowth. Sectored recombinants are colonies with a mixture of recombinant and non-recombinant cells. Solid recombinants are colonies composed of recombinant cells only. (B) Strand bias in targeting the lagging or leading strand template. Recombination targeting the lagging strand template (oligo 11 × RIK423) was 72 times more efficient than recombination targeting the leading strand template (oligo10 × RIK410). (C) The average recombineering frequency approaches 40% when normalized to transformation frequency. (D) MMR can be a potent inhibitor of recombination. MMR was evaluated in isogenic MMR+ and MMR− (ΔmutS::Kan) strains. A one-tailed unpaired t test with Welch’s correction was performed for each of the MutS+ and MutS− pairs. Additional details are given in the Supporting Information.
Following electroporation, cells are grown in liquid medium to recover before plating (14). We saw that different outgrowth times produced different apparent recombination frequencies (Fig. 2a). Shortly after electroporation, Beta is thought to bind an oligo and anneal it to the complementary target sequence as the target is replicated (15), creating a heteroduplex recombination intermediate with the mutation present on one of the two DNA strands. If bacteria are plated before mutant and nonmutant DNA molecules segregate, recombinant colonies are “sectored” with part of the colony comprised of mutant cells and the remainder composed of nonmutant cells (Supporting Fig. 2b and (14)). Plating during the first hour of outgrowth allows one to accurately assess recombination frequencies and also to most efficiently identify mutants. On the other hand, if bacteria are plated after segregation, pure mutant colonies appear at a frequency equal to the initial recombination frequency divided by the number of DNA chains in the original recombinant cells (16), increasing the number of colonies one must screen to recover mutants. We found that plating cells 30 min after electroporation is an optimal balance of high viability and high recovery of recombinant (mutant) colonies.
Previous reports (e.g. (17)) indicate that there is a DNA strand bias in recombineering that results from the direction of replication fork travel across the target gene. To examine strand bias in our system, oligos were designed to anneal to one target strand or to its complement. Reproducibly, oligos predicted to hybridize to the lagging strand template produced ~70 times more mutant recombinants than oligos targeted to the leading strand template (an example is shown in Fig. 2b). Therefore, we recommend using oligos complementary to the lagging strand template.
The heteroduplex recombination intermediate contains one or more mispaired bases. DNA mismatch repair (MMR) can be a potent inhibitor of recombination, depending on the identity and number of mispaired bases in the heteroduplex (17). We evaluated inhibition of FP engineering by MMR using oligos that created different heteroduplex mispairs. The mutS gene was deleted to inactivate the MMR complex (18) and recombineering experiments were carried out in isogenic MutS+ and MutS− strains (Fig. 2d and Supporting Table 3). The highest recombineering rates were achieved with single nucleotide mismatches when MMR was avoided. For example, the C•C mispair escapes MMR and is the most efficient intermediate for mutagenesis in MutS+ strains. Interestingly, mispairs of 2 to 9 nucleotides are equally good substrates if MMR is avoided (the median recombination frequencies are not different by Kruskal-Wallis ANOVA; P=0.4366). We recommend oligos designed to produce the least MMR-correctable heteroduplex. Alternatively, one may inactivate the MMR system during mutagenesis and then recover the mutated gene into an MMR+ strain to reduce the chance of picking up additional mutations from the “mutator” phenotype of MMR− strains.
The efficiency of introducing oligos into cells determines the amount of substrate available for recombineering. We saw that recombination frequencies fluctuated with the transformation efficiency of individual cell preparations. We saw a positive correlation (by a one-tailed nonparametric correlation test; P=0.0215) between the efficiency of plasmid uptake (to estimate transformation) and recombineering. We found that working with cells in early log phase improved both transformation and recombineering efficiencies.
eGFP variants created in a lentiviral plasmid
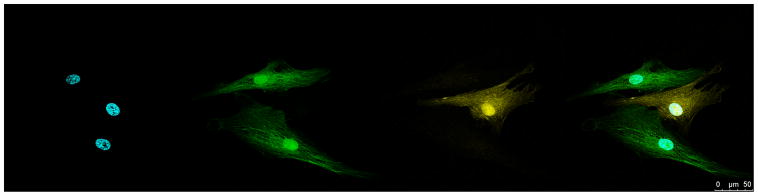
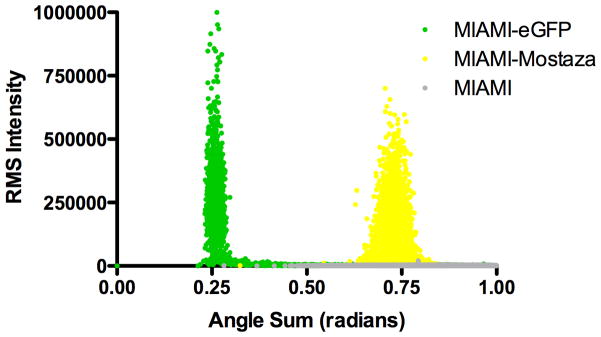
To extend the utility of FP engineering to mammalian cells, we modified egfp carried in a lentiviral plasmid propagated in E. coli. Recombineering in vivo or site-directed mutagenesis in vitro was performed as described in Supporting Methods and FP variants identified by altered bacterial colony fluorescence. We obtained eGFP(L64T65H66…T203), with a pronounced blue spectral shift (Azure), eGFP(L64T65W66…T203) with cyan spectra (Celeste) and eGFP(L64T65Y66…Y203), with yellow spectra (Mostaza) as shown in Fig. 1. We generated lentiviral stocks of the engineered FP variants and used them to transduce human marrow-isolated adult multilineage inducible (MIAMI) stem cells. As shown in Fig. 3, MIAMI cells expressing Mostaza FP are easily distinguished from cells expressing eGFP by microscopy and flow cytometry. The differences allow excellent quantitation over several orders of magnitude in mixtures (Supporting Fig. 3).
Figure 3. Discrimination of MIAMI human stem cells expressing eGFP and a Mostaza variant of eGFP.
(A) Confocal image of MIAMI cell variants. The bar in white at the lower right corner corresponds to 50 μM. (B) Flow cytometry of MIAMI cell variants. Excitation was from a 488 nm laser. Fluorescence emission spectra were collected using 510 ± 7.5 nm and 540 ± 10 nm filters. Fluorescence intensity was transformed from Cartesian to polar coordinates. Each dot represents one cell.
CONCLUSION
We have demonstrated that FP sequence changes easily introduced in vivo via recombineering produce significant spectral shifts. We showed that this method is useful for generating multi-color reporters and novel fluorescent proteins. The Bronze variant looks especially promising for imaging studies, as the excitation maximum (468 nm) overlaps a common laser wavelength. This approach is not restricted to modifying FP genes in the E. coli genome, but also works for altering FP reporter genes in plasmid and viral sequences propagated in E. coli.
Using the optimized protocol, the efficiency of FP engineering is ~40% of transformed cells (Fig. 2c and Supporting Table 3). Therefore, the optimized protocol for in vivo mutagenesis via recombineering is nearly as efficient as in vitro site-directed mutagenesis but without size limits on target DNA molecules. A detailed protocol with explanations is provided in the online Supporting Methods. The most critical parameters for successful mutagenesis via recombineering are summarized in Table 1.
Table 1.
Rules for high efficiency in vivo site-directed mutagenesis by Recombineering
| Rule | Advice |
|---|---|
| One | Use dividing cells to enrich for replication intermediates. |
| Two | Use oligos complementary to the lagging strand template. |
| Three | Use oligos that incorporate a C•C mismatch in the paired recombination intermediate. Alternatively, use a 2+ nucleotide mismatch or knock out MMR. |
| Four | Plate transformants within 30–60 minutes of outgrowth. |
Supplementary Material
Acknowledgments
We thank Dr. George McNamara of the UM Analytical Imaging Core Facility and Dr. Pedro Salas for advice and assistance. We thank Drs Donald L. Court, Jakob Reiser, Chun Chau Sze, Priya Rai and Kenneth E. Rudd for cell lines, plasmids or bacterial strains and Dr Guy Howard for other materials and advice. Support for this work was provided by NIH (IMSD and F31-GM089125-01 fellowships to MVC), the UM Interdisciplinary Stem Cell Institute (QH), from the VA GRECC (PS) and from the Leadership Alliance, Eli Lilly, and the NIH UM Developmental Center for AIDS Research (RSM).
Abbreviations
- BAC
bacterial artificial chromosome
- CMV
cauliflower mosaic virus promoter
- DMEM
Dulbecco’s modified eagle medium
- E. coli
Escherichia coli
- FBS
fetal bovine serum
- FP
fluorescent protein
- MIAMI
marrow-isolated adult multi-lineage inducible adult human stem cells
- MMR
DNA mismatch repair
- oligo
oligonucleotide
- PAGE
polyacrylamide gel electrophoresis
- ssDNA
single-stranded deoxyribonucleic acid
References
- 1.Heim R, Prasher D, Tsien R. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:12501–12504. doi: 10.1073/pnas.91.26.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orm M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Crystal Structure of the Aequorea victoria Green Fluorescent Protein. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 3.Ai HW, Olenych SG, Wong P, Davidson MW, Campbell RE. Hue-shifted monomeric variants of Clavularia cyan fluorescent protein: identification of the molecular determinants of color and applications in fluorescence imaging. BMC Biol. 2008;6:13. doi: 10.1186/1741-7007-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu D, Ellis H, Lee E, Jenkins N, Copeland N, Court D. An efficient recombination system for chromosome engineering in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomason L, Myers R, Oppenheim A, Costantino N, Sawitzke J, Datta S, Bubunenko M, Court D. Recombineering in Prokaryotes. In: Waldor M, Friedman D, Adhya S, editors. Phages: Their Role in Bacterial Pathogenesis and Biotechnology. Waldor MK, Friedman DI, Adhya. ASM Press; Washington, D.C: 2005. pp. 383–399. [Google Scholar]
- 6.Ellis HM. High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proceedings of the National Academy of Sciences. 2001;98:6742–6746. doi: 10.1073/pnas.121164898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geffin R, Myers R. WO Patent WO/2009/146,150 US2009039660 Viral recombineering and uses thereof. 2009
- 8.Britt WJ, Jarvis M, Seo JY, Drummond D, Nelson J. Rapid genetic engineering of human cytomegalovirus by using a lambda phage linear recombination system: demonstration that pp28 (UL99) is essential for production of infectious virus. J Virol. 2004;78:539–543. doi: 10.1128/JVI.78.1.539-543.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Copeland NG, Jenkins NA, Court DL. Recombineering: a powerful new tool for mouse functional genomics. Nat Rev Genet. 2001;2:769–779. doi: 10.1038/35093556. [DOI] [PubMed] [Google Scholar]
- 10.Miao H, Ratnasingam S, Pu CS, Desai MM, Sze CC. Dual fluorescence system for flow cytometric analysis of Escherichia coli transcriptional response in multi-species context. J Microbiol Methods. 2009;76:109–119. doi: 10.1016/j.mimet.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 11.Reiser J, Lai Z, Zhang XY, Brady RO. Development of multigene and regulated lentivirus vectors. J Virol. 2000;74:10589–10599. doi: 10.1128/jvi.74.22.10589-10599.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D’Ippolito G, Diabira S, Howard GA, Menei P, Roos BA, Schiller PC. Marrow-isolated adult multilineage inducible (MIAMI) cells, a unique population of postnatal young and old human cells with extensive expansion and differentiation potential. J Cell Sci. 2004;117:2971–2981. doi: 10.1242/jcs.01103. [DOI] [PubMed] [Google Scholar]
- 13.Thomason LC, Costantino N, Shaw DV, Court DL. Multicopy plasmid modification with phage lambda Red recombineering. Plasmid. 2007;58:148–158. doi: 10.1016/j.plasmid.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sawitzke JA, Costantino N, Li XT, Thomason LC, Bubunenko M, Court C, Court DL. Probing Cellular Processes with Oligo-Mediated Recombination and Using the Knowledge Gained to Optimize Recombineering. J Mol Biol. 2011;407:45–59. doi: 10.1016/j.jmb.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maresca M, Erler A, Fu J, Friedrich A, Zhang Y, Stewart AF. Single-stranded heteroduplex intermediates in lambda Red homologous recombination. BMC Mol Biol. 2010;11:54. doi: 10.1186/1471-2199-11-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sergueev K, Court D, Reaves L, Austin S. E. coli Cell-cycle Regulation by Bacteriophage Lambda. J Mol Biol. 2002;324:297–307. doi: 10.1016/s0022-2836(02)01037-9. [DOI] [PubMed] [Google Scholar]
- 17.Nina Costantino DLC. Enhanced levels of λ Red-mediated recombinants in mismatch repair mutants. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:15748–15753. doi: 10.1073/pnas.2434959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.