Maintenance of genetic integrity is of foremost importance to the cell. Thus, cells are well equipped with a number of repair mechanisms for different types of DNA damage. All DNA polymerases are very sensitive to pyrimidine dimers, a major DNA lesion of UV irradiation. Inhibition of DNA replication by dimers, if not recovered, causes cell death. Escherichia coli cells have at least five different ways to deal with dimers (1). Dimers can be directly reversed by a reaction (photoreactivation) catalyzed by photolyase or can be removed by UvrABC-catalyzed excision repair. The remaining dimers can be tolerated by the replication machinery in different mechanisms. A modified DNA polymerase is able to replicate past noninstructive dimers by a mechanism termed translesion replication (1). Major mechanisms of dimer tolerance, however, involve two recombinational repair activities. The first of the two is RecF-dependent postreplication gap repair. It was demonstrated that DNA replication stalls at a dimer but can restart downstream of the dimer (2, 3). Thus, newly synthesized DNA after UV irradiation contains single-strand (ss) gaps opposite dimers (2). These gaps are subsequently filled by a mechanism involving homologous recombination (4). The second recombinational repair activity is RecBCD-dependent double-strand break (DSB) repair. It is known that a significant fraction of UV-induced lesions is converted to DSBs (5), which can be repaired by a mechanism that requires homologous recombination functions (6). A crucial aspect of recombinational repair is the extent to which the repair process involves DNA replication. Recently, RecBCD-dependent DSB repair was shown to require extensive semiconservative DNA replication that initiates at a D-loop, a recombination intermediate (6). Now, the paper by Courcelle et al. (7) in this issue of the Proceedings suggests that RecF protein, previously identified as a recombination protein, may assist replisome reassembly after UV irradiation. The proposal opens the door to the possibility that RecF-dependent postreplication gap repair also might involve extensive DNA replication.
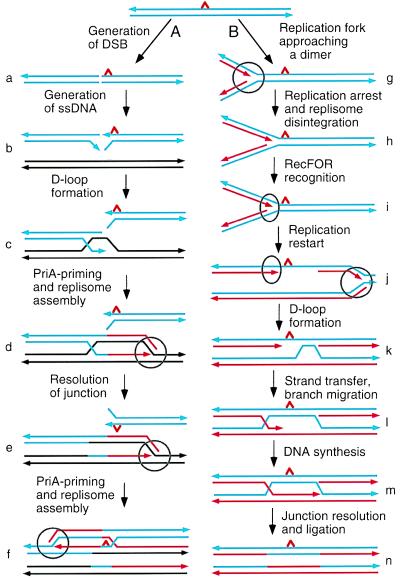
The two mechanisms of recombinational repair are illustrated in Fig. 1. The double-strand (ds) DNA ends generated as a result of DSB that is induced near a dimer are recognized by the RecBCD enzyme (Fig. 1a) (6). This enzyme is a powerful nuclease that efficiently degrades dsDNA. Under conditions of SOS induction, however, the degrading nuclease activity is inhibited. The helicase activity of RecBCD, on the other hand, is not inhibited under these conditions and is expected to unwind the duplex to yield ssDNA tails (Fig. 1b). RecA protein assimilates the 3′-OH ssDNA tail of one end, forming a D-loop (Fig. 1c). PriA protein then recognizes the D-loop and binds to it. This triggers a series of reactions leading to loading of DnaB helicase and DnaG primase onto the structure. This process, known as priming, is essential for establishing a replication fork that requires assembly of a dimeric replisome (Fig. 1d). At the replication fork, DnaB helicase unwinds the duplex ahead of a replisome (DNA polymerase III holoenzyme) and DnaG primase synthesizes RNA primers for lagging-strand DNA synthesis. The junction created as a result of D-loop formation must be resolved by the junction-specific nuclease, RuvC, with aid of the RuvAB and RecG branch-migration helicases (Fig. 1e). While the replication fork thus established replicates in one direction, the ssDNA tail of the other end invades one of the two duplexes that are just produced. The D-loop formation subsequently leads to establishment of a second replication fork that replicates in the opposite direction (Fig. 1f). Repeating the process would help generate one intact dimer-free chromosome.
Figure 1.
Two pathways for recombinational repair. (A) RecBCD-dependent DSB repair (6). (B) RecF-dependent postreplication gap repair (9). Blue parallel lines designate duplex DNA with a pyrimidine dimer (a red triangle) and black lines a homolog duplex. Small arrowheads indicate the 3′ ends of strands. Red lines, newly synthesized strands. Circles, replisomes. Ovals, RecFOR protein complexes. See the text for detail.
Another way to generate dimer-free chromosome is postreplication gap repair (Fig. 1b) (8, 9). When a replisome encounters a dimer, replication is arrested (Fig. 1h). However, replication can resume downstream of the site, leaving a gap opposite the dimer (Fig. 1j). RecA protein then polymerizes on ssDNA at the gap and promotes homologous pairing and strand exchange with the intact strand of the daughter duplex, forming a D-loop (Fig. 1k). Extended strand exchange allows the 3′-OH end of the newly synthesized DNA strand to invade the structure, creating a Holliday junction (Fig. 1l). The gap now can be successfully filled in by DNA synthesis (Fig. 1m), and subsequent junction resolution and ligation would complete gap filling (Fig. 1n).
RecF protein forms a complex (RecFOR) with RecR and RecO, which is involved in homologous recombination in the RecF pathway activated in recBC sbcBC mutants (ref. 10 and references therein). recF, recO, and recR mutants are hypersensitive to UV radiation. The notion that these gene products are involved in repair of UV damage in a manner other than a recombinational capacity was inferred from the fact that in otherwise wild-type genetic backgrounds, despite their UV hypersensitivity, these mutants are as proficient in homologous recombination as recF+, recO+, and recR+ cells. The effect of recF mutations on repair of UV damage appears to be 2-fold. One effect is a delay in the restart of DNA replication after UV irradiation (7, 11); the other is inhibition of gap filling (11). Courcelle et al. (7) suggest that when DNA replication is arrested at a dimer, the replisome disintegrates (Fig. 1h). A RecFOR complex recognizes the replication fork structure and protects it from nuclease degradation (Fig. 1i). The authors further propose that RecFOR helps reassembly of a replisome, which reestablishes a replication fork at the site. For subsequent replication to occur, of course, the dimer must be removed by excision repair.
I would suggest that RecFOR might be involved in assembly of a replisome at the D-loop for initiation of semiconservative DNA replication, just as such replication is thought to be initiated at a D-loop in the DSB repair process (Fig. 1d). It is known that postreplication recombinational gap repair requires DnaB helicase (12), DnaG primase (13), and DNA polymerase III (14), all of which are essential components of a replication fork. Furthermore, priA null mutants, which are defective in priming at a D-loop for semiconservative DNA replication (15), are hypersensitive to UV radiation (6, 16). These observations are consistent with such a proposal.
Two replication forks established at the time of initiation at oriC carry out bidirectional replication and are presumed to each replicate about 2.4 mega base pairs of the chromosome, demonstrating an extraordinary degree of processivity. There are some indications, however, that a replication fork stalls at some frequency and disintegrates during a normal course of replication (17). Courcelle et al. (7) report results that suggest that RecFOR may catalyze reassembly of a replisome, should it disintegrate. Previously, Marians and coworkers (18) suggested that PriA-mediated priming is needed to reestablish a replication fork at a site of fork collapse during chromosome replication. Thus, RecFOR and PriA-mediated priming appear to have overlapping functions in reassembly of a replisome. This notion is strongly supported by the observation that a combination of recF and priA null mutations is lethal (10). Further, the recF priA lethality can be suppressed by a mutation in the dnaC gene, which was shown to rectify the defect of the priA null mutant in priming at D-loop (10). The role of RecF and PriA proteins in reassembly of a replisome during chromosome replication is in line with the above proposal that postreplication recombinational gap repair might involve extensive semiconservative DNA replication.
Homologous recombination in the RecBCD pathway was recently demonstrated to require extensive DNA replication (19). The studies by Courcelle et al. (7) raise the possibility that homologous recombination in the RecF pathway also involves extensive DNA replication. It should be noted that the priA null mutation severely inhibits homologous recombination in recBC sbcBC cells, suggesting an involvement of PriA-mediated priming in the process (6). It is not unlikely that involvement of extensive DNA replication in homologous recombination and repair of DNA damage is a strategy commonly used in many organisms.
References
- 1.Sancar A, Sancar G B. Annu Rev Biochem. 1988;57:29–67. doi: 10.1146/annurev.bi.57.070188.000333. [DOI] [PubMed] [Google Scholar]
- 2.Rupp W D, Howard-Flanders P. J Mol Biol. 1968;31:291–304. doi: 10.1016/0022-2836(68)90445-2. [DOI] [PubMed] [Google Scholar]
- 3.Khidhir M A, Casaregola S, Holland I B. Mol Gen Genet. 1985;199:133–140. doi: 10.1007/BF00327522. [DOI] [PubMed] [Google Scholar]
- 4.Rupp W D, Wilde C E, III, Howard-Flanders P. J Mol Biol. 1971;61:25–44. doi: 10.1016/0022-2836(71)90204-x. [DOI] [PubMed] [Google Scholar]
- 5.Bonura T, Smith K C. J Bacteriol. 1975;121:511–517. doi: 10.1128/jb.121.2.511-517.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kogoma T, Cadwell G C, Barnard K G, Asai T. J Bacteriol. 1996;178:1258–1264. doi: 10.1128/jb.178.5.1258-1264.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Courcelle J, Carswell-Crumpton C, Hanawalt P C. Proc Natl Acad Sci USA. 1997;94:3714–3719. doi: 10.1073/pnas.94.8.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.West S C, Cassuto E, Howard-Flanders P. Nature (London) 1981;290:29–33. doi: 10.1038/290029a0. [DOI] [PubMed] [Google Scholar]
- 9.Mandal T N, Mahdi A A, Sharples G J, Lloyd R G. J Bacteriol. 1993;175:4325–4334. doi: 10.1128/jb.175.14.4325-4334.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sandler S J. Mol Microbiol. 1996;19:871–880. doi: 10.1046/j.1365-2958.1996.429959.x. [DOI] [PubMed] [Google Scholar]
- 11.Rothman R H, Clark A J. Mol Gen Genet. 1977;155:279–286. doi: 10.1007/BF00272806. [DOI] [PubMed] [Google Scholar]
- 12.Johnson R C. In: Molecular Mechanisms for Repair of DNA. Hanawalt P C, Setlow R B, editors. New York: Plenum; 1975. pp. 325–329. [Google Scholar]
- 13.Johnson R H. Biochem Biophys Res Commun. 1976;70:791–796. doi: 10.1016/0006-291x(76)90661-6. [DOI] [PubMed] [Google Scholar]
- 14.Sedgwick S G, Bridges B A. Nature (London) 1974;249:348–349. doi: 10.1038/249348a0. [DOI] [PubMed] [Google Scholar]
- 15.Masai H, Asai T, Kubota Y, Arai K, Kogoma T. EMBO J. 1994;13:5338–5346. doi: 10.1002/j.1460-2075.1994.tb06868.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee E H, Kornberg A. Proc Natl Acad Sci USA. 1991;88:3029–3032. doi: 10.1073/pnas.88.8.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bierne H, Michel B. Mol Microbiol. 1994;13:17–23. doi: 10.1111/j.1365-2958.1994.tb00398.x. [DOI] [PubMed] [Google Scholar]
- 18.Nurse P, Zavitz K H, Marians K J. J Bacteriol. 1991;173:6686–6693. doi: 10.1128/jb.173.21.6686-6693.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kogoma T. Cell. 1996;85:625–627. doi: 10.1016/s0092-8674(00)81229-5. [DOI] [PubMed] [Google Scholar]