Abstract
During sporulation, Bacillus subtilis redeploys the division protein FtsZ from midcell to the cell poles, ultimately generating an asymmetric septum. Here, we describe a sporulation-induced protein, RefZ, that facilitates the switch from a medial to a polar FtsZ ring placement. The artificial expression of RefZ during vegetative growth converts FtsZ rings into FtsZ spirals, arcs, and foci, leading to filamentation and lysis. Mutations in FtsZ specifically suppress RefZ-dependent division inhibition, suggesting that RefZ may target FtsZ. During sporulation, cells lacking RefZ are delayed in polar FtsZ ring formation, spending more time in the medial and transition stages of FtsZ ring assembly. A RefZ-green fluorescent protein (GFP) fusion localizes in weak polar foci at the onset of sporulation and as a brighter midcell focus at the time of polar division. RefZ has a TetR DNA binding motif, and point mutations in the putative recognition helix disrupt focus formation and abrogate cell division inhibition. Finally, chromatin immunoprecipitation assays identified sites of RefZ enrichment in the origin region and near the terminus. Collectively, these data support a model in which RefZ helps promote the switch from medial to polar division and is guided by the organization of the chromosome. Models in which RefZ acts as an activator of FtsZ ring assembly near the cell poles or as an inhibitor of the transient medial ring at midcell are discussed.
INTRODUCTION
Bacteria need to remain highly organized in order to respond rapidly and dynamically to internal and external cues yet must also be extremely efficient due to their small size and small genomes (55). How do microorganisms organize cellular processes without the extensive internal compartmentalization and highways of motor-driven traffic of their larger eukaryotic counterparts? Part of the answer seems to be by evolving intimate spatial relationships between critical processes and the major “structures” of the bacterial cell: the cell envelope and the chromosome (called the nucleoid). Here, we describe a DNA binding protein, RefZ, that appears to take advantage of the nucleoid to facilitate the redeployment of the division apparatus from a medial to a polar position during the developmental process of sporulation in the bacterium Bacillus subtilis.
Differentiating bacteria often commandeer the cell division apparatus to carry out specialized functions. For example, during growth on solid surfaces, the pathogen Proteus mirabilis inhibits cell division and sprouts peritrichous flagella. These highly elongated cells associate along their longitudinal axes, creating rafts of motile cells that swarm in a synchronized fashion (28). Similarly, during competence development in Bacillus subtilis, cell division is arrested at the time of DNA uptake. It is thought that division inhibition provides a pause that permits the cell to repair the chromosome following the integration of transforming DNA (13). Spore formation in B. subtilis provides an even more dramatic example of the regulation of cell division (26, 36, 57). During this developmental process, B. subtilis switches its division site from the middle of the cell to a position close to one of the poles. Asymmetric division gives rise to two daughters of unequal size and different cell fates.
The first known protein to arrive at the site of cell division in bacteria is the tubulin-like protein FtsZ (11). FtsZ polymerizes to form filaments that assemble into a ringlike structure (the Z ring). The localization of FtsZ at midcell in B. subtilis and Escherichia coli is governed by two mechanisms: Min regulation (45) and nucleoid occlusion (10, 63). The Min system of B. subtilis and E. coli prevents division from occurring in the DNA-free regions of the cell near the poles. The nucleoid occlusion proteins of B. subtilis (Noc) and E. coli (SlmA) help ensure that the Z ring does not form on top of the DNA. Noc and SlmA bind site specifically to sequence elements distributed throughout the genome and inhibit division from these locations (21, 59, 66). In both organisms, these binding sites are markedly underrepresented in the terminus region of the chromosome, which is located at midcell at the time of division. This arrangement allows the Z ring to assemble at midcell at a time when DNA replication is nearly complete. Thus, the nucleoid, one of the major structures in the cell, provides key positional information to the division apparatus. Nucleoid occlusion and Min have complementary functions, with both ensuring that cell division occurs at midcell during normal growth (10, 63).
The switch from medial to polar division during sporulation involves the redistribution of FtsZ from the middle of the cell to the cell poles. Following the initiation of this developmental program, FtsZ first forms a transitory ring at midcell (7). The FtsZ ring is then converted into a spiral-like structure that culminates in the re-deployment of FtsZ to the cell poles (7). Ultimately, one of the two polar Z rings becomes the site of asymmetric division (25, 51). The shift in the site of Z ring assembly is driven, in part, by increasing levels of FtsZ and by the induction of SpoIIE, a sporulation-specific protein which interacts with FtsZ and appears to stabilize the FtsZ rings at the cell pole (7, 39, 44).
Here, we describe RefZ, a protein which is synthesized at the onset of sporulation and facilitates the switch from medial to polar division. We discuss models in which RefZ functions at midcell to destabilize the medial FtsZ ring or near the poles to facilitate polar FtsZ ring assembly.
MATERIALS AND METHODS
General methods.
B. subtilis strains were derived from prototrophic strain PY79 (67). The E. coli strains used were TG1 and DH5α. To visualize the localization of fluorescent fusions during vegetative growth, strains were grown in CH medium (35) at 37°C. Sporulation was induced by resuspension at 37°C (unless otherwise indicated) according to the Sterlini-Mandelstam method (35). Except when indicated otherwise, RefZ was induced with 1.0 mM isopropyl-β-d-thiogalactopyranoside (IPTG) in liquid cultures and on LB agar plates. The inhibition of RefZ-dependent cell division occurred with concentrations of IPTG as low as 10 μM. This concentration of inducer resulted in levels of vegetative RefZ-green fluorescent protein (GFP) (assayed with anti-GFP antibodies) that were comparable to the levels of RefZ-GFP during sporulation. MciZ was induced with 10 mM xylose. To test if cells possessing a plasmid-borne RefZ binding motif (RBM) were more sensitive to RefZ, IPTG was added to a final concentration of 7.5 μM. The localization of RefZ-GFP during vegetative growth was visualized 60 min following the addition of IPTG (10 μM final concentration). HPUra [6-(p-hydroxy-phenyl-azo)-uracil] was used at a final concentration of 40 mg/ml. For the medial-to-polar-shift experiments, cells were grown and induced to sporulate by resuspension at 30°C and mounted onto agarose pads just prior to image capture. The data shown in Fig. 3 and Fig. S4 in the supplemental material were derived from two independent experiments. A description of strains (Table S4), plasmids (Table S5), and oligonucleotide primers (Table S6) can be found in supplemental material.
Fig 3.
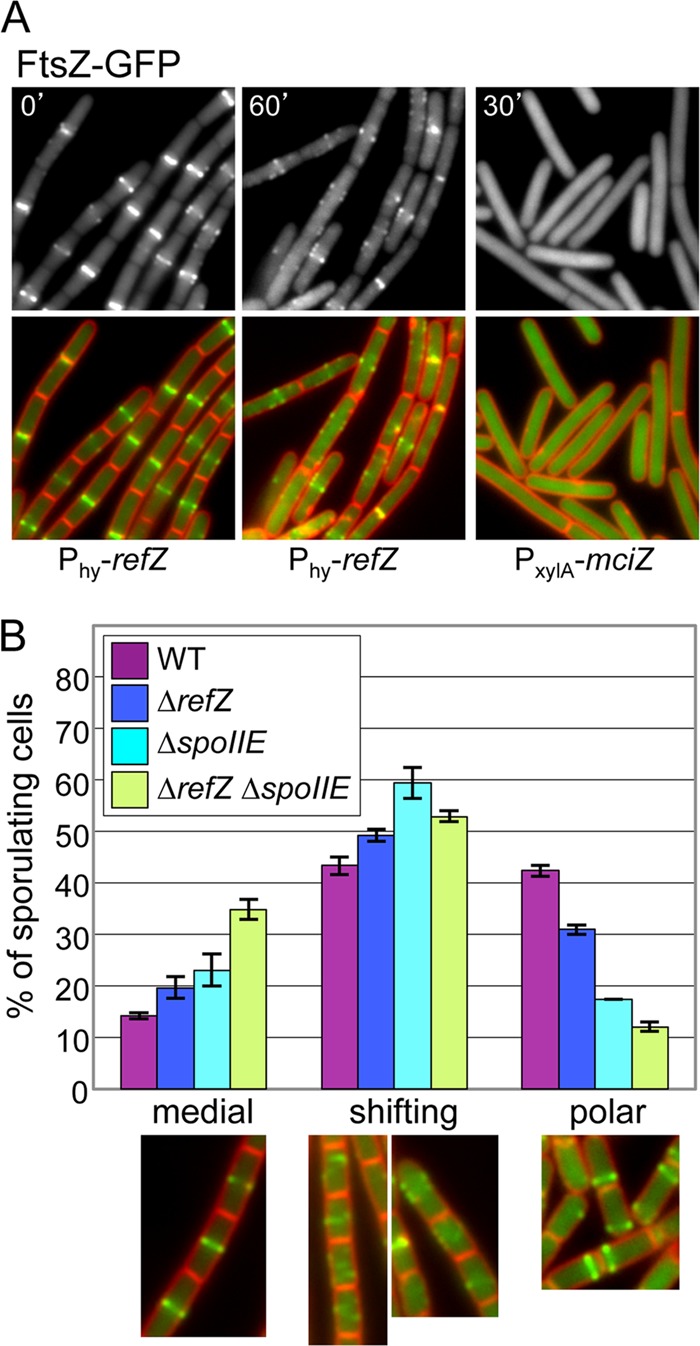
Expression of RefZ during vegetative growth disrupts FtsZ rings. (A) Localization of FtsZ-GFP before induction of refZ (BJW429), 60 min after refZ induction (BJW429), and 30 min after induction of mciZ (BJW481). (Top) FtsZ-GFP images. (Bottom) Overlay of membranes stained with FM4-64 (red) and FtsZ-GFP (green). (B) RefZ facilitates the switch of the FtsZ ring from medial to polar positions during sporulation. Histograms show the quantification of FtsZ-GFP localizations (medial, shifting, or polar) from the wild type (purple) (strain RL3056), a refZ mutant (dark blue) (strain BRB455), a spoIIE mutant (light blue) (strain BRB457), and a refZ spoIIE double mutant (green) (strain BRB459) 120 min after the induction of sporulation. Representative images of FtsZ-GFP localization are shown below the histogram. Values are averages ± standard deviations from 2 independent experiments. More than 300 cells were scored for each strain in each experiment.
Fluorescence microscopy.
Fluorescence microscopy was performed as described previously (23). Exposure times were typically 500 to 1,000 ms for GFP and cyan fluorescent protein (CFP) fusions. Membranes were stained with either TMA-DPH [1-(4-trimethylammoniumphenyl)-6-phenyl-1,3,5-hexatriene p-toluenesulfonate; 0.02 mM] or FM4-64 (3 μg/ml) (Molecular Probes) and imaged with exposure times of 200 to 800 ms. DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI; 2 μg/ml) (Molecular Probes) and imaged with a typical exposure time of 250 ms. Fluorescence images were captured and manipulated by using Metamorph v6.1 software (Molecular Devices). Cells were mounted onto 1% agarose pads made with 1× phosphate-buffered saline (PBS), just prior to imaging. Strains expressing RefZ-GFP or harboring the tetO array and TetR-CFP were mounted onto glass slides with polylysine-treated coverslips, as the localization was found to be similar to that with agarose pads.
Genetic screens.
To identify mutants that had defects in chromosome organization, we used a DNA-trapping assay described previously (58). Briefly, to monitor chromosome organization during sporulation, we used a mutant of the SpoIIIE DNA translocase (spoIIIE36) that engages the forespore chromosome after polar division but is blocked in DNA transport (65). To assess which regions of DNA are trapped in the spore compartment by the polar septum, we fused cfp and yfp to a promoter (PspoIIQ) that is recognized by a forespore-specific transcription factor. These two reporters were inserted at different positions on the B. subtilis chromosome. Accordingly, depending on their location in the axial filament, the spore compartment contained one, both, or neither of the fluorescent reporters. This assay monitors every cell in the field and reveals subtle perturbations in the chromosome organization. The results provide a “snapshot” of the organization of the axial filament at the time of polar division. For the genetic screen, we introduced mutations in Spo0A-controlled genes into our tester strain that contained a yellow fluorescent protein (YFP) reporter at +28° (trapped in the forespore ∼90% of the time) and a CFP reporter at −61° (trapped ∼20% of the time).
To identify point mutants of FtsZ that were resistant to RefZ expression, a plasmid library containing ∼60,000 independent ftsZ mutations generated by mutagenic PCR was transformed into AB88 [zapA::tet yhdG::Phy-refZ (phleo)]. Suppressors were selected on LB agar plates containing tetracycline (10 μg/ml) and IPTG (500 μM), conditions which kill all cells bearing wild-type FtsZ. We recovered 3 × 103 colonies out of 3 × 106 transformants, for a suppressor frequency of ∼0.1%. Genomic DNAs from 90 suppressors were used in back-crosses to the parental strain to determine whether the mutants were linked to ftsZ. Of the 55 suppressors that were linked to ftsZ, 39 were mapped by sequencing.
Immunoblot analysis.
During logarithmic growth or sporulation, the optical density at 600 nm (OD600) was measured (for equivalent loading), and samples (1.0 ml) were collected by centrifugation. Whole-cell extracts were prepared by the resuspension of cell pellets in 50 μl lysis buffer (20 mM Tris [pH 7.0], 10 mM EDTA, 1 mg/ml lysozyme, 10 μg/ml DNase I, and 100 μg/ml RNase A, with 1 mM phenylmethylsulfonyl fluoride [PMSF]). This was followed by incubation at 37°C for 10 min and then the addition of 50 μl sodium dodecyl sulfate (SDS) sample buffer (0.25 M Tris [pH 6.8], 4% SDS, 20% glycerol, 10 mM EDTA) containing 10% 2-mercaptoethanol. Samples were heated for 5 min at 80°C prior to loading. Proteins were separated by SDS-PAGE on 10% polyacrylamide gels, electroblotted onto an Immobilon-P membrane (Millipore), and blocked in 5% nonfat milk in PBS–0.5% Tween 20. Primary antibodies (anti-GFP [53] or anti-σF [49] antibody) were detected by using horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (Bio-Rad) with Supersignal substrate, according to the manufacturer's instructions (Perkin-Elmer).
Chromatin immunoprecipitation (ChIP).
Formaldehyde was added to 20 ml of sporulating cells (75 min after resuspension) to a final concentration of 1%, and samples were incubated for 10 min at room temperature. Cross-linking was quenched by the addition of glycine to a final concentration of 125 mM. Cell pellets were then washed twice with 1× PBS and treated with lysozyme (1-mg/ml final concentration) for 15 min in buffer A (12.5 mM Tris [pH 8], 12.5 mM EDTA [pH 8], 62.5 mM NaCl, 25% sucrose, 1 mM PMSF), followed by the addition of Triton X-100 to a final concentration of 1%. Chromosomal DNA was sheared to an average size of 300 bp by sonication using a Misonix Ultrasonic Liquid S-4000 instrument. After the removal of cell debris by centrifugation, 50 μl was removed to serve as an input control. The lysate was then incubated overnight at 4°C with affinity-purified anti-GFP antibodies (19). The lysate was then incubated with protein A-Sepharose beads (GE Healthcare) for 1 h at 4°C. The beads were washed 3 times with immunoprecipitation (IP) buffer (50 mM Tris [pH 7.5], 5 mM EDTA, 150 mM NaCl, 1% Triton), 3 times with wash buffer (50 mM Tris [pH 7.5], 1 mM EDTA, 500 mM NaCl, 1% Triton), and once with TE (50 mM Tris [pH 8], 10 mM EDTA). The beads were then resuspended in elution buffer (50 mM Tris [pH 8], 10 mM EDTA [pH 8], 1% SDS) and incubated at 65°C for 15 min. The recovered supernatants and the input control samples were placed at 65°C overnight to reverse the cross-links. The samples (input and IP) were treated with RNase A (200-μg/ml final concentration) at 37°C for 1 h, followed by incubation with proteinase K (0.2-mg/ml final concentration). The DNA was purified by phenol-chloroform extraction and ethanol (EtOH) precipitation, followed by the use of the Qiagen minElute PCR purification kit.
ChIP followed by deep sequencing (ChIP-seq).
Control DNAs from a mock IP using wild cells as well as input and IP DNAs were polyadenylated by using terminal transferase (NEB) according to the manufacturer's instructions. After blocking the reaction using 200 μM biotin-ddATP, the DNA samples were submitted for single-molecule Helicos sequencing at the Molecular Biology Core Facility of the Dana-Farber Cancer Institute. This direct sequencing method does not involve PCR amplification and should therefore be less prone to artifacts. The numbers of sequencing reads for the input and IP DNAs were normalized to each other, and the reads were aligned to the GenBank genome reference sequence for Bacillus subtilis 168 (accession number AL009126; NCBI), using CLC genomics software (CLCbio). ChIP peaks were considered further if they were enriched above the input DNA control in at least two biological replicates, given a false-discovery-rate cutoff of 0.001. Of these peaks, we also excluded those that were present in the mock IP control, did not contain roughly equal sequencing coverage in the forward and reverse directions, contained A-T-rich strings, or were shorter than a window size of 100 bp.
Quantitative PCR.
Quantitative PCR (qPCR) was performed by using a Kapa SYBR Fast universal qPCR kit (Kapa Biosystems) and the Mastercycler Ep Realplex real-time PCR system (Eppendorf). ChIP fold enrichment values were calculated as described previously (2) and represent the relative abundance of the sequence of interest compared with a negative-control locus. The PCR efficiency was controlled to be 99% or higher for all primer pairs. All ChIP fold enrichment values represent the averages of data from three biological replicates.
RESULTS
Genetic screen for genes involved in chromosome organization during sporulation.
The refZ gene was identified in a genetic screen for mutants impaired in chromosome organization during sporulation. Upon the initiation of sporulation, the replicated chromosomes are reorganized into an elongated structure that extends from one cell pole to the other, called the axial filament. Within this serpentine structure, the origins of replication are located at the cell poles, while the replication termini reside near midcell. As a result of axial filament formation, the polar division plane traps the origin-proximal ∼25% of the forespore chromosome in the forespore compartment (58, 65). After cytokinesis, a DNA translocase pumps the remaining 75% of the chromosome into the forespore (62). A sporulation-induced protein called RacA plays a central role in axial filament formation by anchoring the origin regions to the cell poles (6, 8, 64). To search for additional factors that function in the remodeling of the chromosome into the axial filament, we performed a candidate-based, genetic screen in which we assayed a subset of a knockout collection of Spo0A-induced genes (29, 48) for defects in chromosome organization (see Materials and Methods) (58). Spo0A is the master transcriptional regulator for entry into sporulation. Genes under its control (like racA) are required at early and late stages of sporulation (50). One of the mutants (in the yttP gene) had a subtle but reproducible defect in chromosome organization. Specifically, a locus on the left arm of the chromosome (at position −61°) that was not usually polarly localized at the time of asymmetric division (58) was more frequently trapped in the forespore compartment. YttP is a protein of unknown function that is annotated as a TetR-like DNA binding protein and contains a conserved helix-turn-helix (HTH) DNA binding motif at its amino terminus (46; this study). Homologs of yttP are present in most endospore-forming Bacillus species, as assessed by synteny with the upstream gene ezrA (data not shown). Cells lacking YttP sporulate to near-wild-type levels (data not shown). Based on our characterization of YttP, we have renamed the protein RefZ, for regulator of FtsZ.
Expression of RefZ during vegetative growth inhibits cell division.
The functions of other proteins that act early during spore formation have been revealed by inducing their expression during vegetative growth (7, 8, 39, 52, 60). Accordingly, we placed refZ under the control of an IPTG-inducible promoter (Phy-refZ) and inserted the fusion into the B. subtilis chromosome in a single copy. Surprisingly, the induction of RefZ led to decreased growth on plates and a dramatic and rapid inhibition of cell division (Fig. 1). Cell filamentation was observed within 1 cell doubling (30 min) and continued unabated until lysis. Partial division inhibition was observed with concentrations of IPTG as low as 7.5 to 10 μM. This concentration of inducer resulted in levels of vegetative RefZ-GFP (assayed with anti-GFP antibodies) that were comparable to the levels of RefZ-GFP expressed under its native promoter during sporulation (see Fig. S1 in the supplemental material). The appearance of the DNA by DAPI staining after RefZ induction was indistinguishable from that of the wild type (Fig. 1B), suggesting that the block to cell division was not due to a gross defect in chromosome organization (9). These results and our characterization of RefZ described below suggest that the chromosome organization defect that led us to RefZ could be indirect (see Discussion).
Fig 1.
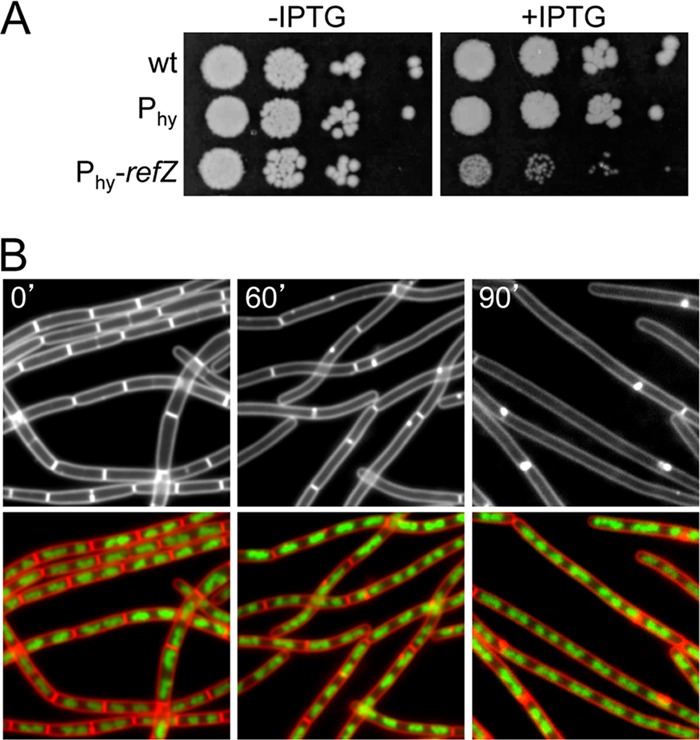
Expression of RefZ during vegetative growth blocks cell division. (A) The vegetative expression of RefZ impairs growth. Tenfold serial dilutions of early-logarithmic-phase cultures were spotted onto LB agar plates in the presence (+) and absence (−) of 1 mM IPTG. The wild type (wt) (PY79) and strains harboring the IPTG-inducible promoter Phy (BJW304) and Phy fused to refZ (BJW123) are shown. (B) Expression of RefZ inhibits cell division. Representative images of filamenting cells (BJW123) after the induction of RefZ are shown. The time (minutes) following the addition of IPTG is indicated. Membranes were visualized with FM4-64 (white in top images and red in bottom images), and DNA was visualized with DAPI (green).
To determine whether or not the division inhibition caused by RefZ was triggered by replication stress (38), which might not be detectable by DAPI staining, we monitored the SOS response in a strain in which the promoter for the SOS-inducible gene yneA was fused to a fluorescent reporter (9). Consistent with the idea that division inhibition was not due to replication stress caused by the induction of RefZ, the level of fluorescence from the SOS reporter did not increase above background levels (see Fig. S2 in the supplemental material). In contrast, the addition of the replication inhibitor HPUra (16) triggered a strong induction of the yneA promoter (see Fig. S2 in the supplemental material). In further support of the idea that RefZ induction does not interfere with DNA replication, the ratio of origin-to-terminus DNA was unperturbed upon the expression of RefZ (data not shown).
RefZ is homologous to proteins of the TetR superfamily of transcriptional regulators (46). Accordingly, we wondered whether RefZ-dependent filamentation was due to the expression of an inhibitor of cell division under RefZ control. To investigate this, we performed transcriptional profiling on vegetative cells before and after RefZ induction and separately on wild-type and RefZ mutant sporulating cells. Only one gene (ytwF) was differentially regulated under both conditions; however, subsequent analysis excluded it as the mediator of cell division inhibition (data not shown). Collectively, these results suggest that the cell division inhibition resulting from RefZ expression is neither a consequence of perturbations in DNA replication nor due to RefZ-dependent gene expression.
FtsZ mutants suppress RefZ-dependent filamentation.
Next, we investigated whether RefZ itself negatively regulates the cell division machinery. Most characterized regulators of cell division act on the tubulin-like protein FtsZ, which assembles into a cytokinetic ring at midcell (1). For many inhibitors of FtsZ, an increase in the FtsZ concentration has a suppressive effect on the activity of the negative regulator (22). Accordingly, we introduced a second copy of the ftsAZ operon into the strain harboring the IPTG-inducible promoter fusion to refZ and spotted serial dilutions onto LB agar plates with and without an inducer. Consistent with the idea that RefZ targets FtsZ, the additional copy of the ftsAZ operon resulted in improved growth in the presence of IPTG (Fig. 2A). Moreover, the extra copy of ftsAZ largely suppressed the cell division defect (Fig. 2B). The expression of an ftsZ-gfp fusion (in the absence of ftsA) under the control of a xylose-inducible promoter also had a suppressive effect on division inhibition (data not shown). To further investigate whether RefZ targets FtsZ, we sought FtsZ point mutants that were resistant to RefZ expression. To do this, we used a strain lacking a positive regulator of FtsZ called ZapA (32). Cells lacking ZapA are exquisitely sensitive to RefZ induction, leading to a complete loss of viability in the presence of an inducer. We introduced a library of ftsZ mutants into this strain and selected for growth on agar plates containing IPTG (see Materials and Methods). Of the 90 suppressor mutants obtained, 55 were linked to ftsZ. Of these, 18 had unique mutations in ftsZ and were further characterized (see Table S1 in the supplemental material). Importantly, all of these mutants suppressed the cell division defect caused by the induction of RefZ. Moreover, most of the mutants had little to no impact on normal cell division in the absence of RefZ (data not shown).
Fig 2.
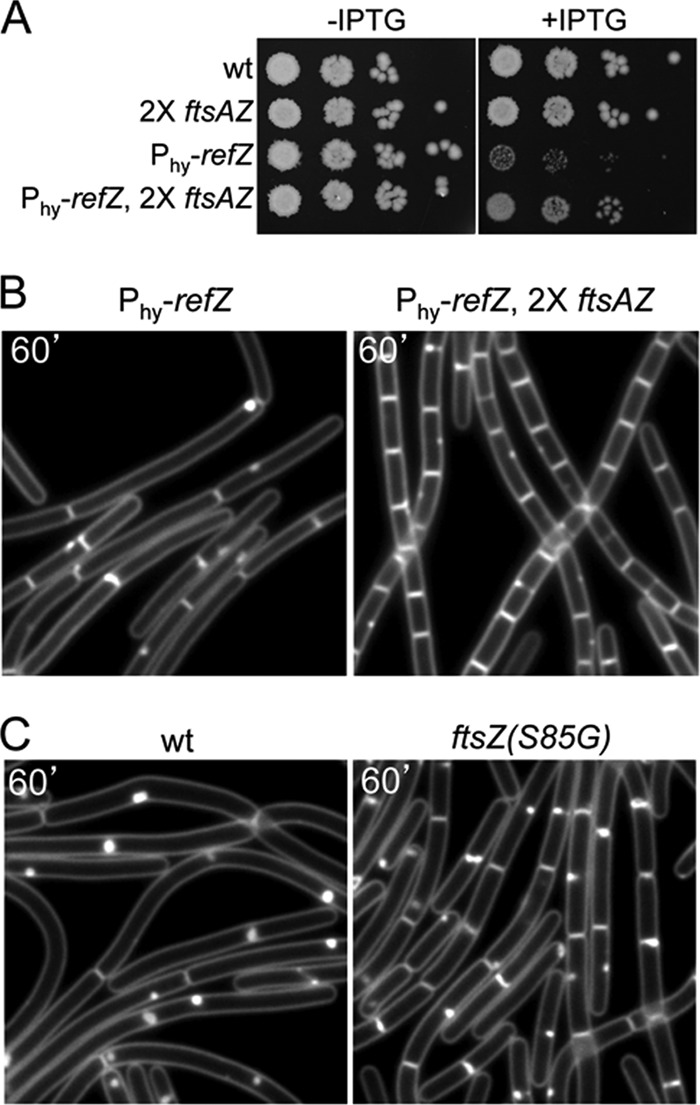
RefZ targets the cell division protein FtsZ. (A) An additional copy of the ftsAZ operon suppresses the growth inhibition caused by RefZ. Tenfold serial dilutions of early-logarithmic-phase cultures were spotted onto LB agar plates in the presence (+) and absence (−) of 1 mM IPTG. The wild type (PY79) and strains harboring an additional copy of the ftsAZ operon (RL3063), Phy-refZ (BJW144), or both (BJW147) are shown. (B) Representative images of cells overexpressing RefZ in the absence (left) or presence (right) of an extra copy of the ftsAZ operon. Membranes were visualized with FM4-64. The time (minutes) following the addition of IPTG is indicated. (C) Cells harboring a point mutant of FtsZ (S85G) are resistant to RefZ-dependent cell division inhibition. Representative images show cells overexpressing RefZ that have wild FtsZ (BJW472) or the FtsZ(S85G) mutant (BJW479).
We further classified the FtsZ point mutants based on their abilities to suppress the lethality caused by the expression (or overexpression) of three other FtsZ-interacting proteins: MciZ, MinC, and ZapA-MTS (ZapA fused to a membrane-targeting signal) (33, 37; F. J. Gueiros-Filho, unpublished data). As expected, some of the suppressor mutants were able to support growth upon the induction of these proteins. These “general” suppressors were observed previously and are thought to improve FtsZ polymerization (A. W. Bisson-Filho and F. J. Gueiros-Filho, unpublished data). In support of the idea that RefZ directly targets FtsZ, four of the suppressor mutants (N24I, A71V, S85G, and M105L) were uniquely resistant to RefZ (Fig. 2C and data not shown). Interestingly, RefZ shared the most cross-resistance with MinC (see Table S1 in the supplemental material), suggesting that RefZ and MinC may share an interaction surface on FtsZ. Attempts to detect an interaction between FtsZ and RefZ using bacterial and yeast two-hybrid assays were unsuccessful. Moreover, biochemical approaches to investigate the interaction between RefZ and FtsZ in vitro were not possible due to the inability to obtain soluble RefZ protein. Thus, we tentatively conclude that RefZ targets FtsZ, but we cannot exclude the possibility that the interaction is indirect.
Expression of RefZ during vegetative growth disrupts FtsZ rings.
To observe the consequences of RefZ expression on FtsZ localization and polymerization, we monitored FtsZ-GFP before and after RefZ induction during vegetative growth. In the absence of an inducer, FtsZ-GFP localized as rings at evenly spaced intervals along the cell length at sites of current or future cell divisions, as reported previously (42) (Fig. 3A; see also Fig. S3 in the supplemental material). However, upon RefZ induction, FtsZ-GFP lost its ringlike structure and instead appeared as spirals, arcs, and foci along the length of the cell (Fig. 3A; see also Fig. S3 in the supplemental material). This phenotype is in striking contrast to the induction of MciZ, an unrelated inhibitor of FtsZ which is induced at a later stage of sporulation (33). The expression of MciZ during vegetative growth resulted in a fully dispersed FtsZ-GFP signal in the cell cytoplasm (33) (Fig. 3A). Cells harboring two copies of Phy-refZ also formed FtsZ-GFP spirals, arcs, and foci albeit more quickly after induction than cells containing one copy (data not shown). Thus, it appears that RefZ, even at high levels, does not cause a wholesale depolymerization of FtsZ filaments. These results suggest that RefZ could function by destabilizing higher-order interactions between FtsZ polymers. Alternatively, RefZ could function as a positive regulator of FtsZ, promoting filament formation at multiple sites throughout the cell and thus preventing the maturation of the cytokinetic ring.
RefZ facilitates the switch from medial to polar division during sporulation.
Previous work indicated that upon the initiation of sporulation, an FtsZ ring is first assembled at midcell (7). Subsequently, as a consequence of the increased expression level of FtsZ and the induction of the sporulation protein SpoIIE, the medial ring is converted into spiral-like intermediates that are redeployed to the cell poles (7, 39). Ultimately, one of the two polar FtsZ rings becomes the asymmetric septum. RefZ is induced as cells enter sporulation (48), and our data suggest that it regulates FtsZ. Accordingly, we hypothesized that RefZ participates in the switch from medial to polar division during the early stages of spore development. To test this, we analyzed the localization of FtsZ-GFP in sporulating cells lacking RefZ, SpoIIE, or both. These experiments were conducted at 30°C to slow down sporulation and more clearly delineate the different stages of FtsZ assembly (7). At various time points after the initiation of sporulation, FtsZ-GFP and cell membranes were imaged by fluorescence microscopy. Individual cells at each time point (n ≥ 300) were classified as medial, shifting, or polar, as described previously (7). As can be seen in Fig. 3B, 120 min after the initiation of sporulation, 42% of wild-type cells had polar FtsZ rings, compared to 31% for the RefZ mutant. This phenotype was even more pronounced in cells lacking SpoIIE, in which 17% had polar FtsZ rings. Finally, the double mutant had the most severe phenotype, with only 12% of the sporulating cells harboring polar rings. The reduction in polar septa in the single and double mutants was statistically significant with a confidence interval of 95% (56) (see Table S2 in the supplemental material). Concomitant with the decrease in polar FtsZ rings, we observed an increase in medial and shifting rings in the mutants. At 120 min, only 14% of wild-type cells had medial FtsZ rings, compared to the RefZ, SpoIIE, and double mutants, which had 20%, 23%, and 35%, respectively. Similar trends were observed at 90 min and 150 min after the onset of sporulation (see Fig. S4 in the supplemental material). These results support the hypothesis that RefZ participates in the repositioning of the FtsZ ring from a medial to a polar location during sporulation. Since the shifting of FtsZ can still take place in the absence of RefZ and SpoIIE, other factors, including the previously characterized role of increased FtsZ levels (7), likely contribute to FtsZ repositioning during sporulation.
RefZ localizes at midcell and the poles during sporulation.
To gain insight into how RefZ facilitates the switch from medial to polar division, we examined the subcellular localization of RefZ during sporulation. We generated a refZ-gfp fusion and inserted it into the chromosome under the control of the native refZ promoter and native ribosome binding site. The fusion was partially functional, as assayed by expression during vegetative growth (under IPTG control); Phy-refZ-gfp caused cell filamentation albeit to a lesser extent than Phy-refZ alone (data not shown). Cells harboring refZ-gfp were induced to sporulate, and the subcellular localization of the fusion was examined by fluorescence microscopy. Consistent with previous studies that showed that refZ is under the control of Spo0A (48), RefZ-GFP was barely detectable before the 60-min time point; furthermore, in accordance with the heterogeneous activity of Spo0A in a sporulating population (20), RefZ-GFP levels were not uniform across the population. At 60 min, the time of axial filament formation but prior to the onset of polar division, RefZ-GFP localized diffusely and in weak foci near the cell poles (Fig. 4C). At the time of polar septation (∼75 min), RefZ-GFP localized principally as a bright focus at midcell, usually at or near the cell membrane (Fig. 4B). Weaker polar foci and diffuse signals were also present at this time point in some cells (Fig. 4B). These results are consistent with a model in which RefZ inhibits FtsZ ring formation near the poles early in sporulation and then shifts to midcell to aide in destabilizing the medial FtsZ ring. These observations are also consistent with an alternative model in which polarly localized RefZ is the active species and functions to promote polar FtsZ ring assembly at these sites (see Discussion). Attempts to colocalize RefZ and FtsZ during the early stages of sporulation could not distinguish between these models due to the weak RefZ-GFP signal in this genetic background (not shown).
Fig 4.
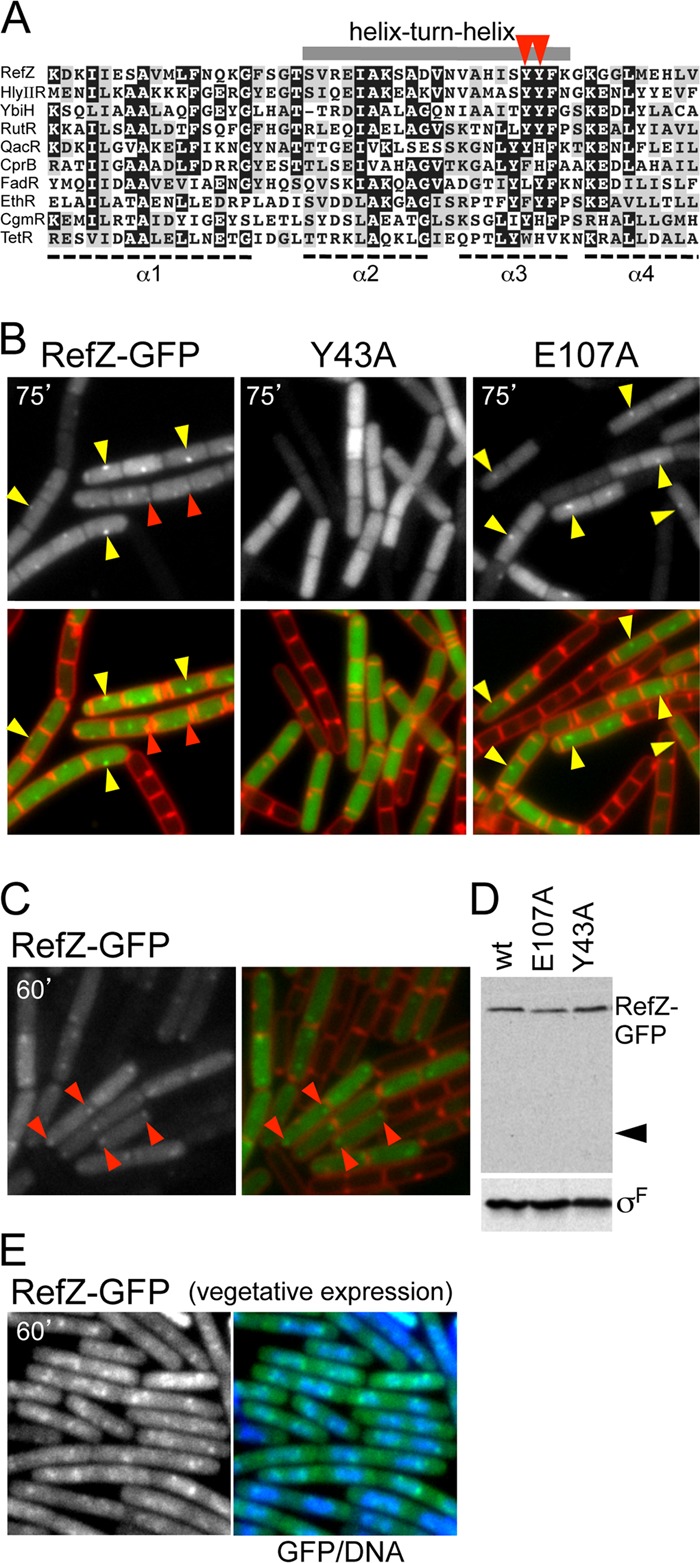
RefZ localization requires an intact helix-turn-helix motif. (A) Alignment of the amino-terminal domain of RefZ with DNA binding domains from TetR superfamily protein members for which structures have been determined. The alignment was made by using ClustalW and cartooned by using BoxShade. Conserved (gray boxes) and highly conserved (black boxes) residues are indicated. The gray bar above the alignment highlights the helix-turn-helix (HTH) motif. The positions of tyrosine 43 and tyrosine 44 in the recognition helix are highlighted (red carets). Aligned proteins are HlyIIR (Bacillus cereus), FadR (B. subtilis), YbiH (Salmonella enterica serovar Typhimurium), RutR (E. coli), EthR (Mycobacterium smegmatis), QacR (Staphylococcus aureus), CprB (Streptomyces coelicolor), CgmR (Corynebacterium glutamicum), and TetR (E. coli). (B) RefZ-GFP localizes in discrete foci that require an intact HTH motif. Cells harboring refZ-gfp (strain BRB672), refZ(Y43A)-gfp (strain BRB698), or refZ(E107A)-gfp (strain BRB697) were visualized by fluorescence microscopy 75 min after the induction of sporulation. Images show RefZ-GFP (top) and an overlay (bottom) of RefZ-GFP (green) and membranes stained with TMA-DPH (red). Midcell foci (yellow carets) and more subtle foci present near the cell poles (red carets) are indicated. (C) RefZ-GFP localizes as weak polar foci at early stages of sporulation. Cells harboring refZ-gfp (strain BJW190) were visualized by fluorescence microscopy 60 min after the induction of sporulation. Images show RefZ-GFP (left) and an overlay of RefZ-GFP (green) and membranes stained with FM4-64 (red) (right). RefZ-GFP foci (red carets) present near the cell poles are indicated. (D) Immunoblot analysis shows that RefZ-GFP and point mutants remain intact during sporulation. RefZ-GFP was analyzed by using anti-GFP antibodies, and the caret identifies the predicted size of free GFP. σF was used to control for loading. RefZ-GFP forms foci that colocalize with the nucleoid when expressed during vegetative growth. (E) RefZ-GFP was induced in cells (strain BJK001) harboring Phy-refZ-gfp. Images show representative fields obtained 60 min after the induction of RefZ-GFP with 10 μM IPTG. Shown is RefZ-GFP alone and merged with DAPI-stained DNA (blue).
Next, we examined the localization of RefZ-GFP induced during vegetative growth. At high concentrations of IPTG, RefZ-GFP appeared as a diffuse haze in the cytosol (not shown). When we expressed the fusion at levels approximately equivalent to those of RefZ-GFP under the control of its native promoter during sporulation (see Fig. S1 in the supplemental material), discrete fluorescent foci that colocalized with the nucleoid were revealed (Fig. 4E; see also Fig. S5 in the supplemental material). These results are consistent with the idea that RefZ foci are a consequence of DNA binding. We suspect that the differences in the localization patterns of the RefZ-GFP foci in vegetatively growing cells compared to sporulating cells likely reflect differences in the organization of the chromosome under these two conditions.
Mutations in the TetR DNA binding motif abrogate RefZ focus formation.
RefZ is homologous to the TetR superfamily of DNA binding proteins and is most similar to the transcriptional regulator HlyIIR from Bacillus cereus (17, 40) (Fig. 4A). The crystal structure of HlyIIR has the canonical TetR family helix-turn-helix (HTH) DNA binding motif, and many conserved residues in this motif are shared with RefZ (Fig. 4A). Mutational analyses of several members of the TetR superfamily have defined critical residues for site-specific binding in the second helix of the HTH motif (the so-called recognition helix) (4, 5, 34). These residues could be identified in RefZ based on the HlyIIR structure. To investigate whether the RefZ-GFP foci which we observed by fluorescence microscopy depended on these conserved residues, we separately mutated two key residues in the putative DNA binding motif to alanine (Y43A and Y44A) and analyzed the fusions by fluorescence microscopy. Both RefZ(Y43A)-GFP and RefZ(Y44A)-GFP localized as a diffuse cytoplasmic haze during sporulation (Fig. 4B and data not shown) and when expressed during vegetative growth under IPTG control (data not shown; see also Fig. S5 in the supplemental material). Discrete fluorescent foci were virtually absent under both conditions. A mutation in another residue conserved among RefZ homologs, but located outside the HTH motif (E107A), formed foci that were indistinguishable from those formed by the wild type during sporulation (Fig. 4B) and vegetative growth (data not shown). The mutants were expressed at levels similar to those of the wild type, and the GFP fusions remained intact (Fig. 4D and data not shown), suggesting that the point mutations did not lead to protein instability. Notably, the expression of the HTH mutants as GFP fusions or untagged proteins during vegetative growth did not lead to filamentation (data not shown). Collectively, these results are consistent with the idea that the localization of RefZ at midcell and to polar foci is mediated by DNA binding. These results further suggest that the ability of RefZ to influence FtsZ depends on DNA binding.
Identification of RefZ binding sites.
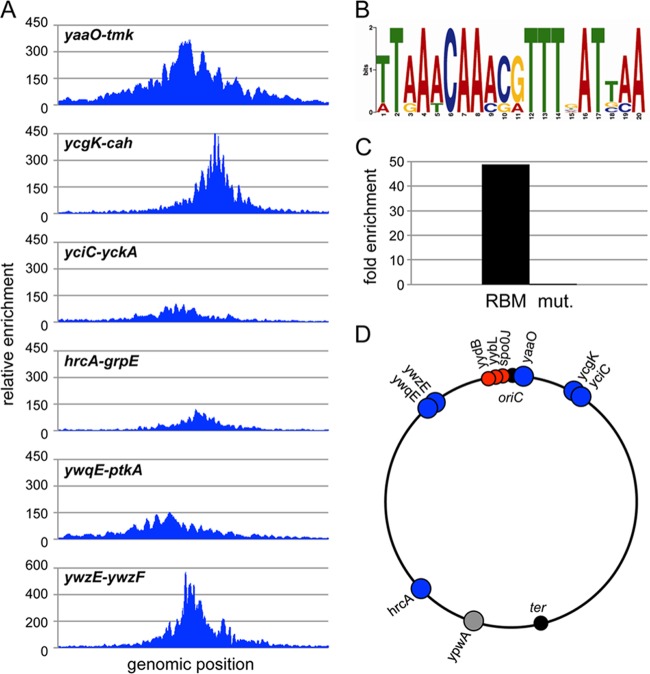
To identify RefZ binding sites, we performed chromatin immunoprecipitation followed by deep sequencing (ChIP-seq). We used a strain harboring the RefZ-GFP fusion (expressed under the control of its native promoter) and cross-linked nucleoprotein complexes at 75 min after the induction of sporulation. After immunoprecipitation with anti-GFP antibodies and the reversal of the cross-links, the purified DNA was subjected to Helicos sequencing (see Materials and Methods). Deep sequencing of two independent samples identified nine chromosomal regions with significant enrichment over the input control (Fig. 5A; see also Fig. S6 and Table S3 in the supplemental material). Six of these sites were more highly enriched (105- to 560-fold over the control) and produced sharp sequencing peaks (Fig. 5A) spanning approximately 1 to 2.5 kb each. All six peaks were confirmed by quantitative PCR (qPCR) on independently collected samples (see Fig. S7 in the supplemental material). Five of the six most enriched peaks were located in the origin-proximal region (Fig. 5D; see also Table S3 in the supplemental material). Only one site, located in the hrcA gene at 225°, was in a more terminus-proximal position. The remaining three binding sites had broad, relatively flat sequencing peaks with 30- to 40-fold enrichment compared to the wild type (see Fig. S6 in the supplemental material). Although these broad peaks were identified in two ChIP-seq experiments with independent biological isolates, significant enrichment using qPCR was observed for only one of these loci (the yydB region) (see Fig. S7 in the supplemental material). Due to the sensitivity of Helicos sequencing, we favor the idea that all three regions are bound by RefZ in vivo.
Fig 5.
RefZ binds to specific sites on the chromosome. (A) RefZ binding sites identified by ChIP-seq. Plots show relative enrichment values (normalized over the input control DNA) for regions (1 to 3 kb) of interest. (B) Position-weighted matrix of the RefZ binding motif (RBM) identified in the RefZ binding regions. The height of each letter represents the relative degree of conservation of each base in all the sequences identified. (C) RefZ binds to the 20-bp RBM when moved to an ectopic location. The RBM from the ywzF locus (BRB789) and a mutated version (RBM mut.) (BRB790) were inserted at 87° (into the yhdG locus) on the B. subtilis chromosome. Binding was assessed by using ChIP-qPCR. Plots show relative enrichment values for a site adjacent to the RBM. (D) Schematic diagram showing the locations of the RefZ binding sites on the B. subtilis chromosome. Sharp peaks (blue circles) and broad peaks (red circles) are indicated. An additional RBM in the ypwA locus (gray circle) identified through bioinformatics is also shown. The replication origin (oriC) and terminus (ter) are indicated.
To identify potential RefZ binding motifs, 1 to 3 kb of the enriched sequence surrounding the six confirmed peaks was analyzed by using MEME (3). The strongest motif identified was a largely palindromic 20-bp sequence, which we have named the RBM (for RefZ binding motif). The position-weighted matrix generated from the analysis is shown in Fig. 5B. Strikingly, in almost all cases, the motif was present close to the center of the six sharp peaks. Only degenerate motifs could be identified in the three broad peaks. To determine whether the motif that we identified was sufficient for RefZ binding, a 28-bp sequence centered on the RBM sequence in ywzF (one of the more highly enriched regions) and a mutant version (see Materials and Methods) were inserted at an ectopic locus (yhdG) at 87° on the chromosome. Chromatin immunoprecipitation followed by qPCR revealed that the wild-type site but not the mutant was sufficient to confer RefZ-GFP binding, resulting in a 45-fold enrichment of this chromosomal position (Fig. 5C). These results are consistent with the idea that RefZ is a DNA binding protein and that it recognizes the RBM in vivo.
To investigate whether there were other potential RefZ binding sites in the chromosome, we used the identified motif to search the B. subtilis genome for additional occurrences of the sequence using a generic DNA pattern search (http://genolist.pasteur.fr/SubtiList/). Allowing for 4 mismatches, the search identified 17 additional sites. Since palindromic sequences are often critical for DNA binding proteins, the remaining pool was further narrowed to include only candidates that possessed the central AAACGTTT palindromic sequence. Only one site passed all of these criteria. This putative RBM is located within the terminal-proximal ypwA gene at 198° (Fig. 5D).
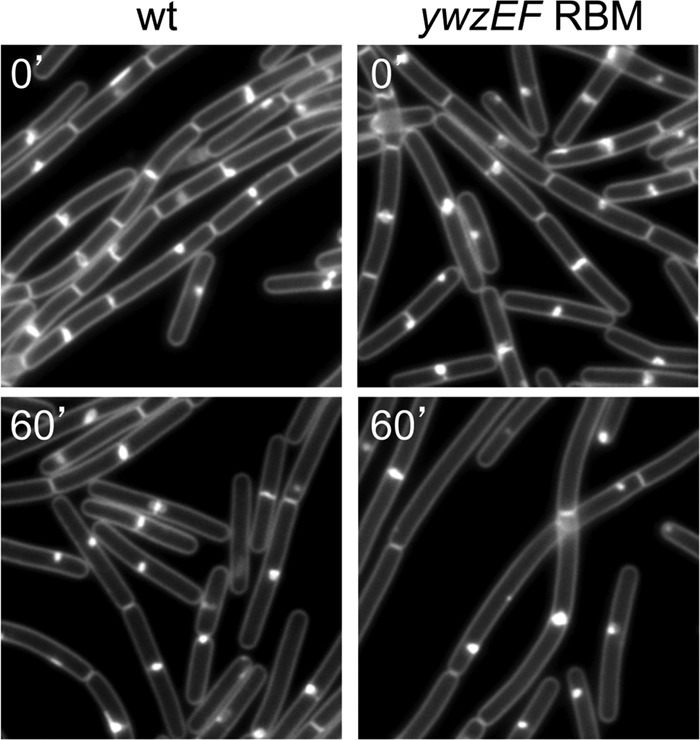
Having identified RefZ binding sites, we next attempted to determine how they relate to the RefZ-GFP localization that we observed during sporulation. Since the replication termini reside near midcell in the axial filament (12, 61), we hypothesized that the terminus-proximal RBMs were responsible for localizing RefZ in the midcell focus. The deletion of the RBMs in hrcA and ypwA, singly or together, did not completely disrupt the midcell foci observed during sporulation (data not shown; see also Fig. S8 in the supplemental material). In general, the midcell foci were weaker, and fewer sporulating cells had them in the mutants lacking the hrcA RBM (see Fig. S8 in the supplemental material). However, since the mutant appeared to enter sporulation less efficiently than the wild type, it is unclear whether this defect was due completely to the loss of the RBMs.
We next determined the cellular location of one of the origin-proximal RefZ binding sites (in the ywzF region) by inserting a tetO array adjacent to the RBMs and monitoring its localization using a TetR-CFP fusion. Consistent with previously reported work on chromosome organization during sporulation (43, 58, 61, 65), the ywzF locus was localized near the cell poles upon the initiation of sporulation (data not shown). These data suggest that RefZ localization at the cell poles is due to binding to the origin-proximal RBMs. Conversely, the presence of some midcell RefZ foci following the deletion of the terminal-proximal RBMs suggests that we either failed to identify by ChIP all of the RefZ-DNA interactions that mediate the midcell localization or that this localization is mediated, at least in part, by another mechanism (see Discussion).
A plasmid-borne RBM is a more potent inhibitor of cell division.
RefZ harboring mutations in its putative DNA recognition helix fails to form RefZ-GFP foci during sporulation and does not block cell division when artificially expressed during vegetative growth. These results suggest that DNA binding is important for the efficient RefZ-dependent regulation of FtsZ. To further test this hypothesis, we introduced the RBM from the ywzF locus into an autonomously replicating plasmid with a copy of number of 10 to 15 per cell (15), thus providing more binding sites for RefZ in vivo. We used a concentration of IPTG (7.5 μM) that led to only a slight increase in cell length and occasional filamentation in wild-type cells containing Phy-refZ. We compared cells harboring the empty plasmid to cells harboring the RBM-containing plasmid. Following 60 min of growth in the presence of an inducer, cells harboring the empty plasmid behaved similarly to the wild type, increasing slightly in average cell length from 3.2 μm before induction to 4.0 μm after induction (Fig. 6; see also Fig. S9 in the supplemental material). In contrast, the cells harboring the RBM-containing plasmid were more sensitive to RefZ, with the cell length increasing from an average of 3.8 μm before induction to 6.0 μm after induction (Fig. 6; see also Fig. S9 in the supplemental material). These results provide further support for the idea that RefZ DNA binding serves to localize the protein and stimulate its activity.
Fig 6.
RefZ is a more potent division inhibitor when bound to DNA. Shown are representative images of cells expressing low levels of RefZ harboring an empty plasmid (left) (strain BJW538) or a plasmid harboring the RBM from the ywzF locus (right) (strain BJW537). Cells were grown to the mid-log phase and induced with 7.5 μM IPTG. The time (in minutes) before and after the addition of IPTG is indicated. Images show membranes stained with TMA-DPH. Before induction, strains harboring the empty plasmid and the RBM-containing plasmid had average cell lengths of 3.2 μm and 3.8 μm, respectively. Following induction, the average cell lengths were 4.0 μm and 6.0 μm, respectively.
DISCUSSION
Entry into the developmental pathway of sporulation is governed by the transcriptional regulator Spo0A. The activation of Spo0A is triggered by phosphorylation in response to starvation through a multicomponent signaling pathway (18, 50). Interestingly, genes under Spo0A control fall into at least two classes: those that require low levels of activated Spo0A (so-called low-threshold genes) and those that require high levels of phosphorylated Spo0A (Spo0A∼P) (high-threshold genes) (30). This hierarchy results in a graded response to starvation. Consistent with the idea that the commitment to sporulation is the last choice for a starving cell, all of the key genes that orchestrate entry into this developmental pathway require high levels of activated Spo0A. These include the genes that encode the mother cell- and forespore-specific transcription factors and their regulators (50, 57); RacA, required for axial filament formation (8); SirA, involved in preventing additional rounds of DNA replication, thereby maintaining the sporulating cell in a diploid state (52, 60); and SpoIIE, required for both efficient polar septation and the activation of the spore-specific transcription factor σF (7, 24, 27, 47). Here, we have defined the role of refZ (formerly yttP), the last uncharacterized high-threshold Spo0A gene defined previously by Fujita and Losick (30).
We have shown that the artificial induction of RefZ during vegetative growth leads to the conversion of midcell FtsZ rings into spirals, arcs, and foci and, consequently, to a block in cell division. Under native control, RefZ is synthesized during the onset of sporulation, where it promotes the repositioning of FtsZ from midcell toward the cell poles. In a refZ mutant, FtsZ spends more time at midcell and in the spiral-like intermediate before shifting to the pole. These effects are even more pronounced in cells lacking both refZ and spoIIE. These data suggest that during sporulation, FtsZ exists in a positional equilibrium between midcell and the pole and that RefZ functions to shift this equilibrium toward the pole. RefZ-GFP forms faint polar foci at the onset of sporulation. The protein then concentrates as a midcell focus at the time of polar division. RefZ has a TetR family DNA binding domain, and our chromatin immunoprecipitation experiments indicate that it binds DNA in vivo. Furthermore, point mutations in the conserved recognition helix of RefZ disrupt RefZ focus formation and abrogate cell division inhibition. Collectively, these data indicate that RefZ functions to promote the switch from medial to polar division during sporulation. We propose that it does so by taking advantage of the structure and organization of the replicated chromosomes within the sporulating cell.
Our data are consistent with two models for how RefZ facilitates the switch in FtsZ positioning. In the first model, RefZ functions as a negative regulator of FtsZ, disrupting higher-order structures in the midcell ring and thereby promoting the redeployment of FtsZ to polar sites. Alternatively, RefZ acts as an activator of FtsZ rings. In the second model, RefZ bound to the RBMs near the origin during sporulation targets the protein to the poles, where it promotes polar Z ring assembly. In this model, the expression of RefZ during vegetative growth promotes FtsZ assembly at multiple sites, preventing the maturation at any one of them and leading to division inhibition. This phenotype was observed previously for cells lacking both the Min system and the nucleoid occlusion protein Noc or SlmA (10, 66). The more potent cell division inhibition observed in the presence of the RBM-containing plasmid is consistent with either model. In the first model, RefZ bound to the multicopy plasmid would more effectively destabilize Z rings. Alternatively, these nucleoprotein complexes could stimulate FtsZ assembly at more sites along the cell length, leading to a loss of FtsZ ring maturation.
Our findings that RefZ binding sites (RBMs) are enriched in the origin-proximal region of the chromosome and that the sites near the terminus are not critical for the midcell RefZ focus favor a model in which RefZ promotes polar FtsZ assembly (model 2). However, the timing and intensity of the RefZ-GFP foci as cells enter sporulation are more consistent with model 1, in which RefZ destabilizes the midcell ring. Resolving whether RefZ acts as a positive or negative regulator of FtsZ awaits biochemical reconstitution in the future.
If RefZ promotes FtsZ ring formation at the cell poles (model 2), one possibility is that it functions by counteracting the polar inhibitor MinC. Our genetic data suggesting that RefZ and MinC have overlapping binding sites on FtsZ are consistent with the idea that RefZ could occlude MinC binding to FtsZ.
If RefZ does indeed function as a midcell inhibitor of FtsZ (model 1), what is the role of the origin-proximal RBMs? One possibility is that these binding sites serve as a reservoir for RefZ as it accumulates under the control of increasing levels of Spo0A∼P. Once RefZ levels saturate these sites or a binding site at midcell is created, RefZ would then localize at midcell at a time when its activity is required. The redistribution of RefZ to the midcell could also be actively driven by its displacement from the sites near the origin as the origin-proximal DNA is anchored to the cell poles (8).
Two possible explanations could account for our inability to identify RefZ binding sites near the terminus region, even though DNA binding appears to be required for the formation of the midcell foci. The first possibility is that the interaction between RefZ and DNA is too transient to be captured by formaldehyde cross-linking (54). For example, RefZ might require a transient DNA interaction to form the initial midcell focus but maintains this localization through a higher-affinity interaction with another protein. We observed that all RefZ-GFP foci (polar and midcell) were lost upon the addition of formaldehyde (data not shown). A second possibility is that RefZ is localized at midcell through an interaction with DNA that is not sequence specific. RefZ-GFP does not localize at midcell during vegetative growth, which suggests that the context for the midcell localization is created by sporulation itself and therefore could be more complex than the presence or absence of a specific binding motif. This context could relate to a DNA structure formed at the time of termination and/or chromosome decatenation as sporulating cells complete the final round of DNA replication. The DNA sequence present in these structures might vary from cell to cell, resulting in an undetectable enrichment by ChIP.
RefZ and chromosome trapping.
We originally identified RefZ in a screen for mutants impaired in chromosome organization during sporulation. Cells lacking RefZ have a subtle increase in the amount of the left chromosomal arm trapped in the forespore by the cell division plane. The identification of RefZ binding sites in the origin-proximal region provides a plausible explanation for the original phenotype. It is possible that the absence of RefZ binding to these sites leads to a modest difference in the organization and/or compaction of the chromosome. This would, in turn, affect how much of each arm is polarly localized at the time of asymmetric division. Alternatively, if RefZ functions to promote polar FtsZ ring assembly, it is possible that it also helps specify the cellular position of the ring. In its absence, the localization of the asymmetric septum could be subtly altered, resulting in more of the arm becoming trapped in the forespore. Unfortunately, the position of the polar division plane was too variable to assess a difference in placement between the wild type and the RefZ mutant.
The chromosome is a key organizer of the bacterial cell.
Cytological studies over the past decade have revealed that the bacterial cell has a complex underlying architecture in which proteins are exquisitely and precisely localized (55). In most cases, the spatial cues that anchor these proteins at specific sites remain unknown. Previous work on the nucleoid occlusion proteins Noc and SlmA highlighted the role that the nucleoid plays in dictating protein localization (21, 59, 66). Several proteins have been implicated in organizing the chromosome in B. subtilis during growth and sporulation (8, 14, 31, 41, 58, 64). This organization is quite robust and ensures extremely faithful chromosome segregation. Our data raise the possibility that RefZ, like Noc and SlmA, takes advantage of this organization to facilitate the shift in the FtsZ ring from midcell to polar positions. We suspect that future studies will reveal that the nucleoid plays an even greater role than we currently appreciate in specifying the organization of the bacterial cytoplasm and the envelope that contains it.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Rudner laboratory for advice and encouragement; Bryan Davies for expertise, advice, and discussions regarding ChIP-seq and qPCR; Cathy Lee for advice about chromatin immunoprecipitations; Meriem El Karoui for help with motif searches; Alan Grossman and Bill Burkholder for reagents; Rich Losick for providing the Spo0A regulon deletion collection; Nick Ratterman for help with statistical analysis; and Xindan Wang for expert fluorescence imaging.
Support for this work comes from National Institutes of Health grant GM086466, the Giovanni Armenise-Harvard Foundation, the Hellman Family Faculty Fund, Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) grant 08/58821-1, and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) grant 478019/2009-2. D.Z.R. and F.J.G.-F. thank the David Rockefeller Center for Latin American Studies faculty grants program.
Footnotes
Published ahead of print 22 June 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Adams DW, Errington J. 2009. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat. Rev. Microbiol. 7:642–653 [DOI] [PubMed] [Google Scholar]
- 2. Aparicio O, Geisberg JV, Struhl K. 2004. Current protocols in molecular biology. Wiley, New York, NY [Google Scholar]
- 3. Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2:28–36 [PubMed] [Google Scholar]
- 4. Baumeister R, Helbl V, Hillen W. 1992. Contacts between Tet repressor and tet operator revealed by new recognition specificities of single amino acid replacement mutants. J. Mol. Biol. 226:1257–1270 [DOI] [PubMed] [Google Scholar]
- 5. Baumeister R, Muller G, Hecht B, Hillen W. 1992. Functional roles of amino acid residues involved in forming the alpha-helix-turn-alpha-helix operator DNA binding motif of Tet repressor from Tn10. Proteins 14:168–177 [DOI] [PubMed] [Google Scholar]
- 6. Ben-Yehuda S, et al. 2005. Defining a centromere-like element in Bacillus subtilis by identifying the binding sites for the chromosome-anchoring protein RacA. Mol. Cell 17:773–782 [DOI] [PubMed] [Google Scholar]
- 7. Ben-Yehuda S, Losick R. 2002. Asymmetric cell division in B. subtilis involves a spiral-like intermediate of the cytokinetic protein FtsZ. Cell 109:257–266 [DOI] [PubMed] [Google Scholar]
- 8. Ben-Yehuda S, Rudner DZ, Losick R. 2003. RacA, a bacterial protein that anchors chromosomes to the cell poles. Science 299:532–536 [DOI] [PubMed] [Google Scholar]
- 9. Bernard R, Marquis KA, Rudner DZ. 2010. Nucleoid occlusion prevents cell division during replication fork arrest in Bacillus subtilis. Mol. Microbiol. 78:866–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bernhardt TG, de Boer PA. 2005. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol. Cell 18:555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bi EF, Lutkenhaus J. 1991. FtsZ ring structure associated with division in Escherichia coli. Nature 354:161–164 [DOI] [PubMed] [Google Scholar]
- 12. Bogush M, Xenopoulos P, Piggot PJ. 2007. Separation of chromosome termini during sporulation of Bacillus subtilis depends on SpoIIIE. J. Bacteriol. 189:3564–3572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Briley K, Jr, Prepiak P, Dias MJ, Hahn J, Dubnau D. 2011. Maf acts downstream of ComGA to arrest cell division in competent cells of B. subtilis. Mol. Microbiol. 81:23–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Britton RA, Lin DC, Grossman AD. 1998. Characterization of a prokaryotic SMC protein involved in chromosome partitioning. Genes Dev. 12:1254–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bron S, et al. 1998. Protein secretion and possible roles for multiple signal peptidases for precursor processing in bacilli. J. Biotechnol. 64:3–13 [DOI] [PubMed] [Google Scholar]
- 16. Brown NC. 1970. 6-(p-Hydroxyphenylazo)-uracil: a selective inhibitor of host DNA replication in phage-infected Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 67:1454–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Budarina ZI, et al. 2004. A new Bacillus cereus DNA-binding protein, HlyIIR, negatively regulates expression of B. cereus haemolysin II. Microbiology 150:3691–3701 [DOI] [PubMed] [Google Scholar]
- 18. Burbulys D, Trach KA, Hoch JA. 1991. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell 64:545–552 [DOI] [PubMed] [Google Scholar]
- 19. Campo N, Marquis KA, Rudner DZ. 2008. SpoIIQ anchors membrane proteins on both sides of the sporulation septum in Bacillus subtilis. J. Biol. Chem. 283:4975–4982 [DOI] [PubMed] [Google Scholar]
- 20. Chastanet A, et al. 2010. Broadly heterogeneous activation of the master regulator for sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 107:8486–8491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cho H, McManus HR, Dove SL, Bernhardt TG. 2011. Nucleoid occlusion factor SlmA is a DNA-activated FtsZ polymerization antagonist. Proc. Natl. Acad. Sci. U. S. A. 108:3773–3778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. de Boer PA, Crossley RE, Rothfield LI. 1990. Central role for the Escherichia coli minC gene product in two different cell division-inhibition systems. Proc. Natl. Acad. Sci. U. S. A. 87:1129–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Doan T, Marquis KA, Rudner DZ. 2005. Subcellular localization of a sporulation membrane protein is achieved through a network of interactions along and across the septum. Mol. Microbiol. 55:1767–1781 [DOI] [PubMed] [Google Scholar]
- 24. Duncan L, Alper S, Arigoni F, Losick R, Stragier P. 1995. Activation of cell-specific transcription by a serine phosphatase at the site of asymmetric division. Science 270:641–644 [DOI] [PubMed] [Google Scholar]
- 25. Eichenberger P, Fawcett P, Losick R. 2001. A three-protein inhibitor of polar septation during sporulation in Bacillus subtilis. Mol. Microbiol. 42:1147–1162 [DOI] [PubMed] [Google Scholar]
- 26. Errington J. 2003. Regulation of endospore formation in Bacillus subtilis. Nat. Rev. Microbiol. 1:117–126 [DOI] [PubMed] [Google Scholar]
- 27. Feucht A, Magnin T, Yudkin MD, Errington J. 1996. Bifunctional protein required for asymmetric cell division and cell-specific transcription in Bacillus subtilis. Genes Dev. 10:794–803 [DOI] [PubMed] [Google Scholar]
- 28. Fraser GM, Hughes C. 1999. Swarming motility. Curr. Opin. Microbiol. 2:630–635 [DOI] [PubMed] [Google Scholar]
- 29. Fujita M, Gonzalez-Pastor JE, Losick R. 2005. High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J. Bacteriol. 187:1357–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fujita M, Losick R. 2005. Evidence that entry into sporulation in Bacillus subtilis is governed by a gradual increase in the level and activity of the master regulator Spo0A. Genes Dev. 19:2236–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gruber S, Errington J. 2009. Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell 137:685–696 [DOI] [PubMed] [Google Scholar]
- 32. Gueiros-Filho FJ, Losick R. 2002. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 16:2544–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Handler AA, Lim JE, Losick R. 2008. Peptide inhibitor of cytokinesis during sporulation in Bacillus subtilis. Mol. Microbiol. 68:588–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hansen D, Altschmied L, Hillen W. 1987. Engineered Tet repressor mutants with single tryptophan residues as fluorescent probes. Solvent accessibilities of DNA and inducer binding sites and interaction with tetracycline. J. Biol. Chem. 262:14030–14035 [PubMed] [Google Scholar]
- 35. Harwood CR, Cutting SM. 1990. Molecular biological methods for Bacillus. Wiley, New York, NY [Google Scholar]
- 36. Hilbert DW, Piggot PJ. 2004. Compartmentalization of gene expression during Bacillus subtilis spore formation. Microbiol. Mol. Biol. Rev. 68:234–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hu Z, Mukherjee A, Pichoff S, Lutkenhaus J. 1999. The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc. Natl. Acad. Sci. U. S. A. 96:14819–14824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kawai Y, Moriya S, Ogasawara N. 2003. Identification of a protein, YneA, responsible for cell division suppression during the SOS response in Bacillus subtilis. Mol. Microbiol. 47:1113–1122 [DOI] [PubMed] [Google Scholar]
- 39. Khvorova A, Zhang L, Higgins ML, Piggot PJ. 1998. The spoIIE locus is involved in the Spo0A-dependent switch in the location of FtsZ rings in Bacillus subtilis. J. Bacteriol. 180:1256–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kovalevskiy OV, Lebedev AA, Surin AK, Solonin AS, Antson AA. 2007. Crystal structure of Bacillus cereus HlyIIR, a transcriptional regulator of the gene for pore-forming toxin hemolysin II. J. Mol. Biol. 365:825–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee PS, Grossman AD. 2006. The chromosome partitioning proteins Soj (ParA) and Spo0J (ParB) contribute to accurate chromosome partitioning, separation of replicated sister origins, and regulation of replication initiation in Bacillus subtilis. Mol. Microbiol. 60:853–869 [DOI] [PubMed] [Google Scholar]
- 42. Levin PA, Losick R. 1996. Transcription factor Spo0A switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Genes Dev. 10:478–488 [DOI] [PubMed] [Google Scholar]
- 43. Lin DC, Levin PA, Grossman AD. 1997. Bipolar localization of a chromosome partition protein in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 94:4721–4726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lucet I, Feucht A, Yudkin MD, Errington J. 2000. Direct interaction between the cell division protein FtsZ and the cell differentiation protein SpoIIE. EMBO J. 19:1467–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lutkenhaus J. 2007. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu. Rev. Biochem. 76:539–562 [DOI] [PubMed] [Google Scholar]
- 46. Marchler-Bauer A, et al. 2011. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 39:D225–D229 doi:10.1093/nar/gkq1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Margolis P, Driks A, Losick R. 1991. Establishment of cell type by compartmentalized activation of a transcription factor. Science 254:562–565 [DOI] [PubMed] [Google Scholar]
- 48. Molle V, et al. 2003. The Spo0A regulon of Bacillus subtilis. Mol. Microbiol. 50:1683–1701 [DOI] [PubMed] [Google Scholar]
- 49. Pan Q, Garsin DA, Losick R. 2001. Self-reinforcing activation of a cell-specific transcription factor by proteolysis of an anti-sigma factor in B. subtilis. Mol. Cell 8:873–883 [DOI] [PubMed] [Google Scholar]
- 50. Piggot PJ. 1996. Spore development in Bacillus subtilis. Curr. Opin. Genet. Dev. 6:531–537 [DOI] [PubMed] [Google Scholar]
- 51. Pogliano J, et al. 1999. A vital stain for studying membrane dynamics in bacteria: a novel mechanism controlling septation during Bacillus subtilis sporulation. Mol. Microbiol. 31:1149–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rahn-Lee L, Gorbatyuk B, Skovgaard O, Losick R. 2009. The conserved sporulation protein YneE inhibits DNA replication in Bacillus subtilis. J. Bacteriol. 191:3736–3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rudner DZ, Losick R. 2002. A sporulation membrane protein tethers the pro-sigmaK processing enzyme to its inhibitor and dictates its subcellular localization. Genes Dev. 16:1007–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schmiedeberg L, Skene P, Deaton A, Bird A. 2009. A temporal threshold for formaldehyde crosslinking and fixation. PLoS One 4:e4636 doi:10.1371/journal.pone.0004636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shapiro L, McAdams HH, Losick R. 2009. Why and how bacteria localize proteins. Science 326:1225–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sokal RR, Rohlf FJ. (ed). 2012. Biometry, 4th ed WH Freeman & Company, New York, NY [Google Scholar]
- 57. Stragier P, Losick R. 1996. Molecular genetics of sporulation in Bacillus subtilis. Annu. Rev. Genet. 30:297–341 [DOI] [PubMed] [Google Scholar]
- 58. Sullivan NL, Marquis KA, Rudner DZ. 2009. Recruitment of SMC by ParB-parS organizes the origin region and promotes efficient chromosome segregation. Cell 137:697–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tonthat NK, et al. 2011. Molecular mechanism by which the nucleoid occlusion factor, SlmA, keeps cytokinesis in check. EMBO J. 30:154–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wagner JK, Marquis KA, Rudner DZ. 2009. SirA enforces diploidy by inhibiting the replication initiator DnaA during spore formation in Bacillus subtilis. Mol. Microbiol. 73:963–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Webb CD, et al. 1997. Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B. subtilis. Cell 88:667–674 [DOI] [PubMed] [Google Scholar]
- 62. Wu LJ, Errington J. 1994. Bacillus subtilis spoIIIE protein required for DNA segregation during asymmetric cell division. Science 264:572–575 [DOI] [PubMed] [Google Scholar]
- 63. Wu LJ, Errington J. 2004. Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell 117:915–925 [DOI] [PubMed] [Google Scholar]
- 64. Wu LJ, Errington J. 2003. RacA and the Soj-Spo0J system combine to effect polar chromosome segregation in sporulating Bacillus subtilis. Mol. Microbiol. 49:1463–1475 [DOI] [PubMed] [Google Scholar]
- 65. Wu LJ, Errington J. 1998. Use of asymmetric cell division and spoIIIE mutants to probe chromosome orientation and organization in Bacillus subtilis. Mol. Microbiol. 27:777–786 [DOI] [PubMed] [Google Scholar]
- 66. Wu LJ, et al. 2009. Noc protein binds to specific DNA sequences to coordinate cell division with chromosome segregation. EMBO J. 28:1940–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Youngman PJ, Perkins JB, Losick R. 1983. Genetic transposition and insertional mutagenesis in Bacillus subtilis with Streptococcus faecalis transposon Tn917. Proc. Natl. Acad. Sci. U. S. A. 80:2305–2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.