Abstract
Macromolecular structures such as the bacterial flagellum in Gram-negative bacteria must traverse the cell wall. Lytic transglycosylases are capable of enlarging gaps in the peptidoglycan meshwork to allow the efficient assembly of supramolecular complexes. We have previously shown that in Rhodobacter sphaeroides SltF, the flagellar muramidase, and FlgJ, a flagellar scaffold protein, are separate entities that interact in the periplasm. In this study we show that the export of SltF to the periplasm is dependent on the SecA pathway. A deletion analysis of the C-terminal portion of SltF shows that this region is required for SltF-SltF interaction. These C terminus-truncated mutants lose the capacity to interact with themselves and also bind FlgJ with higher affinity than does the wild-type protein. We propose that this region modulates the interaction with the scaffold protein FlgJ during the assembly process.
INTRODUCTION
Motility has given bacteria an evolutionary advantage and is therefore widespread in nature. The assembly of the cell-spanning flagellum is costly for the bacterial cell and is tightly regulated (2, 5). The bacterial flagellum can be divided into three substructures: a basal body that acts as a motor, a filament that acts as a propeller, and a universal joint also known as the hook, which links the basal body and the filament. The electrochemical gradient drives rotation of the motor that generates the thrust needed to propel the bacterial cell. Biogenesis of the bacterial flagellum requires the hierarchical expression of more than 50 genes, and the assembly of this structure proceeds outwardly from the proximal to the distal end (6, 22, 34). The basal body is composed in Gram-negative bacteria of an axial rod and three ring-like structures named the MS, P, and L rings. The MS ring is embedded in the cytoplasmic membrane and interacts with the C ring that houses the export apparatus, which acts as a switch and interacts with the stator proteins (28, 40). The rod is a heterogeneous filamentous structure composed by four proteins: FlgB, FlgC, FlgF, and FlgG (14). The P and L rings are attached to the peptidoglycan (PG) layer and outer membrane, respectively, and also surround the axial rod. At a certain point in the assembly process, the PG layer, a mesh-like structure of glycan chains of alternating N-acetyl muramic acid and N-acetylglucosamine cross-linked by interpeptide bonds (36) that has an average mesh diameter of ca. 4 nm (23), must be penetrated by the rod, which has a diameter of ca. 11 nm (33). FlgJ, which is required for rod assembly (19) in beta- and gammaproteobacteria, has a dual function: acting as a scaffold for rod assembly and also acting as a muramidase degrading the PG layer to facilitate rod penetration (13, 27). Although the muramidase activity of FlgJ is important for flagellum biogenesis, it is not absolutely required for the assembly process, given that mutants that lack this activity show a leaky phenotype (13). This was explained based on the idea that, occasionally, the nascent structure finds a gap in the peptidoglycan layer; when this happens, a complete flagellum is assembled.
It has been reported that only beta- and gammaproteobacteria possess FlgJ homologues that contain a muramidase domain at the C terminus (25). We have previously reported that FlgJ from Rhodobacter sphaeroides lacks the muramidase domain (11). At least three studies have been performed that analyzed FlgJ homologues from various bacteria, and it was noticed that various alphaproteobacteria, including R. sphaeroides, possess FlgJ homologues that lack the C-terminal muramidase domain (8, 25, 39). In R. sphaeroides, FlgJ is similar to the N-terminal part of FlgJ from Salmonella enterica, suggesting that this polypeptide of 100 residues accomplishes the same function as the N-terminal domain of FlgJ from S. enterica, which acts as a scaffolding rod-capping protein (11, 13). In accordance with this, an flgJ mutant in R. sphaeroides has a Fla− phenotype (13), indicating that the canonical domains of S. enterica FlgJ can act separately in other species. We have previously demonstrated the presence of a flagellum-specific muramidase in R. sphaeroides, SltF, that is encoded within the flgG operon (8). It is possible that this protein is exported to the periplasm via the SecA pathway, where it would interact with FlgJ and would open a gap in the PG layer. In the present study, we investigated whether the C-terminal region of SltF is involved in the interaction with FlgJ.
MATERIALS AND METHODS
Strains, plasmids, and oligonucleotides.
The bacterial strains, plasmids, and oligonucleotides used in the present study are listed in Table 1.
Table 1.
Bacterial strains, plasmids, and oligonucleotides
| Strain, plasmid, or oligonucleotide | Relevant characteristics or sequence (5′–3′)a | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| JM103 | hsdR4 Δ(lac-pro) F′ traD36 proAB lacIqZΔM15 | 1 |
| M15[pREP4] | thi lac ara+ gal mtl F′ recA+ uvr+ lon+; pREP4 plasmid; Kanr | Qiagen |
| BL21(DE3)/pLysS | F′ ompT hsdSB(rB− mB−) gal dcm(DE3)/pLysS | Novagen |
| DH5α | supE44 ΔlacU169(ϕ80dlacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 12 |
| S17-1 | recA endA thi hsdR RP4-2-Tc::Mu::Tn7; Tpr Smr Kan::Tn7 | 30 |
| R. sphaeroides | ||
| WS8-N | Wild type; spontaneous; Nalr | 32 |
| SltF1 | WS8-N derivative ΔsltF(1–336)::aadA Fla−; Spcr Nalr | 8 |
| RsgJ-np | WS8-N derivative flgJ::aadA Fla−; Spcr Nalr | 11 |
| S. cerevisiae AH109 | Yeast reporter strain, for HIS3, ADE2, and lacZ | Clontech |
| Plasmids | ||
| pQE30 | Expression vector; Ampr N-terminal His6 tag | Qiagen |
| pQE60 | Expression vector; Ampr C-terminal His6 tag | Qiagen |
| pRSJ | flgJ cloned into the NcoI/BglII sites of pQE60 | 8 |
| pRS1sltF | sltF cloned into the SacI/HindIII sites of pQE30 | This study |
| pRsltFΔ4 | pQE30 derivative carrying, sltFΔ(723–795) | This study |
| pRsltFΔ5 | pQE30 derivative, carrying sltFΔ(651–723) | This study |
| pRsltFΔ6 | pQE30 derivative, carrying, sltFΔ(651–795) | This study |
| pRKsltF | 1.4-kb PstI/PstI fragment carrying sltF cloned into the EcoRI/HindIII sites of pRK415 | 8 |
| pRKsltFE57A | sltF2(E57A) cloned into the EcoRI/HindIII sites of pRK415 | This study |
| pRKsltFΔ4 | sltFΔ(723–795) cloned into the EcoRI/HindIII sites of pRK415 | This study |
| pRKsltFΔ5 | sltFΔ(651–723) cloned into the EcoRI/HindIII sites of pRK415 | This study |
| pRKsltFΔ6 | sltFΔ(651–795) cloned into the EcoRI/HindIII sites of pRK415 | This study |
| pTZ19R/18R | pUC derivative multifunctional plasmid carrying lacZ gene for screening; Ampr | Pharmacia |
| pRK415 | pRK404 derivative; used for expression on R. sphaeroides under the lac promoter; lacZ mob+; Tcr | 18 |
| pIND4 | IPTG-inducible expression vector for R. sphaeroides; Kanr | 16 |
| pINSltF | sltF wild type cloned into the NcoI/BglII sites of pIND4 | This study |
| pINSltFΔSP | sltF wild type without SEC signal cloned into the NcoI/BglII sites of pIND4 | This study |
| pINSltFΔ4 | sltFΔ(723–795) cloned into the NcoI/HindIII sites of pIND4 | This study |
| pINSltFΔ5 | sltFΔ(651–723) cloned into the NcoI/HindIII sites of pIND4 | This study |
| pINSltFΔ6 | sltFΔ(651–795) cloned into the NcoI/HindIII sites of pIND4 | This study |
| psltFΔ5 | 1.4-kb PstI fragment carrying sltFΔ(651–723) cloned into pTZ19R; Ampr | This study |
| pGT001 | 1.4-kb PstI fragment from pBG313 subcloned into pTZ19R; Ampr | 11 |
| pGADT7 | GAL4 activation domain, LEU2 | Clontech |
| pGBKT7 | GAL4 DNA binding domain TRP1 | Clontech |
| pGAD-SltF | pGADT7 derivative expressing GAL4 AD-sltF | This study |
| pGAD-SltFΔ4 | pGADT7 derivative expressing GAL4 AD-sltFΔ(723–795) | This study |
| pGAD-SltFΔ5 | pGADT7 derivative expressing GAL4 AD-sltFΔ(651–723) | This study |
| pGAD-SltFΔ6 | pGADT7 derivative expressing GAL4 AD-sltFΔ(651–795) | This study |
| pGBD-SltF | pGBKT7 derivative expressing GAL4 BD-sltF | This study |
| pGBD-SltFΔ4 | pGBKT7 derivative expressing GAL4 BD-sltFΔ(723–795) | This study |
| pGBD-SltFΔ5 | pGBKT7 derivative expressing GAL4 BD-sltFΔ(651–723) | This study |
| pGBD-SltFΔ6 | pGBKT7 derivative expressing GAL4 BD-sltFΔ(651–795) | This study |
| pGADT7-FlgJ | pGADT7 derivative expressing GAL4 AD-flgJ | This study |
| pGBKT7-FlgJ | pGBKT7 derivative expressing GAL4 BD-flgJ | This study |
| Oligonucleotides | ||
| sltFfwsec+ | CATGCCATGGCACGGCCCTTGCCCGCG | This study |
| sltFfwsec− | CATGCCATGGCGGACGAGGGCTGCGAGACG | This study |
| fw1 | CTGATCTAGACCCTCCGGCCCCGGCCACGGTG | This study |
| sltFrv | GGAAGATCTCGGTTGCATTGCGAGCAGGTC | This study |
| orf72fw | CATGGAGCTCGCGGACGAGGGCTGCGAGACG | This study |
| orf72rv | CCCGAAGCTTTCACGGTTGCATTGCGAGCAG | This study |
| orf72rvΔ24 | CCCGAAGCTTTCACGCCTCGGCGAGGAGGTCCGG | This study |
| orf72rvΔ48 | CCCGAAGCTTTCACCGGGGCAGCCCTCCCTGCGT | This study |
| orf72fwΔ24int | CGCAGGGAGGGCTGCCCCGGGACCTTCCGCAGCGCCTGCG | This study |
| orf72rvΔ24int | CGCAGGCGCTGCGGAAGGTCCCGGGGCAGCCCTCCCTGCG | This study |
| orf72rv | CCCGAAGCTTTCACGGTTGCATTGCGAGCAG | This study |
| orf72fw57A | GGCGATTGCCCGCGTGGCGTCGGGCCGGGGCGGGC | This study |
| orf72rv57A | GCCCGCCCCGGCCCGACGCCACGCGGGCAATCGCC | This study |
| ADBDsltFfw | CCTGCATATGGCGGACGAGGGCTGCGAG | This study |
| ADDBsltFrv | CCTGGGATCCTCACGGTTGCATTGCGAG | This study |
| ADBDflgJfw | CCTGCATATGGAACTGAAGCTTCAGGCC | This study |
| ADBDFlgJrv | CCTGGGATCCTCACGACTTGCCGTCCCT | This study |
| ADBDsltF24rv | CCTGGGATCCTCACGCCTCGGCGAGGAG | This study |
| ADBDsltF48rv | CCTGGGATCCTCACCGGGGCAGCCCTCC | This study |
Smr, streptomycin resistance; Nalr, nalidixic acid resistance; Tpr, trimethoprim resistance; Tcr, tetracycline resistance; Ampr, ampicillin resistance; Spcr, spectinomycin resistance; Kanr, kanamycin resistance.
Media and growth conditions.
Sistrom's culture medium (31) was used to grow R. sphaeroides at 30°C under constant illumination in completely filled screw-cap tubes. When required, the following antibiotics were added at the indicated concentrations: nalidixic acid, 20 μg/ml; spectinomycin, 50 μg/ml; tetracycline, 1 μg/ml; and kanamycin, 25 μg/ml. Strains of Escherichia coli were grown in Luria-Bertani medium (1). When needed, antibiotics were added at the following concentrations: spectinomycin, 50 μg/ml; kanamycin, 50 μg/ml; tetracycline, 25 μg/ml; and ampicillin, 200 μg/ml. Saccharomyces cerevisiae was grown in YPDA medium (1) at 30°C or in minimal synthetic defined (SD) medium (Clontech, Mountain View, CA). Standard molecular biology techniques were used for the isolation and purification of chromosomal DNA from R. sphaeroides WS8-N (1). Plasmid DNA and PCR fragments were purified with QIAprep spin and QIAquick PCR kits, respectively (Qiagen, GmbH). The products were cloned either in pTZ19R or pTZ18R as required. DNA sequence was carried out in an ABS-Prism automatic sequencer. PCRs were carried out with PfuTurbo (Invitrogen, Carlsbad, CA), and the oligonucleotides were synthesized by Sigma-Aldrich.
Motility assays.
A 5-μl sample of a stationary-phase culture was placed on the surface of swarm plates (1), followed by aerobic incubation in the dark at 30°C. Swarming ability was recorded as the ability of bacteria to move away from the inoculation point after 24 to 36 h of incubation. Soft agar (0.25%) swimming plates were prepared with Sistrom's minimal medium devoid of succinic acid, to which 100 μM sodium propionate and 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) were added when indicated.
Overexpression and purification of SltF, SltFΔ4, SltFΔ5, SltFΔ6, and FlgJ.
For the wild-type sltF and flgJ genes, we used previously described plasmids (8). To obtain the mutants SltFΔ4 and SltFΔ6, the following oligonucleotides were used: orf72fw, orf72rvΔ24, and orf72rvΔ48. sltFΔ5 mutagenesis was carried out using the QuikChange method (Stratagene) with plasmid pGT001 as a template and the oligonucleotides orf72fwΔ24int and orf72rvΔ24int, followed by a second PCR with orf72fw and orf72rv. All of the PCR products were cloned in the overexpression vector pQE30. Overexpression and purification was carried out as described previously (8). Protein quantification was carried out by the method described by Lowry (21). Specific polyclonal gamma globulins for SltF and FliC, respectively, were produced as described previously (8, 11).
Site-directed mutagenesis and complementation assays.
The SltF point mutant (E57) was obtained using the QuikChange method (Stratagene). Glutamic acid was replaced by alanine using orf72fw57A and orf72rv57A oligonucleotides and plasmid pGT001 as a template. The resulting product was subcloned into pRK415 (18). Mutants SltFΔ4 and SltFΔ6 were obtained by PCR using the oligonucleotides fw1 or sltFfwsec+ and the oligonucleotides orf72rvΔ24 or orf72rvΔ48, respectively. SltFΔ5 was obtained by PCR from plasmid psltFΔ5. All of the products were cloned in pTZ19R and further subcloned in pRK415 and pIND4. The plasmids were introduced into a ΔsltF strain by diparental conjugation using E. coli strain S17-1 as described previously (30). The presence of SltF was verified by Western blot analysis.
Protein export analysis.
Two versions of the wild-type sltF gene were amplified by PCR; one contained the export signal sequence, and the other lacked it. For this purpose, the oligonucleotide sltFfwsec+ or sltFfwsec− was used, with sltFrv as a reverse oligonucleotide. Both products were cloned in the pIND4 plasmid and introduced into a ΔsltF mutant. Proteolysis of the exported protein was carried out as follows. Cell cultures (15 ml) were grown aerobically at 30°C to an optical density at 600 nm (OD600) of 0.35. At this point, 1 mM IPTG was added, and growth continued for 3 h. Harvested cells were washed two times with 5 ml of a buffer containing 10 mM Tris-Cl (pH 8.0) and suspended in 0.3 ml of a buffer containing 100 mM Tris-Cl (pH 8.0) and 20% sucrose. The cells were divided into two aliquots; both were treated with lysozyme (100 μg/ml) and EDTA (10 mM) for 5 min at 37°C. To one of the vials, 1.5 μl of a stock solution of proteinase K (10 mg/ml) was added. The tubes were incubated for 15 min at 37°C, and the reactions were arrested by the addition of 1.5 μl of a 1 mM solution of phenylmethylsulfonyl fluoride. The samples were analyzed by Western blotting as described previously (20).
SltF stability assays.
Wild-type and mutant strains expressing the truncated versions of SltF were grown at 30°C to an OD600 of 0.35. At this point, 1 mM IPTG was added, and growth continued for 1 h. RNA synthesis was arrested by the addition of rifampin from a stock solution (50 mg/ml) to a final concentration of 800 μg/ml. Growth was continued, and samples were drawn at different times and analyzed by Western blotting. The density of the corresponding bands was determined and plotted as a function of time. The data were adjusted to fit a first-order decay function in order to establish the degradation constant and the half-lives of the various versions of SltF (3).
FliC secretion.
Cells were grown photoheterotrophically for 16 h until reaching an OD600 of 2.0 and then centrifuged for 15 min at 16,000 × g. The supernatant was centrifuged again, and the filtered through a Millipore membrane (pore size, 22 μm). Soluble proteins in the supernatant were precipitated with chloroform-methanol, centrifuged, and suspended in sample buffer as described earlier (38). The pellet fraction was suspended in 200 μl of sample buffer. The protein concentration was determined by the method of Lowry (21), and the samples were analyzed by Western blotting.
Muramidase activity assays.
A technique known as zimodot or lisoplates was used to determine muramidase activity. Petri dishes filled with soft 1% agarose containing a cell lysate from Micrococcus lysodeikticus to be used as a substrate at a concentration of 50 mg/ml in phosphate buffer (50 mM NaH2PO4 [pH 6.5]) as described previously (2a). The lisoplates were inoculated with 20 μg of protein and incubated for 12 h at 30°C.
Immunoprecipitation.
Sepharose CL-4B coupled to protein-A (20 μl) (Sigma Chemicals) was incubated with 8 μg of anti-SltF gamma globulin in 1 ml of phosphate buffer (50 mM; pH 7.5) for 12 h at 4°C; the tube was then centrifuged, and the supernatant was discarded. To evaluate the interaction of SltF, SltFE57A, SltFΔ4, SltFΔ5, and SltFΔ6 with FlgJ, 0.07 or 0.14 μM concentrations of each protein were incubated for 1 h at 4°C before the addition of the specific anti-SltF gamma globulins attached to protein A-Sepharose. The mixture was then incubated for 60 min at 4°C and washed five times with 1 ml of phosphate buffer. The resulting pellet was suspended in 30 μl of sample buffer and boiled for 10 min. The samples were then subjected to SDS-PAGE and transferred to nitrocellulose membranes treated as described above and developed using HisProbe-HRP at a 1:10,000 dilution (Pierce Chemicals, Rockford, IL).
Yeast two-hybrid assays.
The Matchmaker GAL4 Two-Hybrid System 3 (Clontech) was used to test the interaction between SltF with FlgJ and the various versions of truncated mutants. The genes were amplified using oligonucleotides ADBDsltFfw, ADBDsltFrv, ADBDsltF24rv, ADBDsltF48rv, ADBDflgJfw, and ADBDflgJrv. For the sltFΔ5 cells, mutant plasmid psltFΔ5 was obtained by using oligonucleotides ADBDsltFfw and ADBDsltFrv. The products were cloned into pGADT7 (encoding the DNA activation domain [AD] of GAL4 and LEU2 as a selection marker) and pGBKT7 (encoding the DNA binding domain [BD] of GAL4 and TRP1 as selection marker). Different combinations of plasmids (AD+BD) were cotransformed into yeast strain AH109. Double transformants were selected as tryptophan and leucine prototrophs. After the initial selection, the different transformant strains were grown overnight in 3 ml of SD minimal medium lacking tryptophan and leucine and supplemented with histidine. Aliquots of the cultures were washed once with SD minimal medium lacking Trp, Leu, and His and then normalized to an OD600 of 0.5. Next, 10-fold serial dilutions were made in the same medium. Aliquots (10 μl) drawn from these dilutions were seeded onto selection plates lacking Trp, Leu, and His. A control plate lacking Trp and Leu was also included. The yeast strain AH109 was transformed with plasmids provided with the kit that represent positive and negative interactions. These plasmids encode the DNA BD or the AD of GAL4 fused either to p53 (murine) or to the simian virus 40 large T antigen. These plasmids were used as a control for positive interaction. As a negative control, the plasmids encoding the fusion AD-T antigen and the plasmid BD-lamin C (human) were cotransformed into strain AH109.
RESULTS
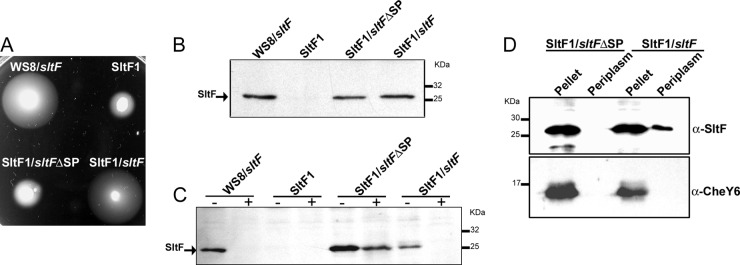
In silico analysis suggests that SltF is secreted into the periplasm via the SecA pathway (8). To obtain experimental evidence to support this prediction, we deleted the nucleotide sequence on the wild-type sltF gene that codes for the Sec export signal peptide. The protein lacking the signal peptide expressed from plasmid pINSltFΔSP was unable to complement an SltF1 mutant. On the other hand, swimming was recovered when the SltF1 mutant was complemented with the wild-type sltF allele (Fig. 1A). We carried out Western blot analyses in order to rule out the possibility that the lack of motility in the SltF1 mutant expressing the SltF Sec− protein was due to a difference in protein concentration. We found that SltF, with or without the Sec export sequence, is present in equivalent amounts in the cell (Fig. 1B). We also determined the location of the two versions of SltF by testing the accessibility of proteinase K to SltF in cells that were preincubated with lysozyme. We found that the mutant version (SltFΔSP) is preserved intact after the addition of the protease, whereas the wild-type form (SltF), which is exported to the periplasm, is completely degraded (Fig. 1C). We also carried out cell fractionation experiments that confirmed that SltF is exported to the periplasm, whereas SltFΔSP was not (Fig. 1D). For this experiment, we also verified that both protein versions were soluble in the cytoplasm and hence competent to be exported (see Fig. S1 in the supplemental material). In addition, we determined that CCCP and NaN3 inhibited the export of SltF, as expected for a protein dependent on SecA activity (see Fig. S2 in the supplemental material).
Fig 1.
(A) Phenotypic analysis of various strains of R. sphaeroides: WS8 (wild-type), SltF1 (ΔsltF), SltF1/sltFΔSP, and SltF1/sltF. The sltF alleles were cloned in vector pIND4 (for details, see Materials and Methods). Swimming assays on soft agar plates were carried out in 0.25% agar supplemented with 100 μM sodium propionate and 1 mM IPTG. The cells were grown under aerobic conditions at 30°C for 48 h. (B) Western blot analysis of the same strains shown in panel A, using anti-SltF gamma globulins. (C) Sensitivity to proteinase K of SltF with or without signal peptide analyzed by Western blotting with anti-SltF gamma globulins. Proteinase K was added where indicated (+). (D) Cell fractionation and osmotic shock were performed as described previously (10). Pellet and periplasmic fractions were analyzed by Western blotting with specific anti-SltF and CheY6 gamma globulins at dilutions of 1:2,500 and 1:300,000, respectively. SDS–12.5% PAGE was used in all cases.
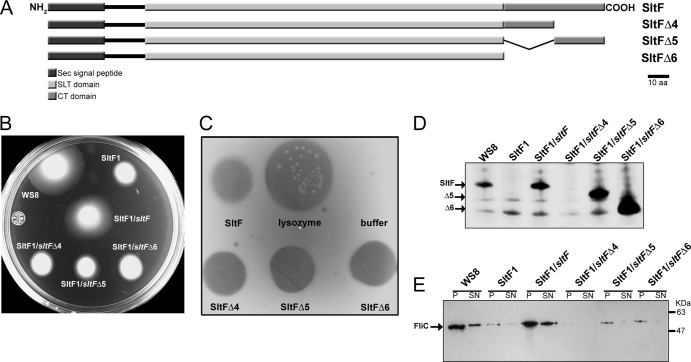
The amino acid sequence alignment of various membrane-lytic transglycosylases that belong to family 1D (4) shows a conserved domain of 171 amino acids. This conserved putative transglycosylase domain spans residues 47 to 217 of SltF from R. sphaeroides (Fig. 2). The rest of the sequence does not share homology with any known protein. This led us to carry out an analysis of the remaining 48 residues of the C terminus of SltF (Fig. 2B). Furthermore, secondary structure analysis of SltF does not show with certainty the presence of coil-coiled structures within this domain that could account for protein-protein interactions. We therefore dissected the C-terminal region of SltF by deletion of either the first 24 residues (SltFΔ4), the last 24 residues (SltFΔ5), or the complete removal of the 48 nonconserved residues of the C terminus domain (SltFΔ6) (Fig. 3A). Analysis of this region was performed by complementation of SltF1 mutants with each of the three C-terminal truncated mutant alleles. The three C-terminal deletions were unable restore swimming in soft agar plates (Fig. 3B). We therefore determined whether the three overexpressed and purified C terminus mutants were catalytically active on lysoplates containing a cell lysate from M. lysodeikticus. It was found that the three isolated mutant proteins (SltFΔ4, SltFΔ5, and SltFΔ6) showed a transglycosylase activity similar to that of the wild-type protein (Fig. 3C). In addition, we observed that these mutant versions of SltF did not exert a negative dominance effect on the swimming behavior of the wild-type strain (data not shown).
Fig 2.
(A) Schematic representation of the different functional domains proposed for SltF. (B) Amino acid alignment of various family 1D membrane lytic transglycosylases. The alignment was carried out using the program MUSCLE (9) and the following sequences: SltF R. sphaeroides (Rs), Pseudomonas aeruginosa (Pa), Vibrio cholerae (Vc), Yersinia pestis (Yp), Escherichia coli (Ec), and Klebsiella pneumoniae (Kp). Arrow indicates the conserved catalytic glutamic residue among these lytic transglycosylases. An asterisk indicates the start of the nonconserved C terminus of SltF analyzed here.
Fig 3.
(A) Schematic representation of SltF showing the Sec signal domain, the transglycosylase (SLT) domain, the C terminus domain (CT), and the three C-terminus deletions. (B) Swimming assays on soft agar plates were carried out in 0.25% agar supplemented with 100 μM sodium propionate. The strains analyzed included WS8 (wild-type), SltF1 (ΔsltF), SltF1/sltF, SltF1/sltFΔ4, SltF1/sltFΔ5, and SltF1/sltFΔ6. The various alleles of sltF were cloned in pRK415 (for details, see Materials and Methods). (C) Zimodot analysis of wild-type SltF and the three C-terminal mutant proteins. Lysozyme activity was included as a control. (D) Western blot analysis with anti-SltF gamma globulins (1:2,500 dilution) of the same strains shown in panel B. (E) Western blot analysis of the same strains with anti-flagellin gamma globulin (1:10,000 dilution). SDS–12.5% PAGE was used in all cases.
We also analyzed whether the C-terminal deletions affected the stability of the protein in R. sphaeroides. For this, the presence of the modified proteins in cell extracts was determined by means of Western blot analyses with specific anti-SltF polyclonal antibodies. As shown in Fig. 3D, the presence of either wild-type or mutant SltF was confirmed in strains WS8, SltF1/sltF, SltF1/sltFΔ5, and SltF1/sltFΔ6. It should be noted that two of the mutant proteins (SltF1/sltFΔ5 and SltF1/sltFΔ6) are very abundant, as revealed by Western blot analysis. In contrast, the deletion of the last 24 residues (SltF1/sltFΔ4) renders the protein unstable and was therefore undetectable (Fig. 3D). This finding led us to determine the half-life of the various forms of SltF, i.e., with or without the Sec export sequence, as well as the three C terminus mutants. The half-life of wild-type SltF was evaluated in a strain devoid of the scaffold protein FlgJ. The different versions of SltF were tested in assays wherein the expression of sltF was induced with IPTG and subsequently arrested by the addition of rifampin. Table 2 shows that the native SltF protein has an average half-life of 4.62 min, whereas SltF expressed in an flgJ genetic background displayed an average half-life of 7.32 min. On the other hand, the Sec− version of SltF showed an average half-life of 6.42 min, which is longer than that of the wild-type protein. The three C-terminal mutants displayed different half-life values; these ranged from 3.45 min for SltFΔ4 to 11.03 min for SltFΔ5 and 15.8 min for SltFΔ6. It should be noted that the increased stability of SltFΔ5 and SltFΔ6 is consistent with the increased abundance of protein observed for these two mutants in Fig. 3D. It was also determined that these C-terminal mutants of SltF were competent for export to the periplasm (data not shown).
Table 2.
Half-lives of different versions of SltF
| SltF version | Mean half-life (min) ± SD (n = 3) |
|---|---|
| SltF | 4.62 ± 0.34 |
| SltF (Δsec) | 6.42 ± 0.21 |
| SltF (ΔflgJ) | 7.32 ± 0.69 |
| SltFΔ4 | 3.45 ± 0.242 |
| SltFΔ5 | 11.03 ± 0.87 |
| SltFΔ6 | 15.80 ± 0.37 |
The lack of complementation observed in swimming assays (Fig. 3B) was correlated with the absence of flagellin (FliC) in exconjugant cells. FliC was present in either pellet or supernatant fractions of wild-type WS8 and SltF1/sltF cells, whereas it was barely detectable in the pellets of SltF1/sltFΔ5 and SltF1/sltFΔ6 cells and absent in SltF1/sltFΔ4 (Fig. 3E).
It was important to determine whether the three SltF C-terminal mutants retained the ability to interact with FlgJ. Therefore, we carried out pulldown and yeast double-hybrid assays. Figure 4A shows that both SltF and the three C-terminal mutants (SltFΔ4, SltFΔ5, and SltFΔ6) bound to FlgJ. It should be noted that the signal obtained for the three mutants is stronger than the signal observed for the wild-type protein. The interaction of the various mutant proteins was further confirmed in a yeast double-hybrid assay. For this assay, the activation domain or the DNA BD of GAL4 was fused to SltF or FlgJ or to the C-terminal mutants of SltF. A positive interaction was detected as prototrophy for histidine compared to the growth of yeast expressing pairs of proteins that interact or not (see Material and Methods for details). Figure 4B shows that two of the C-terminal mutants (SltFΔ5 and SltFΔ6) interacted with FlgJ with a higher affinity than the interaction displayed by wild-type SltF with FlgJ. This assay also showed that SltF readily interacts with itself. This finding is in contrast to the lack of interaction between these mutants. We analyzed by size-exclusion chromatography whether SltF was able to form multimeric complexes. The results showed under our experimental conditions that SltF exists in solution mainly as a dimer (data not shown).
Fig 4.
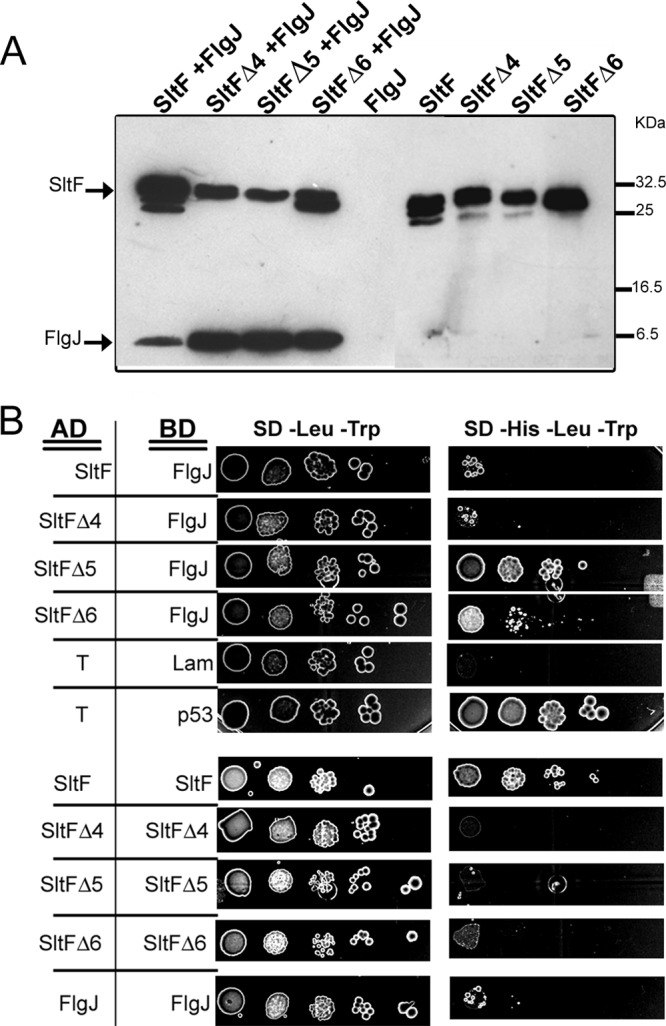
Interaction of various SltF versions with FlgJ. (A) Coimmunoprecipitation of wild-type SltF and three C-terminal mutants in the presence or absence of FlgJ. Anti-SltF gamma globulins were coupled to Protein A-Sepharose beads and tested for binding with the proteins that are indicated. The SDS-PAGE gel used for this analysis was 17.5%. Proteins were detected using HisProbe-HRP at a 1:10,000 dilution (for details, see Materials and Methods). (B) Yeast double-hybrid assays. Different versions of SltF and FlgJ were cloned in plasmids pGADT7 and pGBKT7. pGADT7-T and pGBKT7-Lam were used as negative controls, and PGADT7-T and pGBKT7-p53 were used as positive controls. Serial dilutions of cells were inoculated on culture dishes containing media lacking leucine and tryptophan (−Leu −Trp), and lacking tryptophan, leucine, and histidine. The plates were incubated for 14 days at 30°C.
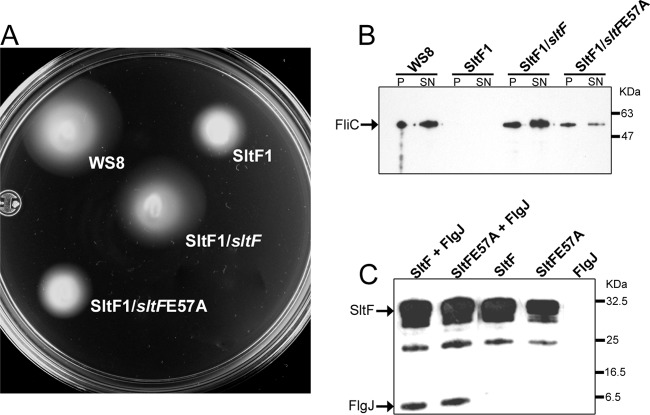
We had previously found that a single point mutation of a glutamic acid (E57A) in SltF inhibits transglycosylase enzymatic activity in an in vitro assay (8). We carried out in vivo analyses of this point mutant (SltFE57A) by complementation of the SltF1 mutant. Figure 5A shows that SltFE57A is unable to restore motility in an sltF mutant. We also determined the presence of flagellin (FliC) in the pellets and supernatants of strains expressing SltFE57A by Western blot analysis (Fig. 5B). It should be noted that the mutant strain SltFE57A, which was unable to complement swimming, shows a reduced amount of FliC in the pellet and supernatant compared to the wild-type or complemented strains. Given that the phenotype of this point mutant is Fla−, it was important to test the ability of SltFE57A to interact with FlgJ. Figure 5C shows that the interaction of either the wild-type or the mutant SltF (SltFE57A) with FlgJ is equivalent for the two SltF proteins.
Fig 5.
Characterization of a point mutation of SltF. (A) Swimming assay on 0.25% soft agar plates supplemented with 100 μM sodium propionate. Portions (3 μl) of the strains WS8, SltF1, SltF1/sltF, SltF1/sltFE57A were inoculated onto soft agar plates, followed by incubation as described in Materials and Methods. The various versions of sltF were cloned in pRK415 (for details, see Materials and Methods). (B) Flagellin (FliC) secretion analyzed by Western blot analysis. The same strains were grown and fractionated in either pellet (P) or supernatant (SN) and then probed with specific gamma globulins at a 1:10,000 dilution. (C) Coimmunoprecipitation assay. Anti-SltF gamma globulins were coupled to protein A-Sepharose beads, followed by incubation with the indicated proteins. These were detected using HisProbe-HRP at a 1:10,000 dilution as indicated in Materials and Methods.
DISCUSSION
In this study we show that SltF from R. sphaeroides is exported to the periplasm by the SecA pathway. In E. coli and Salmonella, the majority of flagellar proteins are exported by the type III flagellar specific export system (fT3SS) (24). The exceptions are the P-ring scaffolding protein, FlgA, as well as FlgI and FlgH, which are proteins that conform the P and L rings, respectively. These proteins are also exported to the periplasm through the SecA pathway (17, 26). It should also be noted that in these bacteria, FlgJ contains the rod-scaffolding domain at the N-terminal region and a muramidase domain at the C terminus; this arrangement allows its exportation by the fT3SS. Analysis of the genome sequence of other bacteria, such as Silicibacter pomeyori and Bradyrhizobium japonicum, has shown that similar, to our observations in R. sphaeroides, flgJ encodes a single domain scaffolding protein (8, 25). In addition, in these species, a gene encoding a putative lytic muramidase is located in a flagellar context (8). These putative muramidases also show a predictable signal peptide, suggesting that they could be exported to the periplasm using the SecA pathway. However, not all of the genes encoding potential flagellar single domain muramidases contain a coding sequence for a signal peptide, indicating that they are exported either by the fT3SS or through a different export pathway. For example, it has been reported that PleA, the flagellar muramidase of Caulobacter crescentus, is translocated by the ABC pathway (35).
The presence of specific muramidases is not restricted to the flagellar system. These enzymes are also required for pilus formation and for many of the various secretion systems reported in different bacteria (29). Recently, it was shown that EtgA, the specific muramidase of the type III secretion system of enteropathogenic E. coli is secreted to the periplasm via the Sec pathway (10). It should be noted that EtgA interacts with EscI (rOrf8), which constitutes part of the inner rod of the injectisome (7). In this respect, muramidases or lytic transglycosylases usually interact with other proteins that could exert a physical constrain to control enzymatic activity, such as occurs with VirB1 in the type IV secretion system, which interacts with VirB8, VirB9, and VirB11, core components of this secretion system (15).
In a previous study, we reported that the purified mutant version of SltF (SltF E57A) did not show enzymatic activity when tested in vitro (lisoplates) (8). On the other hand, we show here that SltF E57A cannot complement the ΔsltF mutant strain. However, it was still possible to detect a low amount of flagellin in the culture medium, indicating that this mutant protein still retains a low level of activity. In addition, we also show that SltF E57A is still able to interact with FlgJ, as wild-type SltF does, indicating that this interaction is not affected by the change in the catalytic domain of SltF.
Likewise, we have previously shown that SltF interacts with FlgJ in pulldown assays (8). In the present study, we specifically tested whether the C-terminal region of SltF is involved in this interaction. Using two different assays, we found that the deletion of the C-terminal region of SltF improved SltF-FlgJ interaction. Therefore, these results indicate that the C-terminal domain of SltF exerts a negative effect on SltF-FlgJ interaction. Nonetheless, this same region favors SltF-SltF interaction. This apparent contradiction could be explained if the C-terminal region of SltF in R. sphaeroides prevents a strong interaction with FlgJ, thus making this interaction transitory for a successful flagellar assembly to take place. On the other hand, the lack of either one of the C-terminal sections results in a defective export of flagellin; this suggests that SltFΔ5 and SltFΔ6 do not support an efficient flagellar assembly. Given that these versions of SltF have an intact muramidase domain, as shown in the activity assay, we propose that the C-terminal region of SltF modulates the activity of this protein, perhaps through its interaction with FlgJ.
Supplementary Material
ACKNOWLEDGMENTS
We thank Aurora Osorio for helpful technical assistance and the IFC Molecular Biology Unit for sequencing facilities.
This study was supported by grants from CONACyT (grant 106081) and DGAPA/UNAM (IN206811-3).
Footnotes
Published ahead of print 15 June 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Ausubel FM, et al. 1987. Current protocols in molecular biology. John Wiley & Sons, Inc, New York, NY [Google Scholar]
- 2. Bardy SL, Ng SY, Jarrell KF. 2003. Prokaryotic motility structures. Microbiology 149:295–304 [DOI] [PubMed] [Google Scholar]
- 2a. Becktel WJ, Basse WA. 1985. A lysoplate assay for Escherichia coli cell wall-active enzymes. Anal. Biochem. 150:258–263 [DOI] [PubMed] [Google Scholar]
- 3. Belle A, Tanay A, Bitincka L, Shamir R, OŚhea EK. 2006. Quantification of protein half-lives in the budding yeast proteome Proc. Natl. Acad. Sci. U. S. A. 103:13004–13009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blackburn NT, Clarke AJ. 2001. Identification of four families of peptidoglycan lytic transglycosylases. J. Mol. Evol. 52:78–84 [DOI] [PubMed] [Google Scholar]
- 5. Chevance FF, Hughes KT. 2008. Coordinating assembly of a bacterial macromolecular machine. Nat. Rev. Microbiol. 6:455–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chilcott GS, Hughes KT. 2000. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64:694–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Creasey EA, Delahay RM, Daniell SJ, Frankel G. 2003. Yeast two-hybrid system survey of interactions between LEE-encoded proteins of enteropathogenic Escherichia coli. Microbiology 149:2093–2106 [DOI] [PubMed] [Google Scholar]
- 8. de la Mora J, Ballado T, González-Pedrajo B, Camarena L, Dreyfus G. 2007. The flagellar muramidase from the photosynthetic bacterium Rhodobacter sphaeroides. J. Bacteriol. 189:7998–8004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. García-Gómez E, Espinosa N, de la Mora J, Dreyfus G, González-Pedrajo B. 2011. The muramidase EtgA from enteropathogenic Escherichia coli is required for efficient type III secretion. Microbiology 157:145–1160 [DOI] [PubMed] [Google Scholar]
- 11. González-Pedrajo B, de la Mora J, Ballado T, Camarena L, Dreyfus G. 2002. Characterization of the flgG operon of Rhodobacter sphaeroides WS8 and its role in flagellum biosynthesis. Biochim. Biophys. Acta 1579:55–63 [DOI] [PubMed] [Google Scholar]
- 12. Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557–580 [DOI] [PubMed] [Google Scholar]
- 13. Hirano T, Minamino T, Macnab RM. 2001. The role in flagellar rod assembly of the N-terminal domain of Salmonella FlgJ, a flagellum-specific muramidase. J. Mol. Biol. 312:359–369 [DOI] [PubMed] [Google Scholar]
- 14. Homma M, Kutsukake K, Hasebe M, Iino T, Macnab RM. 1990. FlgB, FlgC, FlgF, and FlgG: a family of structurally related proteins in the flagellar basal body of Salmonella typhimurium. J. Mol. Biol. 211:465–477 [DOI] [PubMed] [Google Scholar]
- 15. Hoppner C, Carle A, Sivanesan D, Hoeppner S, Baron C. 2005. The putative lytic transglycosylase VirB1 from Brucella suis interacts with the type IV secretion system core components VirB8, VirB9, and VirB11. Microbiology 151:3469–3482 [DOI] [PubMed] [Google Scholar]
- 16. Ind AC, et al. 2009. Inducible-expression plasmid for Rhodobacter sphaeroides and Paracoccus denitrificans. Appl. Environ. Microbiol. 75:6613–6615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jones CJ, Homma M, Macnab RM. 1989. L-, P-, and M-ring proteins of the flagellar basal body of Salmonella typhimurium: gene sequences and deduced protein sequences. J. Bacteriol. 171:3890–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Keen NT, Tamaki S, Kobayashi D, Trollinger D. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191–197 [DOI] [PubMed] [Google Scholar]
- 19. Kubori T, Shimamoto N, Yamaguchi S, Namba K, Aizawa S. 1992. Morphological pathway of flagellar assembly in Salmonella typhimurium. J. Mol. Biol. 226:433–446 [DOI] [PubMed] [Google Scholar]
- 20. Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 21. Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265–275 [PubMed] [Google Scholar]
- 22. Macnab RM. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol. 57:77–100 [DOI] [PubMed] [Google Scholar]
- 23. Meroueh SO, et al. 2006. Three-dimensional structure of the bacterial cell wall peptidoglycan. Proc. Natl. Acad. Sci. U. S. A. 103:4404–4409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Minamino T, Macnab RM. 1999. Components of the Salmonella flagellar export apparatus and classification of export substrates. J. Bacteriol. 181:1388–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nambu T, Inagaki Y, Kutsukake K. 2006. Plasticity of the domain structure in FlgJ, a bacterial protein involved in flagellar rod formation. Genes Genet. Syst. 81:381–389 [DOI] [PubMed] [Google Scholar]
- 26. Nambu T, Kutsukake K. 2000. The Salmonella FlgA protein, a putativeve periplasmic chaperone essential for flagellar P ring formation. Microbiology 146(Pt 5):1171–1178 [DOI] [PubMed] [Google Scholar]
- 27. Nambu T, Minamino T, Macnab RM, Kutsukake K. 1999. Peptidoglycan-hydrolyzing activity of the FlgJ protein, essential for flagellar rod formation in Salmonella typhimurium. J. Bacteriol. 181:1555–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Oosawa K, Ueno T, Aizawa S. 1994. Overproduction of the bacterial flagellar switch proteins and their interactions with the MS ring complex in vitro. J. Bacteriol. 176:3683–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Scheurwater EM, Burrows LL. 2011. Maintaining network security: how macromolecular structures cross the peptidoglycan layer. FEMS Microbiol. Lett. 318:1–9 [DOI] [PubMed] [Google Scholar]
- 30. Simon R, Priefer U, Puhler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Biotechnology 1:37–45 [Google Scholar]
- 31. Sistrom WR. 1962. The kinetics of the synthesis of photopigments in Rhodopseudomonas sphaeroides. J. Gen. Microbiol. 28:607–616 [DOI] [PubMed] [Google Scholar]
- 32. Sockett RE, Foster JCA, Armitage JP. 1990. Molecular biology of Rhodobacter sphaeroides flagellum. FEMS Symp. 53:473–479 [Google Scholar]
- 33. Sosinsky GE, Francis NR, Stallmeyer MJ, DeRosier DJ. 1992. Substructure of the flagellar basal body of Salmonella typhimurium. J. Mol. Biol. 223:171–184 [DOI] [PubMed] [Google Scholar]
- 34. Thormann KM, Paulick A. 2010. Tuning the flagellar motor. Microbiology 156:1275–1283 [DOI] [PubMed] [Google Scholar]
- 35. Viollier PH, Shapiro L. 2003. A lytic transglycosylase homologue, PleA, is required for the assembly of pili and the flagellum at the Caulobacter crescentus cell pole. Mol. Microbiol. 49:331–345 [DOI] [PubMed] [Google Scholar]
- 36. Vollmer W, Blanot D, de Pedro MA. 2008. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 32:149–167 [DOI] [PubMed] [Google Scholar]
- 37. Reference deleted.
- 38. Wessel D, Flugge UI. 1984. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138:141–143 [DOI] [PubMed] [Google Scholar]
- 39. Zhang K, Tong BA, Liu J, Li C. 2012. A single-domain FlgJ contributes to flagellar hook and filament formation in the Lyme disease spirochete Borrelia burgdorferi. J. Bacteriol. 194:866–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhou J, Lloyd SA, Blair DF. 1998. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc. Natl. Acad. Sci. U. S. A. 95:6436–6441 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.