Abstract
The response regulator Nla28 is a key component in a cascade of transcriptional activators that modulates expression of many important developmental genes in Myxococcus xanthus. In this study, we identified and characterized Nla28S, a histidine kinase that modulates the activity of this important regulator of M. xanthus developmental genes. We show that the putative cytoplasmic domain of Nla28S has the in vitro biochemical properties of a histidine kinase protein: it hydrolyzes ATP and undergoes an ATP-dependent autophosphorylation that is acid labile and base stable. We also show that the putative cytoplasmic domain of Nla28S transfers a phosphoryl group to Nla28 in vitro, that the phosphotransfer is specific, and that a substitution in the predicted site of Nla28 phosphorylation (aspartate 53) abolishes the phosphotransfer reaction. In phenotypic studies, we found that a mutation in nla28S produces a developmental phenotype similar to, but weaker than, that produced by a mutation in nla28; both mutations primarily affect sporulation. Together, these data indicate that Nla28S is the in vivo histidine kinase partner of Nla28 and that the primary function of the Nla28S/Nla28 two-component signal transduction system is to regulate sporulation genes. The results of genetic studies suggest that phosphorylation of Nla28S is important for the in vivo sporulation function of the Nla28S/Nla28 two-component system. In addition, the quorum signal known as A-signal is important for full developmental expression of the nla28S-nla28 operon, suggesting that quorum signaling regulates the availability of the Nla28S/Nla28 signal transduction circuit in developing cells.
INTRODUCTION
The rod-shaped Gram-negative soil bacterium Myxococcus xanthus is an ideal model for studying multicellular behavior in bacteria. In nature, M. xanthus is a microbial predator; large swarms of M. xanthus cells obtain nutrients from prey microbes by secreting pools of digestive enzymes (42). When they are starving, M. xanthus cells stop searching for prey. Instead, they build multicellular fruiting bodies that contain dormant, stress-resistant myxospores, which are well suited for survival during long bouts of starvation.
Fruiting-body development occurs in an ordered series of steps, starting with the starvation-induced accumulation of the intracellular alarmone guanosine-5′(tri)di-phosphate-3′diphosphate [(p)ppGpp] (10, 28, 44). Accumulation of (p)ppGpp informs individual cells that they are starving and that it is time to start the transition from feeding and growth to development. After (p)ppGpp accumulates, cells wait several hours before they begin to aggregate. During this period, referred to as preaggregation, cells ascertain whether the density of the starving cell population is sufficient to construct fruiting bodies. A diffusible quorum signal known as A-signal serves as the cell density indicator (21, 22, 33). If the level of this quorum signal is high enough, M. xanthus cells initiate the morphological changes needed to build fruiting bodies: cells move into aggregation centers, they form dome-shaped fruiting bodies, and they differentiate into myxospores. The execution of these morphological changes is controlled by progressively higher levels of a contact-stimulated, surface-associated cell-cell signaling protein known as C-signal (15, 16, 19, 26).
The behavioral and morphological events that occur during fruiting-body development are accompanied by large-scale changes in gene expression (5, 12, 17). M. xanthus developmental genes are activated sequentially, and in the case of several genes linked to sporulation, expression is spatially localized to the fruiting-body structure (13, 39). It is believed that developmental signals such as A-signal and C-signal coordinate these global changes in gene expression directly or indirectly through signal transduction circuits (18, 20). A number of transcriptional regulators that directly modulate expression of developmental genes have been identified (5, 25, 29, 30, 45). However, the signal transduction circuits that link particular developmental signals to the temporal and spatial expression of developmental genes remain largely undefined. Here, we define a signal transduction system (Nla28S/Nla28) that is expressed in response to A-signal accumulation and that regulates M. xanthus sporulation.
Nla28 was originally identified in a search for enhancer binding proteins (EBPs) that are important for fruiting-body development; a mutation in nla28 causes a slight delay in aggregation and a strong decrease in the number of viable, stress-resistant myxospores (3). Recently, it was shown that Nla28 is a key component in an EBP cascade that regulates expression of important genes during early development (5). EBPs activate transcription of genes with σ54 promoter elements. The signal-activated ATPase function of EBPs allows σ54-RNA polymerase to form an open σ54-promoter complex and initiate transcription (34, 40, 49). EBPs typically activate gene expression in response to a specific interaction with a signal transduction partner that detects a particular environmental cue (46). Indeed, Nla28 is predicted to be a response regulator (RR) in a two-component signal transduction system (TCS) (6). In TCSs that contain EBPs such as Nla28, the EBP activates gene expression in response to phosphorylation by a histidine kinase (HK) protein that detects a particular environmental signal (14). In the case of the Escherichia coli RR NtrC, which is one of the prototype EBPs, phosphorylation induces oligomerization and its ATPase activity, which is essential for transcriptional activation (1, 36, 49, 50).
The goals of this study were to identify and characterize the HK protein that modulates the activity of Nla28. Nla28S was classified as a strong candidate for the HK partner of Nla28 because DNA sequence analysis and expression studies suggest that the nla28 and nla28S genes are part of the same operon (5, 6). Our in vitro studies showed that the purified putative cytoplasmic domain of Nla28S (GST-Nla28S-cyt) has the signature biochemical properties of an HK protein, that it is capable of transferring a phosphoryl group to purified Nla28 (GST-Nla28), and that the phosphotransfer between GST-Nla28S-cyt and GST-Nla28 is specific. In addition, we found that a mutation in nla28S produced a developmental phenotype similar to, but weaker than, that of a mutation in nla28 (3); both mutations primarily affect sporulation. These results are consistent with our in vitro data indicating that Nla28S is the in vivo HK partner of Nla28, and they suggest that the primary developmental function of the Nla28S/Nla28 signal transduction circuit is to regulate sporulation.
MATERIALS AND METHODS
Strains and plasmids.
The strains and plasmids used in this study are listed in Table 1, and the primers used in this study are listed in Table 2.
Table 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Relevant feature | Source or reference |
|---|---|---|
| Strains | ||
| M. xanthus | ||
| DK1622 | Wild type | 13a |
| DK4398 | asgB | 19a |
| AG1400 | DK1622 Δnla28S | This study |
| AG1401 | Δnla28S + nla28S (pZS202) | This study |
| AG1402 | Δnla28S + nla28S H242A (pZS210) | This study |
| E. coli | ||
| TOP10 | Cloning host | Invitrogen |
| BL21(DE3) | Expression host | Lab collection |
| NiCO21(DE3) | Expression host | New England Biolabs |
| Plasmids | ||
| pCR-Blunt | Cloning vector, Kanr | Invitrogen |
| pGEX-4T3 | GST fusion expression vector, Ampr | GE Healthcare |
| pET28b | His tag fusion expression vector, Kanr | EMD Biosciences |
| pBJ114 | Plasmid used for gene replacements and deletions, Kanr galK+ | 47 |
| pZS93b | nla28S deletion cassette in pCR-Blunt | This study |
| pZS93 | nla28S deletion cassette in pBJ114 | This study |
| pZS108b | nla28S-cyt in pCR-Blunt | This study |
| pZS109b | nla28 in pCR-Blunt | This study |
| pZS108 | nla28S-cyt in pGEX-4T3 | This study |
| pZS109 | nla28 in pGEX-4T3 | This study |
| pZS134b | envZ-cyt in pCR-Blunt | This study |
| pZS135b | ompR in pCR-Blunt | This study |
| pZS134 | envZ-cyt in pET28b | This study |
| pZS135 | ompR in pET28b | This study |
| pZS187 | nla28S-cyt H242A in pGEX-4T3 | This study |
| pZS188 | nla28S-cyt D386A in pGEX-4T3 | This study |
| pZS191 | nla28- D53A in pGEX-4T3 | This study |
| pZS202 | 1,000-bp upstream region + nla28S in pCR-Blunt | This study |
| pZS210 | 1,000-bp upstream region + nla28S H242A in pCR-Blunt |
Table 2.
Primers used in this study
| Primer use | Primer name (sequencea) |
|---|---|
| In-frame deletion of nla28S | |
| nla28S upstream 700 bp | ZS92F (GGATGTCCGTGGGGAGAAACTCCGCAGTTGG) |
| ZS92R (CGACGAGGATGCGGGCTGAGCGCCGTTCCTCTATCACGGGGCC) | |
| nla28S downstream 700 bp | ZS93F (GGCCCCGTGATAGAGGAACGGCGCTCAGCCCGCATCCTCGTCGTG) |
| ZS93R (CCGCTGGCCTCCTCGAAGACGCCTCGCCGC) | |
| Cloning of nla28S, nla28, envZ, and ompR | |
| nla28S-cyt | ZS108F (GCGCCGGGATCCCGCGTCACCTCGCTGCTCAAG) |
| ZS108R (GGGCGGGAATTCTCATGTTCCGACCTTGCGCATTTC) | |
| nla28 | ZS109F (GCGCCGGGATCCAGCTCAGCCCGCATCCTC) |
| ZS109R (GGGCGGGAATTCTCACGACTCGGCCTCCGGGGCCTC) | |
| envZ-cyt | ZS134F (GGCCGCCATATGGCGGATGACCGCACGCTGCTG) |
| ZS134R (GCGCCGAAGCTTCCCTTCTTTTGTCGTGCCCTGC) | |
| ompR | ZS135F (GGCCGCCATATGCAAGAGAACTACAAGATTCTGGTG) |
| ZS135R (GCGCCGAAGCTTTGCTTTAGAGCCGTCCGGTAC) | |
| Site-directed mutagenesis | |
| nla28S-cyt H242A | ZS188F (CCTTCACTCGGCGCGTGGCGGCCGACCTCATCTCCCCGCTGG) |
| ZS188R (CCAGCGGGGAGATGAGGTCGGCCGCCACGCGCCGAGTGAAGG) | |
| nla28S-cyt D386A | ZS189F (GGCCGTGCTGGAGGTCGTGGCCAACGGCATCGGCATGGCGC) |
| ZS189R (GCGCCATGCCGATGCCGTTGGCCACGACCTCCAGCACGGCC) | |
| nla28 D53A | ZS192F (CCTTTGACCTGGTCCTCACGGCCATGGCCATGCCCGAGCCGG) |
| ZS192R (CCGGCTCGGGCATGGCCATGGCCGTGAGGACCAGGTCAAAGG) | |
| qPCR | |
| nla28S (1,030 bp–1,209 bp of nla28S) | ZS155F (CAGTTGTTGCAGGTGAGTGC) |
| ZS155R (GAAGGGCTGGAACAGTGAGG) | |
| rpoD (519 bp–687 bp of rpoD) | ZS156F (CGCGGAAGAGAAGGAAGACG) |
| ZS156R (CTTCTCACCATCCTCGATGC) |
Primer sequences are presented in the 5′ to 3′ direction. Restriction endonuclease recognition sites are presented in italics. Mutated codons are presented in bold type.
Media for growth and development.
M. xanthus strains were grown at 32°C in CTTYE broth (1% Casitone, 0.2% yeast extract, 10 mM Tris [pH 8.0], 1 mM potassium phosphate [pH 7.6], 8 mM MgSO4) or on plates containing CTTYE broth and 1.5% agar. CTTYE broth and CTTYE agar plates were supplemented with 50 μg/ml of kanamycin as needed. CTT soft agar contains 1.0% Casitone, 10.0 mM Tris-HCl (pH 8.0), 1.0 mM KH2PO4, 8.0 mM MgSO4, and 0.7% agar. Fruiting-body development was carried out at 32°C on TPM agar plates (10 mM Tris [pH 7.6], 8 mM MgSO4, 1 mM KH2PO4, 1.5% agar), on CF agar plates (10 mM Tris [pH 7.6], 0.015% Casitone, 8 mM MgSO4, 1 mM KH2PO4, 2% sodium citrate, 1% sodium pyruvate, 1.5% agar), or in 6-well microtiter plates containing MC7 buffer (10 mM morpholinepropanesulfonic acid [MOPS; pH 7.0], 1 mM CaCl2). Escherichia coli strains were grown in Luria-Bertani (LB) broth (1% tryptone, 0.5% yeast extract, 1% NaCl) or on plates containing LB broth and 1.5% agar. LB broth and LB agar plates were supplemented with 100 μg/ml of ampicillin or 50 μg/ml of kanamycin as needed. For protein expression and purification, E. coli strains were grown in 2XYT media (1.6% tryptone, 1% yeast extract, 0.5% NaCl) supplemented with 100 μg/ml of ampicillin or 50 μg/ml of kanamycin as needed.
Construction of the nla28S deletion mutant.
The in-frame deletion of nla28S was generated using an adaptation (7) of the homologous recombination and counterselection method developed by Ueki et al. (47). The entire nla28S gene, except the last 4 bp that overlap the nla28 gene, was deleted, yielding strain AG1400. The developmental expression patterns of nla28, which is located immediately downstream of nla28S, were similar in AG1400 and wild-type cells (data not shown), indicating that the in-frame deletion of nla28S does not alter nla28 transcription via a polar effect.
M. xanthus development.
M. xanthus strains were grown to a density of ∼5 × 108 cells/ml in CTTYE broth at 32°C with shaking. The cells were pelleted, the supernatant was removed, and the cells were resuspended to a density of 5 × 109 cells/ml in TPM buffer. Fifteen-microliter aliquots of the concentrated cell suspension were spotted onto TPM agar plates and CF agar plates and incubated in a humidity chamber at 32°C. For development in MC7 buffer-submerged cultures, a 200-μl aliquot of the concentrated cell suspension was added to each well of a 6-well plate and the cells were incubated in a 32°C humidity chamber. The progress of fruiting-body development was monitored visually 0, 12, 24, 48, 72, and 120 h after the aliquots were placed on TPM agar plates or CF agar plates using a Nikon Eclipse model E400 microscope. The images were captured using an Insight FireWire camera system and analyzed using SPOT, version 4.6, software (Diagnostic Instruments).
To determine the sporulation efficiency of M. xanthus strains, cells were allowed to develop for 120 h as described above, the cells were harvested, and the harvested cells were resuspended in 1 ml of TPM buffer. The resuspended cells were first dispersed by a 10-s burst of sonication using a model 100 Sonic Dismembrator (Fisher Scientific) that was set at an intensity of 1.5. Subsequently, the cells were subjected to three 10-s bursts of sonication using an intensity setting of 4, and the sonication-treated cells were incubated at 50°C for 2 h. The number of viable, heat- and sonication-resistant spores was determined by placing aliquots of the treated cells in warm CTT soft agar, pouring the CTT soft agar onto CTTYE agar plates, and counting the colonies that arose after 3 to 4 days of incubation at 32°C.
Protein expression and purification.
PCR-generated DNA corresponding to the putative cytoplasmic domain of the M. xanthus Nla28S protein and the full-length M. xanthus Nla28 protein was cloned into the vector pGEX-4T3 (GE Healthcare) to create the N-terminal glutathione S-transferase (GST) fusion proteins GST-Nla28S-cyt and GST-Nla28, respectively (Fig. 1B). GST-Nla28S-cyt and GST-Nla28 were expressed in E. coli strain BL21(DE3). GST was expressed from the pGEX-4T3 plasmid (GE Healthcare) in E. coli strain BL21(DE3). PCR-generated DNA corresponding to the cytoplasmic domain of the E. coli EnvZ protein and the full-length E. coli OmpR protein were cloned into the vector pET28b (EMD Biosciences) to create N-terminal and C-terminal 6× histidine tags (His-EnvZ-cyt and His-OmpR, respectively). His-EnvZ-cyt and His-OmpR were expressed in E. coli strain NiCo21(DE3) (New England BioLabs). Cells were grown to an optical density at 600 nm (OD600) of ∼0.6 at 37°C with shaking at 210 rpm. Protein expression was induced by the addition of 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and cell cultures were incubated for 12 h at 16°C with shaking at 210 rpm. Cells were harvested, resuspended in lysis buffer (100 mM sodium phosphate [pH 8.0], 300 mM NaCl, 10% glycerol, 1 mg/ml of lysozyme, 5 U/ml of DNase I, 1 μg/ml of pepstatin, 1 μg/ml of leupeptin), and lysed by three 30-s sonication bursts using a model 100 Sonic Dismembrator (Fisher Scientific) set at an intensity of 4. GST and the GST fusion proteins were purified using 5-ml GSTrapFF columns (GE Healthcare) on an AKTA purifier UPC 10 fast-performance liquid chromatography (FPLC) system (GE). GST fusion proteins were eluted from the GSTrapFF columns using GST elution buffer (100 mM Tris [pH 7.4], 300 mM NaCl, 1 mM reduced glutathione). The His-tagged proteins were purified using 5-ml HisPur Cobalt columns (Thermo Scientific) on an AKTA purifier UPC 10 FPLC system (GE Healthcare). The His-tagged proteins were eluted from the HisPur Cobalt columns using a step elution method with elution buffer (100 mM Tris [pH 7.4], 300 mM NaCl) containing 20 mM, 100 mM, 250 mM, and 500 mM imidazole. All purified proteins were concentrated using Amicon Ultra centrifugal filter units (Millipore). Protein expression and purification were monitored visually using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The concentration of purified protein was determined using Bradford assays. Circular-dichroism (CD) spectroscopy was used to monitor the folding of the purified proteins. CD spectra were collected on a model 202 spectropolarimeter (Aviv Biomedical). CD data were recorded from 195 nm to 260 nm at 10°C in a 1-mm-path-length cell. The spectral bandwidth was 1.0 nm, the step size was 1 nm, and the averaging time was 10 s. Each spectrum was recorded in triplicate.
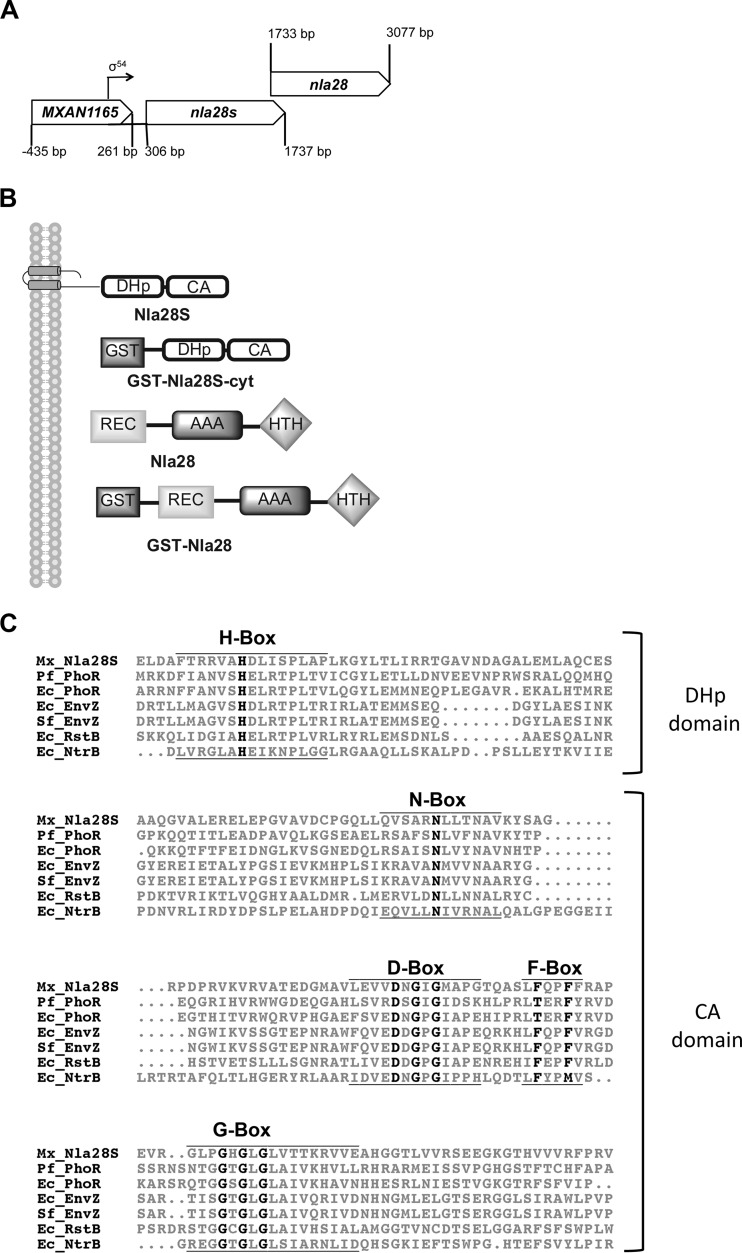
Fig 1.
Organization of the nla28S-nla28 operon, domain organization of relevant proteins, and an Nla28S sequence alignment. (A) Organization of the nla28S-nla28 operon. (B) Domain organization of Nla28S (476 amino acids), GST-Nla28S-cyt (an N-terminal GST fusion to the C-terminal 270 amino acids of Nla28S), Nla28 (447 amino acids), and GST-Nla28 (an N-terminal GST fusion to full-length Nla28). (C) Alignment of the Nla28S amino acid sequence with those of other well-characterized HKs indicates that Nla28S has all of the conserved HK sequence motifs. The alignment was generated using ClustalW2. The H-, N-, D-, F-, and G-boxes are shown, and the conserved residues are in black and bold type. HKs from Escherichia coli (Ec), Pseudomonas fluorescens (Pf), Shigella flexneri (Sf), and Myxococcus xanthus (Mx) are shown. GST, glutathione S-transferase; DHp, dimerization and histidine phosphorylation domain; CA, catalytic and ATPase domain; REC, receiver domain; AAA, ATPase domain; HTH, helix-turn-helix domain.
ATP hydrolysis assay.
The ATP-hydrolyzing activity of purified GST-Nla28S-cyt was investigated using a standard colorimetric assay that couples the hydrolysis of ATP with the oxidation of NADH (23). In this assay, the change in absorbance at 340 nm caused by the oxidation of NADH to NAD is used to monitor the amount of ATP hydrolyzed to ADP. Briefly, 5 μM purified GST-Nla28S-cyt in ATPase buffer (50 mM Tris [pH 7.4], 5 mM MgCl2, 75 mM KCl) was incubated with 1 U of l-lactate dehydrogenase (Roche), 1 U of pyruvate kinase (Roche), 0.5 mM phosphoenolpyruvate, 0.3 mM NADH, and different concentrations of ATP (0.1 mM, 0.3 mM, or 2 mM) at room temperature. Absorbance readings at 340 nm were taken using a Spectronic GENESYS 6 UV-visible spectrophotometer (Thermo Scientific) every 30 s for a total of 5 min to measure the oxidation of NADH to NAD, which is proportional to the conversion of ATP to ADP. His-EnvZ-cyt was used as a positive control and purified GST was used as a negative control for the ATPase assay.
Autophosphorylation assay.
A 5 μM aliquot of purified GST-Nla28S-cyt was incubated with 500 μM ATP and 30 μCi of [γ-32P]ATP in kinase buffer (5 mM MgCl2, 2 mM dithiothreitol [DTT], 100 mM Tris [pH 7.4]) at room temperature. At 0, 0.5, 1, 5, 10, 30, and 60 min after the reaction was started, 10-μl aliquots of the reaction mixture were removed and the reaction was stopped by the addition of 6× SDS-PAGE loading buffer (375 mM Tris-HCl [pH 6.8], 9% SDS, 50% glycerol, 9% β-mercaptoethanol, 0.03% bromophenol blue). Excess [γ-32P]ATP was removed from the samples using Zeba Micro Spin desalting columns (Pierce Protein Research Products, Thermo Scientific). His-EnvZ-cyt was used as a positive control and purified GST was used as a negative control for the autophosphorylation assay. The samples were separated using SDS-PAGE and visualized using a Typhoon 9410 variable-mode imager (GE Healthcare).
To determine the acid and base stability of phosphorylated GST-Nla28S-cyt, a 60-min autophosphorylation reaction was performed as described above. The reaction was stopped by the addition of 6× SDS-PAGE loading buffer, and then the sample was treated with 0.1 M HCl or 1 M NaOH for 20 min. The samples were separated using SDS-PAGE and visualized using a Typhoon 9410 variable-mode imager (GE Healthcare).
Phosphotransfer assay.
Purified GST-Nla28S-cyt was allowed to autophosphorylate for 60 min as described above, and excess [γ-32P]ATP was removed from the reaction mixture using Zeba Micro Spin desalting columns (Pierce Protein Research Products, Thermo Scientific). Subsequently, 10 μl of kinase buffer containing 5 μM phosphorylated GST-Nla28S-cyt and 5 μM purified GST-Nla28 was incubated at room temperature. Samples were removed from the reaction mixture at 0, 0.25, 0.5, 1, 5, 10, 30, and 60 min after the reaction was started, and the reaction was stopped by the addition of 6× SDS-PAGE loading buffer. The samples were separated using SDS-PAGE, and the phosphotransfer from GST-Nla28S-cyt to GST-Nla28 was visualized using a Typhoon 9410 variable-mode imager (GE Healthcare). The phosphotransfer from His-EnvZ-cyt to its cognate response regulator His-OmpR was used as a positive control for the above-described assay. As a negative control for the phosphotransfer assay, a reaction mixture containing phosphorylated GST-Nla28S-cyt was incubated with an equimolar amount of GST. In order to investigate the specificity of phosphotransfer from Nla28S to Nla28, phosphotransfer reactions between GST-Nla28S-cyt and His-OmpR and between His-EnvZ-cyt and GST-Nla28 were carried out and analyzed as described above.
Phosphorylation of response regulators by [32P]acetyl phosphate.
[32P]acetyl phosphate was synthesized as described by Quon et al. (37) and added to kinase buffer containing 5 μM GST-Nla28, and the reaction mixture was incubated at room temperature for 30 min. The reaction was terminated by addition of 6× SDS-PAGE loading buffer. Excess [32P]acetyl phosphate was removed using Zeba Micro Spin desalting columns (Thermo Scientific). The phosphorylation of GST-Nla28 was analyzed by SDS-PAGE and phosphorimaging as described above. His-OmpR was used as a positive control and purified GST was used as a negative control for the acetyl phosphate phosphorylation assay.
Expression analysis of nla28S.
To determine the expression profile of the nla28S gene during early development, DK1622 was allowed to develop in MC7 buffer-submerged cultures as described above and samples were taken at 0, 0.5, 1, 1.5, 2, 2.5, 3, and 4 h poststarvation for RNA isolation. To examine nla28S expression in an asg mutant background, wild-type and asgB cells were allowed to develop in MC7 buffer-submerged culture as described above and samples were removed for RNA isolation at the peak expression time point (2 h poststarvation) of nla28S in wild-type cells. The procedure was repeated 3 times for each strain.
Total RNA was isolated using the RNAprotect Bacteria Reagent (Qiagen) and RNeasy Plus Minikit (Qiagen) according to the manufacturer's protocol. The iScript cDNA synthesis kit (Bio-Rad) was used to generate cDNA from the purified RNA samples. A 10-fold dilution series of the pooled cDNA from the three replicate RNA samples from wild-type or asgB cells was used for the quantitative PCR (qPCR) experiments. The qPCR experiments were also performed in triplicate. The expression of nla28S in wild-type or asgB cells was normalized to that of rpoD, which is expressed at similar levels during growth and development (see Fig. S1 in the supplemental material). Primers for qPCR were designed to produce 180-bp to 200-bp amplicons of the nla28S and rpoD genes. The qPCR mixtures contained 300 mM each primer, 10 μl of the 2× iQ SYBR green Supermix (Bio-Rad), 5 μl of diluted cDNA, and nuclease-free water to a total volume of 20 μl. qPCR was performed on the iCycler iQ system (Bio-Rad) with the following conditions: 1 cycle of 95°C for 2 min and 40 cycles of 95°C for 15 s, 55°C for 30 s, and 72°C for 30 s. Standard-curve R2 values and amplification efficiency values ranged from 0.990 to 1.0 and 90.0% to >100%, respectively. The amplification efficiency was calculated using the program LinRegPC (38).
RESULTS
Identification of a potential HK partner for Nla28.
The putative HK gene nla28S (MXAN1166) is located directly upstream of nla28 (MXAN1167) (6), and the two genes show similar expression patterns during development (5), suggesting that nla28S and nla28 are likely to be part of the same operon (Fig. 1A). Since HK and RR genes that code for TCS partners are often located in the same operon (11), Nla28S was considered a prime candidate for the in vivo HK partner of Nla28. Nla28S has similarity to type I histidine kinase proteins such as PhoR and EnvZ, which have N-terminal domains for binding extracellular signal molecules, membrane-spanning regions, and C-terminal cytoplasmic kinase domains (14). An alignment of the conserved regions in the C termini of known HK proteins and the corresponding regions in the C terminus of Nla28S is shown in Fig. 1C.
Nla28S has the in vitro properties of an HK protein.
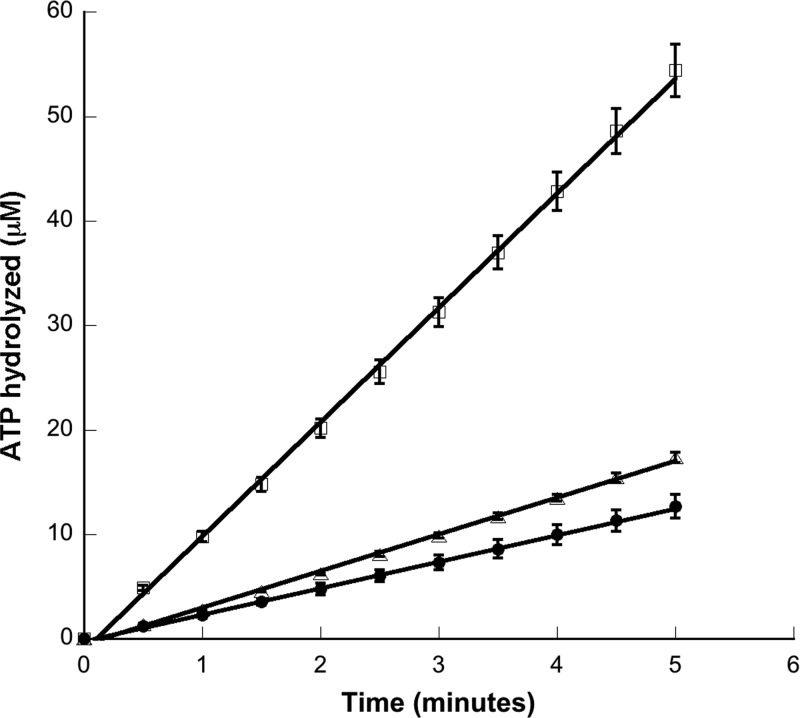
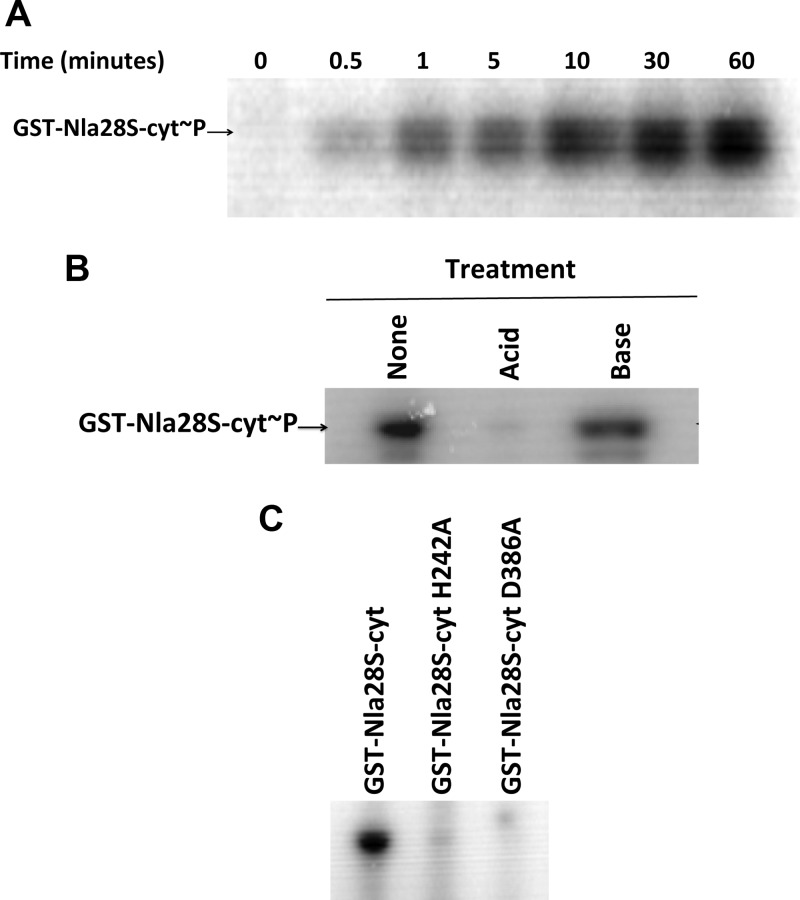
When they detect a specific signal, HKs bind and hydrolyze ATP and autophosphorylate at a conserved histidine residue (14). To confirm that Nla28S was indeed a functional HK protein, we first examined the in vitro ATPase activity of purified GST-Nla28S-cyt using a colorimetric assay that couples ATP hydrolysis to NADH oxidation (23). As shown in Fig. 2, GST-Nla28S-cyt hydrolyzed ATP and the amount of hydrolyzed ATP increased linearly with time. Next, we examined the in vitro autophosphorylation activity of purified GST-Nla28S-cyt. When it was incubated with [γ-32P]ATP, GST-Nla28S-cyt autophosphorylated within 30 s and the level of GST-Nla28S-cyt autophosphorylation increased at every time point up to 60 min (Fig. 3A). Phosphorylated histidine residues are acid labile but base stable (32). To examine whether phosphorylated GST-Nla28S-cyt has such properties, purified GST-Nla28S-cyt was incubated with [γ-32P]ATP for 60 min, and the samples were treated with acid or base for 20 min or left untreated for 20 min. Figure 3B shows that in the sample treated with base, the level of GST-Nla28S-cyt phosphorylation is similar to that in the untreated sample, but little or no phosphorylated GST-Nla28S-cyt was detected after acid treatment. In additional experiments, we examined the kinetics of Nla28S autophosphorylation by performing ATPase assays with different concentrations of ATP. The calculated Km of 0.518 mM and kcat of 0.65 min−1 are comparable to the kinetic parameters of other characterized HKs such as CheA from E. coli (0.77 mM and 3 min−1), CheA from Sinorhizobium melliloti (0.1 mM and 0.48 min−1), and CheA1 and CheA2 from Rhodobacter sphaeroides (0.25 mM and 0.36 min−1 and 0.61 mM and 0.78 min−1, respectively) (35). Thus, the results of all our in vitro biochemical assays support the idea that Nla28S is an HK protein.
Fig 2.
ATP hydrolysis activity of GST-Nla28S. A standard colorimetric assay that couples the hydrolysis of ATP to the oxidation of NADH was used to determine ATP hydrolysis activity of GST-Nla28S-cyt. The plot shows the concentration of ATP hydrolyzed by GST-Nla28S-cyt versus the time of the reaction. The initial ATP concentrations in the reactions were 0.1 mM (filled circles), 0.3 mM (open triangles), and 2 mM (open squares). Each measurement was done in triplicate. Error bars represent standard errors of the means of three replicates.
Fig 3.
In vitro autophosphorylation activity of GST-Nla28S-cyt. (A) Time course of the autophosphorylation activity of GST-Nla28S-cyt (GST-Nla28S-cyt∼P) incubated with [γ-32P]ATP at room temperature. (B) Autophosphorylation activity of GST-Nla28S-cyt incubated with [γ-32P]ATP for 60 min and treated with 0.1 N HCl or 1 N NaOH for 20 min at room temperature. (C) Autophosphorylation activity of GST-Nla28S-cyt, GST-Nla28S-cyt H242A, and GSTNla28S-cyt D386A incubated with [γ-32P]ATP for 60 min at room temperature.
His242 in the H-box of Nla28S is crucial for in vitro autophosphorylation.
The transmitter domain of HKs is divided into the dimerization and histidine phosphorylation domain (DHp) and the catalytic and ATP binding domain (CA). The conserved His residue of HKs that is autophosphorylated is located within the H-box in the DHp domain. Sequence analysis of Nla28S revealed that His242 in the H-box (Fig. 1C) is the putative site of autophosphorylation in Nla28S. To test this finding, we examined the autophosphorylation of the GST-Nla28S-cyt H242A mutant. Figure 3C shows that after 60 min of incubation with [γ-32P]ATP, the level of autophosphorylation of GST-Nla28S-cyt H242A was less than 1% that of GST-Nla28S-cyt. This result suggests that His242 is the likely site of Nla28S autophosphorylation.
The putative CA domain of Nla28S is required for in vitro autophosphorylation.
The CA domain consists of the four conserved sequence motifs (N-, D-, F-, and G-boxes) that form a pocket for the binding and hydrolysis of ATP (4). The conserved D-box Asp in the CA domain of HK proteins plays an important role in ATP binding by directly interacting with ATP via a hydrogen bond with the N6-amine of the adenine moiety (4). In Nla28S, residue 386 is the putative D-box Asp (Fig. 1C). To examine the importance of the Nla28S CA domain in autophosphorylation, we generated the D386A substitution in GST-Nla28S-cyt (GST-Nla28S-cyt D386A). After incubation with [γ-32P]ATP for 60 min, the level of phosphorylation of GST-Nla28S-cyt D386A was less than 1% that of GST-Nla28S-cyt (Fig. 3C). This finding indicates that the putative D-box Asp and, presumably, the CA domain of Nla28S are important for in vitro autophosphorylation.
Nla28 uses acetyl phosphate as an in vitro phosphodonor.
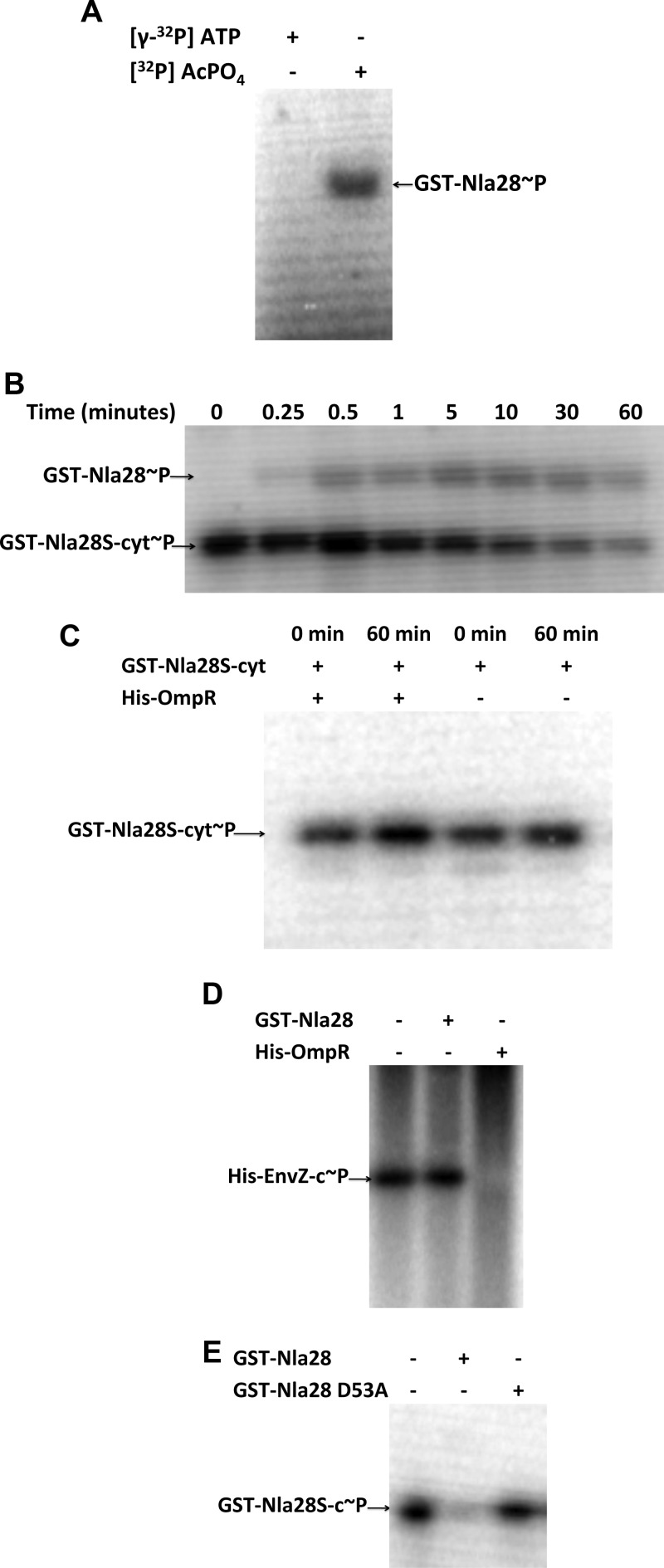
The receiver domain of RRs catalyzes the phosphotransfer from HK proteins (2). In addition, many RRs can catalyze a phosphotransfer from small molecule donors such as acetyl phosphate (27). Nla28 is known to bind to the σ54 promoters of several developmental genes and to be important for expression of these genes in developing cells (5; K. Murphy, T. Li, and A. Garza, personal communication), which is consistent with its proposed function as a transcriptional activator. To investigate whether Nla28 has the in vitro properties of an RR, purified GST-Nla28 was incubated with [32P]acetyl phosphate for 30 min. Figure 4A shows that phosphorylated GST-Nla28 was detected when incubated with [32P]acetyl phosphate but not with [γ-32P]ATP (negative control). This finding indicates that GST-Nla28 is capable of catalyzing a phosphotransfer reaction in vitro, which is a signature property of bacterial RRs.
Fig 4.
In vitro phosphotransfer assays. (A) GST-Nla28 was incubated with [γ-32P]ATP or [32P]acetyl phosphate ([32P]AcPO4) for 30 min. (B) Autophosphorylated GST-Nla28S-cyt (GST-Nla28S-cyt∼32P) was incubated with GST-Nla28 at room temperature. (C) Autophosphorylated GST-Nla28S-cyt was incubated with His-OmpR or in kinase buffer alone at room temperature for 60 min. (D) Autophosphorylated His-EnvZ-cyt was incubated in kinase buffer alone, with GST-Nla28, or with His-OmpR at room temperature for 60 min. (E) Autophosphorylated GST-Nla28S-cyt was incubated in kinase buffer alone, with GST-Nla28, or with GST-Nla28 D53A at room temperature for 60 min.
Nla28S transfers its phosphoryl group to Nla28 in vitro.
To examine whether Nla28S and Nla28 can serve as in vitro phosphotransfer partners, which would be consistent with the idea that they form an in vivo TCS, purified GST-Nla28S-cyt was incubated with [γ-32P]ATP for 60 min, the [γ-32P]ATP was removed, and then the phosphorylated GST-Nla28S-cyt was incubated with an equimolar amount of purified GST-Nla28 for various amounts of time before the reaction was terminated (Fig. 4B). An HK-to-RR phosphotransfer is detected by the appearance of a second band corresponding to the radiolabeled RR or by the depletion of radiolabel from the HK, which indicates that the HK-to-RR phosphotransfer was followed by RR dephosphorylation (24). As shown in Fig. 4B, phosphorylation of GST-Nla28 was detected after only 15 s of incubation with GST-Nla28S-cyt. The level of GST-Nla28 phosphorylation increased after an additional 15 s of incubation with GST-Nla28S-cyt and remained fairly steady with longer incubation periods up to 60 min, when the level of phosphorylated GST-Nla28 decreased. Over the same time course of incubation, the general trend for GST-Nla28S-cyt was a decrease in phosphorylation (Fig. 4B). In contrast, the level of phosphorylated GST-Nla28S-cyt remained relatively constant over time when it was incubated with His-OmpR, which is a polyhistidine-tagged version of the noncognate E. coli RR OmpR (Fig. 4C). Moreover, no band corresponding to phosphorylated His-OmpR was observed, even after 60 min of incubation with phosphorylated GST-Nla28S-cyt (Fig. 4C). In addition, after 60 min of incubation with phosphorylated His-EnvZ-cyt (Fig. 4D), which is a polyhistidine-tagged version of the noncognate E. coli HK EnvZ, no phosphorylated GST-Nla28 was detected. Furthermore, GST-Nla28 did not cause His-EnvZ-cyt radiolabel to become depleted after 60 min of incubation (Fig. 4D). In contrast, after phosphorylated His-EnvZ-cyt and His-OmpR (EnvZ and OmpR are TCS partners in E. coli) were incubated for 60 min, the radiolabel from His-EnvZ-cyt was depleted (Fig. 4D). Presumably, His-EnvZ-cyt successfully transferred a phosphoryl group to His-OmpR and His-OmpR was subsequently dephosphorylated. Taken together, these findings indicate that Nla28S and Nla28 are in vitro phosphotransfer partners and that the Nla28S-to-Nla28 phosphotransfer is specific, suggesting that Nla28S is likely to be the in vivo HK partner of Nla28.
The Asp53 residue of Nla28 is required for in vitro phosphotransfer.
The HK-to-RR phosphotransfer reaction is catalyzed by the receiver domain of the RR, which contains a conserved phosphor-accepting Asp residue. To investigate whether the conserved Asp53 residue in Nla28 is important for its phosphorylation, we generated a D53A substitution in GST-Nla28 (GST-Nla28 D53A) and performed phosphotransfer assays with purified GST-Nla28S-cyt and GST-Nla28 D53A (Fig. 4E). After 60 min of incubation with phosphorylated GST-Nla28S-cyt, no band corresponding to the phosphorylated GST-Nla28 D53A was detected. Furthermore, there was negligible depletion of radiolabel from the phosphorylated GST-Nla28S-cyt. These results indicate that Nla28 requires a conserved Asp residue for phosphotransfer from a cognate HK.
An nla28S mutation primarily affects sporulation.
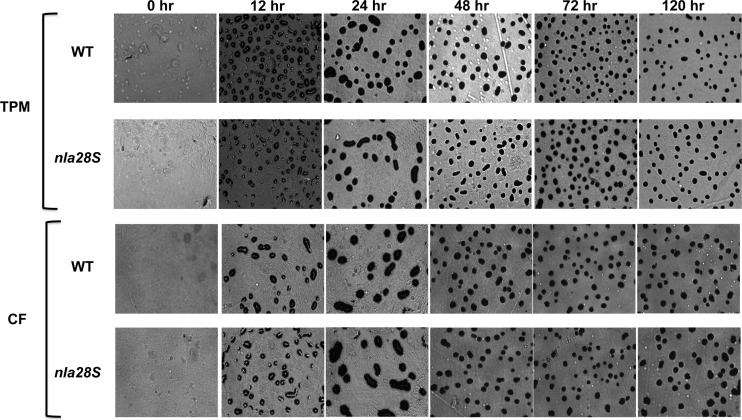
If Nla28S and Nla28 are partners in the same TCS, then a mutation in nla28S is likely to produce developmental defects that are similar to those produced by an nla28 mutation. A mutation in the nla28 gene causes a slight aggregation delay but a strong (>10-fold) decrease in sporulation efficiency relative to that of wild-type cells (3). To examine whether a mutation in nla28S produces similar developmental defects, we constructed a strain of M. xanthus that contains an in-frame deletion of nla28S (see Materials and Methods). When we placed Δnla28S and wild-type cells on TPM starvation agar plates, no differences in aggregation or fruiting-body formation were observed (Fig. 5). However, Δnla28S cells did show a slight (about 1.5-fold) decrease in sporulation efficiency relative to that of wild-type cells (Table 3). When we monitored the development of Δnla28S and wild-type cells on CF agar, which induces a more gradual starvation than TPM agar, we found that aggregation and fruiting-body formation proceeded similarly. The sporulation efficiency of Δnla28S cells was, however, reduced about 4-fold relative to that of wild-type cells (Table 3). These results indicate that the Δnla28S mutation affects sporulation but not aggregation. Thus, it seems that the primary function of Nla28S and Nla28 is to regulate sporulation, which supports our proposal that Nla28S and Nla28 are in vivo TCS partners. A potential explanation for the finding that the Δnla28S mutation produces a much weaker sporulation defect than the nla28 mutation is mentioned in the Discussion.
Fig 5.
Development phenotypes of wild-type (WT) and Δnla28S mutant cells. Wild-type and Δnla28S cells were spotted onto TPM agar plates (top two panels) or CF agar plates (bottom two panels), and the progress of fruiting-body development was monitored for 5 days using a Nikon Eclipse model E400 microscope at a total magnification of ×40. Photographs were taken at 0, 24, 48, 72, and 120 h poststarvation.
Table 3.
Sporulation efficiency of wild-type and Δnla28S mutant cells
| Medium | % sporulation efficiency (mean ± SD)a |
|||
|---|---|---|---|---|
| Wild type (DK1622) | Δnla28S (AG1400) | Δnla28S + nla28S (AG1401) | Δnla28S + nla28S H242A (AG1402) | |
| TPM | 100 ± 9.72 | 65.4 ± 7.13 | ||
| CF | 100 ± 5.15 | 22.6 ± 1.61 | 96.7 ± 6.77 | 26.2 ± 2.14 |
All spore assays were performed in triplicate. The mean sporulation efficiencies of AG1400, AG1401, and AG1402 mutant cells are shown as percentages of that of DK1622 (wild-type) cells.
To examine whether Nla28S phosphorylation is important for the in vivo sporulation function of the Nla28S/Nla28 TCS, two nla28S alleles were introduced into the native nla28S locus in the Δnla28S mutant and placed under the transcriptional control of the native nla28S-nla28 operon promoter (5). One of the nla28S alleles encodes wild-type Nla28S, and the other allele encodes Nla28S H242A, an altered version of Nla28S that is defective for in vitro autophosphorylation (Fig. 3C). The nla28S allele encoding the wild-type Nla28S protein rescued the sporulation defect of the Δnla28S mutant (96.7% ± 6.77% of wild-type sporulation efficiency), whereas the nla28S allele encoding the altered Nla28S protein failed to rescue the sporulation defect of the Δnla28S mutant (26.2% ± 2.14% of wild-type sporulation efficiency). These findings suggest that the in vivo phosphorylation of Nla28S is indeed important for the sporulation function of the Nla28S/Nla28 two-component system.
Developmental expression of nla28S is A-signal dependent.
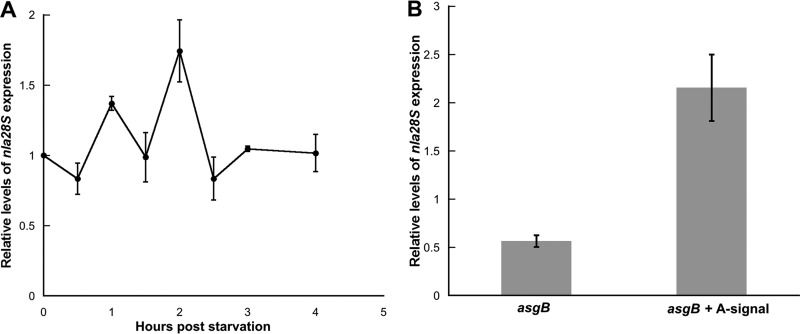
As shown in Fig. 6A, we monitored the developmental expression pattern of nla28S using qPCR. As previously reported (5), the developmental expression pattern of nla28S is similar to that of nla28: nla28S mRNA levels start to increase at 1 h poststarvation (which is early in preaggregation), they peak at 2 h poststarvation, and they return to vegetative levels between 2 and 3 h poststarvation. Since A-signal production is activated about 1 to 2 h poststarvation (13b, 20), we speculated that developmental expression of nla28S might be A-signal dependent. To test this proposal, we examined the levels of nla28S mRNA in asgB cells, which are defective for A-signal production (20), and wild-type cells at 2 h poststarvation. As shown in Fig. 6B, expression of nla28S mRNA was reduced about 2-fold in asgB cells relative to that of wild-type cells. To confirm that expression of nla28S is A-signal dependent, asgB cells were allowed to develop in MC7 buffer that was previously conditioned with exogenous A-signal by wild-type cells. When exogenous A-signal was provided to asgB cells, expression of nla28S mRNA was completely restored (Fig. 6B). These findings indicate that full expression of nla28S and, presumably, nla28 in developing cells requires A-signal; expression of the genes for the Nla28S/Nla28 TCS components is modulated by A-signal.
Fig 6.
Expression of nla28S in wild-type and asgB mutant cells. (A) qPCR was used to examine developmental expression of nla28S in wild-type (DK1622) cells. The nla28S expression levels shown are relative to the levels found in growing wild-type cells (0 h). The values are means derived from three replicates. The error bars indicate standard deviations of the means. (B) qPCR was used to examine developmental expression of nla28S in asgB mutant (DK4398) cells at the time of its peak expression (2 h poststarvation) in wild-type cells. The nla28S expression levels shown are relative to the levels found in wild-type cells at 2 h poststarvation. The values are means derived from three replicates. The error bars are standard errors of the means.
DISCUSSION
The assembly of multicellular structures such as M. xanthus fruiting bodies depends on signal transduction networks that tie extracellular and intracellular signals to stage-specific changes in gene expression. M. xanthus has a plethora of signal transduction genes that can fulfill this need to link developmental signals to changes in gene expression (6). This includes 272 TCS genes, which constitute about 3.7% of the M. xanthus genome (6). Many of these TCS genes are differentially expressed during fruiting-body development (5) (Gene Expression Omnibus [GEO] accession number GSE13523), and 35 TCS genes that are important for the developmental process have been identified (41). However, much of the work on TCS components has concentrated on RRs and their target developmental genes (3, 5, 8, 9, 48; Murphy et al., personal communication); few of the HK and RR signal transduction circuits that modulate developmental gene expression have been defined, and very little is known about the signals that activate the characterized HK and RR circuits.
In this study, we identified and characterized the HK partner of Nla28, a member of the EBP family of RRs (6). A mutation in the nla28 gene causes a slight aggregation delay but a strong (>10-fold) decrease in sporulation efficiency relative to that of wild-type cells (3), suggesting that the primary targets of Nla28-mediated transcription are sporulation genes. Indeed, most of the developmental genes that are known to be directly regulated by Nla28, including nla6 and actB, are primarily involved in sporulation (3, 5, 7; Murphy et al., personal communication). Nla28S was identified as a potential HK partner of the Nla28 RR based on data suggesting that nla28S and nla28 are part of the same developmentally regulated operon (5, 6). The in vitro studies presented here indicate that Nla28S is a functional HK protein, it is capable of transferring a phosphoryl group to Nla28, and the phosphotransfer between the two proteins is specific (Fig. 1 to 4). Thus, all the in vitro experiments that we performed in this study support the idea that Nla28S and Nla28 are partners in a TCS and, based on studies of Nla28 (3, 5; Murphy et al., personal communication), the primary output of this TCS is transcription of sporulation genes.
Since our in vitro work indicated that Nla28S and Nla28 are part of the same TCS and Nla28 appears to be a regulator of sporulation genes (3, 5; Murphy et al., personal communication), we predicted that an nla28S mutation would primarily affect sporulation. Under the stringent starvation conditions of TPM agar and the gradual starvation conditions of CF agar, the Δnla28S mutant showed no obvious aggregation defect (Fig. 5). However, the sporulation efficiency of the Δnla28S mutant was reduced about 1.5-fold and 4-fold relative to that of wild-type cells when placed on TPM and CF agar, respectively (Table 3). These results show that an Δnla28S mutation affects sporulation as predicted, which supports the idea that Nla28S and Nla28 are in vivo signal transduction partners. Why is the sporulation defect of the Δnla28S mutant much weaker than that of the nla28 mutant? One possibility is that in the Δnla28S background, Nla28 is being activated (at least to some degree) by a nonspecific small molecule phosphodonor such as acetyl phosphate (Fig. 4A) or by a noncognate HK protein. Previous studies have reported nonspecific activation of RRs in the absence of their HK partners (27, 28a, 31, 43, 51); there are precedents for nonspecific activation of Nla28. It is also possible that Nla28 has more than one HK partner to modulate its in vivo phosphorylation state and that this partner is active in the Δnla28S mutant. Thus, it seems reasonable to speculate that the developmental function of Nla28 is not completely abolished in the nla28S mutant, leading to a less severe development defect than that observed for the nla28 mutant.
What might Nla28S be sensing? Nla28 begins modulating gene expression at the onset of the preaggregation stage of fruiting-body development (5). Presumably, Nla28S detects its signal during this early stage of development and activates Nla28. At this time in development, cells must closely monitor the nutrient levels in the environment to confirm that they are still starving and that embarking on a developmental process that yields dormant spores is the best course of action. Therefore, Nla28S might sense and respond to particular nutrients in the environment. Since A-signal begins to accumulate during preaggregation (20), it is also possible that Nla28S is involved in sensing and responding to this cell density signal, which is composed of a mixture of amino acids and peptides (21, 33).
Whether or not Nla28S is involved in detecting A-signal, the data presented here indicate that A-signal is important for full expression of nla28S and, presumably, nla28 in developing cells (Fig. 6B). Interestingly, an nla28 mutant fails to fully complement the developmental defect of an A-signal-deficient strain in codevelopment assays (3), suggesting that the nla28 mutant might be defective for A-signal production. This result is consistent with recent data showing that the promoter region of the asgA gene, which is important for A-signal production, contains a tandem repeat that is a good match to the Nla28 consensus binding site (Murphy et al., personal communication). Thus, it seems that A-signal and the Nla28S/Nla28 TCS may have a reciprocal relationship: expression of the genes for the Nla28S/Nla28 TCS components is modulated by A-signal, and the Nla28S/Nla28 TCS may modulate A-signal production. The goal of future work will be to explore this relationship and to examine whether nutrients, A-signal, or some other molecule is responsible for the activation of the Nla28S/Nla28 TCS during early development in M. xanthus.
Supplementary Material
ACKNOWLEDGMENTS
Zaara Sarwar was funded in part by an International Fellowship from the American Association of University Women (AAUW). This work was supported by National Science Foundation grant IOS-0950976 to A. G. Garza.
Footnotes
Published ahead of print 29 June 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Austin S, Dixon R. 1992. The prokaryotic enhancer binding protein NTRC has an ATPase activity which is phosphorylation and DNA dependent. EMBO J. 11:2219–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bourret RB. 2010. Receiver domain structure and function in response regulator proteins. Curr. Opin. Microbiol. 13:142–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Caberoy NB, Welch RD, Jakobsen JS, Slater SC, Garza AG. 2003. Global mutational analysis of NtrC-like activators in Myxococcus xanthus: identifying activator mutants defective for motility and fruiting body development. J. Bacteriol. 185:6083–6094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dutta R, Inouye M. 2000. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem. Sci. 25:24–28 [DOI] [PubMed] [Google Scholar]
- 5. Giglio KM, Caberoy N, Suen G, Kaiser D, Garza AG. 2011. A cascade of coregulating enhancer binding proteins initiates and propagates a multicellular developmental program. Proc. Natl. Acad. Sci. U. S. A. 108:E431–E439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Goldman BS, et al. 2006. Evolution of sensory complexity recorded in a myxobacterial genome. Proc. Natl. Acad. Sci. U. S. A. 103:15200–15205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gronewold TM, Kaiser D. 2001. The act operon controls the level and time of C-signal production for Myxococcus xanthus development. Mol. Microbiol. 40:744–756 [DOI] [PubMed] [Google Scholar]
- 8. Gronewold TMA, Kaiser D. 2002. act operon control of developmental gene expression in Myxococcus xanthus. J. Bacteriol. 184:1172–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gronewold TMA, Kaiser D. 2007. Mutations of the act promoter in Myxococcus xanthus. J. Bacteriol. 189:1836–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Harris BZ, Kaiser D, Singer M. 1998. The guanosine nucleotide (p)ppGpp initiates development and A-factor production in Myxococcus xanthus. Genes Dev. 12:1022–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hoch JA. 2000. Two-component and phosphorelay signal transduction. Curr. Opin. Microbiol. 3:165–170 [DOI] [PubMed] [Google Scholar]
- 12. Jakobsen JS, et al. 2004. σ54 enhancer binding proteins and Myxococcus xanthus fruiting body development. J. Bacteriol. 186:4361–4368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Julien B, Kaiser AD, Garza A. 2000. Spatial control of cell differentiation in Myxococcus xanthus. Proc. Natl. Acad. Sci. U. S. A. 97:9098–9103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13a. Kaiser D. 1979. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc. Natl. Acad. Sci. U. S. A. 76:5952–5956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13b. Kaiser D. 2004. Signaling in myxobacteria. Annu. Rev. Microbiol. 58:75–98 [DOI] [PubMed] [Google Scholar]
- 14. Khorchid A, Ikura M. 2006. Bacterial histidine kinase as signal sensor and transducer. Int. J. Biochem. Cell Biol. 38:307–312 [DOI] [PubMed] [Google Scholar]
- 15. Kim SK, Kaiser D. 1991. C-factor has distinct aggregation and sporulation thresholds during Myxococcus development. J. Bacteriol. 173:1722–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim SK, Kaiser D. 1990. C-factor: a cell-cell signaling protein required for fruiting body morphogenesis of M. xanthus. Cell 61:19–26 [DOI] [PubMed] [Google Scholar]
- 17. Kroos L, Kuspa A, Kaiser D. 1986. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev. Biol. 117:252–266 [DOI] [PubMed] [Google Scholar]
- 18. Kroos L, Kaiser D. 1987. Expression of many developmentally regulated genes in Myxococcus depends on a sequence of cell interactions. Genes Dev. 1:840–854 [DOI] [PubMed] [Google Scholar]
- 19. Kruse T, Lobedanz S, Berthelsen NM, Søgaard-Andersen L. 2001. C-signal: a cell surface-associated morphogen that induces and co-ordinates multicellular fruiting body morphogenesis and sporulation in Myxococcus xanthus. Mol. Microbiol. 40:156–168 [DOI] [PubMed] [Google Scholar]
- 19a. Kuspa A, Kaiser D. 1989. Genes required for developmental signalling in Myxococcus xanthus: three asg loci. J. Bacteriol. 171:2762–2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kuspa A, Kroos L, Kaiser D. 1986. Intercellular signaling is required for developmental gene expression in Myxococcus xanthus. Dev. Biol. 117:267–276 [DOI] [PubMed] [Google Scholar]
- 21. Kuspa A, Plamann L, Kaiser D. 1992. Identification of heat-stable A-factor from Myxococcus xanthus. J. Bacteriol. 174:3319–3326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kuspa A, Plamann L, Kaiser D. 1992. A-signalling and the cell density requirement for Myxococcus xanthus development. J. Bacteriol. 174:7360–7369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lascu I, Pop RD, Porumb H, Presecan E, Proinov I. 1983. Pig heart nucleosidediphosphate kinase. Phosphorylation and interaction with Cibacron blue 3GA. Eur. J. Biochem. 135:497–503 [DOI] [PubMed] [Google Scholar]
- 24. Laub MT, Biondi EG, Skerker JM. 2007. Phosphotransfer profiling: systematic mapping of two-component signal transduction pathways and phosphorelays. Methods Enzymol. 423:531–548 [DOI] [PubMed] [Google Scholar]
- 25. Lee JS, Son B, Viswanathan P, Luethy PM, Kroos L. 2011. Combinatorial regulation of fmgD by MrpC2 and FruA during Myxococcus xanthus development. J. Bacteriol. 193:1681–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li S, Lee BU, Shimkets LJ. 1992. csgA expression entrains Myxococcus xanthus development. Genes Dev. 6:401–410 [DOI] [PubMed] [Google Scholar]
- 27. Lukat GS, McCleary WR, Stock AM, Stock JB. 1992. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc. Natl. Acad. Sci. U. S. A. 89:718–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Manoil C, Kaiser D. 1980. Guanosine pentaphosphate and guanosine tetraphosphate accumulation and induction of Myxococcus xanthus fruiting body development. J. Bacteriol. 141:305–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28a. McCleary WR, Stock JB. 1994. Acetyl phosphate and the activation of two-component response regulators. J. Biol. Chem. 269:31567–31572 [PubMed] [Google Scholar]
- 29. Mittal S, Kroos L. 2009. A combination of unusual transcription factors binds cooperatively to control Myxococcus xanthus developmental gene expression. Proc. Natl. Acad. Sci. U. S. A. 106:1965–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mittal S, Kroos L. 2009. Combinatorial regulation by a novel arrangement of FruA and MrpC2 transcription factors during Myxococcus xanthus development. J. Bacteriol. 191:2753–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Msadek T, Kunst F, Klier A, Rapoport G. 1991. DegS-DegU and ComP-ComA modulator-effector pairs control expression of the Bacillus subtilis pleiotropic regulatory gene degQ. J. Bacteriol. 173:2366–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Parkinson JS, Kofoid EC. 1992. Communication modules in bacterial signaling proteins. Annu. Rev. Genet. 26:71–112 [DOI] [PubMed] [Google Scholar]
- 33. Plamann L, Kuspa A, Kaiser D. 1992. Proteins that rescue A-signal-defective mutants of Myxococcus xanthus. J. Bacteriol. 174:3311–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Popham DL, Szeto D, Keener J, Kustu S. 1989. Function of a bacterial activator protein that binds to transcriptional enhancers. Science 243:629–635 [DOI] [PubMed] [Google Scholar]
- 35. Porter SL, Armitage JP. 2002. Phosphotransfer in Rhodobacter sphaeroides chemotaxis. J. Mol. Biol. 324:35–45 [DOI] [PubMed] [Google Scholar]
- 36. Porter SC, North AK, Wedel AB, Kustu S. 1993. Oligomerization of NTRC at the glnA enhancer is required for transcriptional activation. Genes Dev. 7:2258–2273 [DOI] [PubMed] [Google Scholar]
- 37. Quon KC, Marczynski GT, Shapiro L. 1996. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell 84:83–93 [DOI] [PubMed] [Google Scholar]
- 38. Ramakers C, Ruijter JM, Deprez RHL, Moorman AFM. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339:62–66 [DOI] [PubMed] [Google Scholar]
- 39. Sager B, Kaiser D. 1993. Spatial restriction of cellular differentiation. Genes Dev. 7:1645–1653 [DOI] [PubMed] [Google Scholar]
- 40. Sasse-Dwight S, Gralla JD. 1990. Role of eukaryotic-type functional domains found in the prokaryotic enhancer receptor factor 54. Cell 62:945–954 [DOI] [PubMed] [Google Scholar]
- 41. Shi X, et al. 2008. Bioinformatics and experimental analysis of proteins of two-component systems in Myxococcus xanthus. J. Bacteriol. 190:613–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shimkets LJ. 1990. Social and developmental biology of the myxobacteria. Microbiol. Rev. 54:473–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Silva JC, Haldimann A, Prahalad MK, Walsh CT, Wanner BL. 1998. In vivo characterization of the type A and B vancomycin-resistant enterococci (VRE) VanRS two-component systems in Escherichia coli: a nonpathogenic model for studying the VRE signal transduction pathways. Proc. Natl. Acad. Sci. U. S. A. 95:11951–11956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Singer M, Kaiser D. 1995. Ectopic production of guanosine penta- and tetraphosphate can initiate early developmental gene expression in Myxococcus xanthus. Genes Dev. 9:1633–1644 [DOI] [PubMed] [Google Scholar]
- 45. Son B, Liu Y, Kroos L. 2011. Combinatorial regulation by MrpC2 and FruA involves three sites in the fmgE promoter region during Myxococcus xanthus development. J. Bacteriol. 193:2756–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Studholme DJ, Dixon R. 2003. Domain architectures of σ54-dependent transcriptional activators. J. Bacteriol. 185:1757–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ueki T, Inouye S, Inouye M. 1996. Positive-negative KG cassettes for construction of multi-gene deletions using a single drug marker. Gene 183:153–157 [DOI] [PubMed] [Google Scholar]
- 48. Viswanathan P, Ueki T, Inouye S, Kroos L. 2007. Combinatorial regulation of genes essential for Myxococcus xanthus development involves a response regulator and a LysR-type regulator. Proc. Natl. Acad. Sci. U. S. A. 104:7969–7974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wedel A, Kustu S. 1995. The bacterial enhancer-binding protein NTRC is a molecular machine: ATP hydrolysis is coupled to transcriptional activation. Genes Dev. 9:2042–2052 [DOI] [PubMed] [Google Scholar]
- 50. Weiss DS, Batut J, Klose KE, Keener J, Kustu S. 1991. The phosphorylated form of the enhancer-binding protein NTRC has an ATPase activity that is essential for activation of transcription. Cell 67:155–167 [DOI] [PubMed] [Google Scholar]
- 51. Xu H, et al. 2010. Role of acetyl-phosphate in activation of the Rrp2-RpoN-RpoS pathway in Borrelia burgdorferi. PLoS Pathog. 6:e1001104 doi:10.1371/journal.ppat.1001104 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.