Abstract
Cyclic dimeric GMP (c-di-GMP) regulates numerous processes in Gram-negative bacteria, yet little is known about its role in Gram-positive bacteria. Here we characterize two c-di-GMP phosphodiesterases from the filamentous high-GC Gram-positive actinobacterium Streptomyces coelicolor, involved in controlling colony morphology and development. A transposon mutation in one of the two phosphodiesterase genes, SCO0928, hereby designated rmdA (regulator of morphology and development A), resulted in decreased levels of spore-specific gray pigment and a delay in spore formation. The RmdA protein contains GGDEF-EAL domains arranged in tandem and possesses c-di-GMP phosphodiesterase activity, as is evident from in vitro enzymatic assays using the purified protein. RmdA contains a PAS9 domain and is a hemoprotein. Inactivation of another GGDEF-EAL-encoding gene, SCO5495, designated rmdB, resulted in a phenotype identical to that of the rmdA mutant. Purified soluble fragment of RmdB devoid of transmembrane domains also possesses c-di-GMP phosphodiesterase activity. The rmdA rmdB double mutant has a bald phenotype and is impaired in aerial mycelium formation. This suggests that RmdA and RmdB functions are additive and at least partially overlapping. The rmdA and rmdB mutations likely result in increased local pools of intracellular c-di-GMP, because intracellular c-di-GMP levels in the single mutants did not differ significantly from those of the wild type, whereas in the double rmdA rmdB mutant, c-di-GMP levels were 3-fold higher than those in the wild type. This study highlights the importance of c-di-GMP-dependent signaling in actinomycete colony morphology and development and identifies two c-di-GMP phosphodiesterases controlling these processes.
INTRODUCTION
Cyclic diguanosine monophosphate (c-di-GMP) is a ubiquitous second messenger in bacteria. Since its identification by Moshe Benziman and colleagues and characterization as an allosteric activator of cellulose synthase in Gluconacetobacter xylinus (formerly Acetobacter xylinum) (47), c-di-GMP has been shown to coordinate numerous cellular activities, including biofilm formation, motility, cell cycle progression, and virulence. c-di-GMP regulates these processes by affecting the synthesis and activities of flagella, pili, adhesins, exopolysaccharides, and extracellular DNA, as well as virulence factors. It plays important roles in the cellular transition from a motile to surface-attached lifestyle, from formation to dissolution of biofilms, and, in pathogens, from acute to chronic infections and from one host to another (reviewed in references 26, 45, 52, 55, and 65).
Intracellular c-di-GMP levels are inversely regulated by diguanylate cyclases (DGCs) involved in c-di-GMP synthesis and by c-di-GMP-specific phosphodiesterases (PDEs) involved in c-di-GMP degradation (46, 47). A conserved and widely spread GGDEF domain is responsible for the DGC activity. c-di-GMP formation is catalyzed by a homodimer formed from two GGDEF domains, each of which binds the substrate, GTP (11, 43, 51). The c-di-GMP-specific PDE activity is encoded by the EAL (11, 14, 53, 56) or HD-GYP (23, 34, 49) protein domains. A large number of c-di-GMP-metabolizing enzymes contain both GGDEF and EAL domains arranged in tandem (54). Most of these proteins possess only one enzymatic activity, either DGC or PDE. It is often possible to predict the enzymatic activity of the GGDEF-EAL proteins based on conservation of the amino acids essential for catalysis. However, some enzymes are bifunctional, and their prevailing activity depends on the input signal (20, 32, 57).
Changes in c-di-GMP levels affect activities of diverse c-di-GMP receptors (effectors). Among protein-based c-di-GMP receptors are enzymes, transcription factors, and proteins controlling downstream output activities via protein-protein interactions. c-di-GMP also controls gene expression via two specific classes of riboswitches (reviewed in reference 38).
At present, our knowledge of c-di-GMP signaling pathways is limited almost exclusively to the Gram-negative bacteria (proteobacteria and spirochetes). The c-di-GMP signaling pathways are present in many Gram-positive bacteria (9), but functional studies have been done only in mycobacteria, where c-di-GMP signaling appears to play a relatively limited role and is involved in long-term survival (32). Recent reports uncovered the involvement of c-di-GMP in the development of the Gram-positive actinomycete Streptomyces coelicolor (17, 59).
S. coelicolor is a nonmotile, nonpathogenic soil bacterium characterized by a complex cycle of morphological differentiation and therefore traditionally utilized as a model of bacterial development. The genus Streptomyces has significant pharmacological importance because more than two-thirds of the antibiotics currently in use are produced by its representatives. The S. coelicolor life cycle begins when a free spore germinates to produce long, branching vegetative filaments that grow into and on the substrate surface. These vegetative filaments rarely divide, yielding a network of multinucleated hyphae. Colony maturation leads to the development of aerial hyphae that are erected above the colony surface. These aerial hyphae undergo a sporulation-specific cell division process, resulting in an aerial mycelium comprised of uninucleoid prespores that metamorphose into gray-pigmented mature spores (13, 22).
Here we searched for developmental mutants of S. coelicolor following transposon mutagenesis. One mutant, resulting from transposon insertion in the gene designated rmdA (regulator of morphology and development A), produced a decreased amount of gray spore pigment relative to that produced by the wild-type strain, was delayed in sporulation, and had abnormal colony morphology. The rmdA gene was found to encode a GGDEF-EAL protein. Following genetic and biochemical analysis, we elucidated that RmdA functions as a c-di-GMP PDE. Bioinformatics analysis uncovered a potential second GGDEF-EAL PDE in S. coelicolor. We have shown here that this protein, designated RmdB, also acts as a c-di-GMP PDE and that the rmdB mutant has a phenotype similar to that of the rmdA mutant. The rmdA rmdB double mutant is completely blocked in aerial mycelium formation, which suggests that RmdA and RmdB are critical, partially redundant, signaling enzymes regulating c-di-GMP-dependent development in S. coelicolor.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Strains used in this work are listed in Table 1. The prototrophic SCP1− SCP2− strain MT1110 of S. coelicolor A3(2) was used as the wild type (6, 30). Liquid cultures were grown in baffled flasks containing YEME medium with 10.3% sucrose at 30°C (30). The agar media R2YE and mannitol soya flour (MS) were prepared as described previously (30). For S. coelicolor, neomycin, apramycin, and kanamycin were used at final concentrations of 10 μg/ml, 25 μg/ml, and 160 μg/ml, respectively.
Table 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| S. coelicolor | ||
| MT1110 | Prototrophic, SCP1−, SCP2− | 30 |
| TH1 | SC00928::Tn5 | This work |
| TH100 | SCO0928::Tn5062 | This work |
| TH101 | SCO5495::Tn5062 | This work |
| TH105 | SCO0929::Tn5062 | This work |
| TH106 | SCO5495::Tn5062 SCO0928::Tn5 | This work |
| E. coli | ||
| SURE | Cloning strain | Stratagene |
| ER2-1 | dam dcm mutant to avoid Streptomyces methyl restriction | 5 |
| ET12567 | dam dcm mutant strain with mating helper plasmid pUZ8002 | 30 |
| DH5α | Host for overexpression of MBP fusions | NEB |
| MG1655 | K12 strain used in motility assays | ATCC |
| MG1655 yhjH | MG1655 yhjH::Kanr | 19 |
| Plasmids | ||
| pJA77 | Mini-Tn5 delivery vector, Neor | 4 |
| pMAL-c5x | MBP overexpression vector | NEB |
| pMAL-rmdA | MBP::(PAS9-GGDEF-EAL) from RmdA | This work |
| pMAL-rmdB | MBP::(GGDEF-EAL) from RmdB | This work |
| pMAL-sco1397 | MBP::SCO1397 | This work |
| Cosmids | ||
| SCM10 | SCM10.2.B08 (03071421MU); contains SCO0928 | 7 |
| SCM10 | SCM10.1.G03 (03021117RA); contains SCO0929 | 7 |
| SC8D9 | 8D9.1.F04 (8BF-1 022 2007-04-18); contains SCO5495 | 7 |
| SC1A8A | 1A8A.2.B09; contains SCO1397 | 7 |
Escherichia coli SURE was utilized for the preparation of cosmid DNA. The dam dcm mutant strains ET12567 and ER2-1 were used to circumvent the methyl-specific restriction system of S. coelicolor for propagation of cosmid or plasmid DNA (35). E. coli strains were grown at 37°C in Luria-Bertani medium containing, where necessary, ampicillin (100 μg/ml), apramycin (100 μg/ml), and/or kanamycin (50 μg/ml).
Bioinformatics analysis.
The S. coelicolor genome database was searched for annotated gene and protein sequences. Protein sequences were analyzed by the SMART (33) and Pfam (21) domain databases. BLAST software was employed to identify homologous proteins in other bacteria. Multiple sequence alignments were constructed using ClustalW on the Biology Workbench 3.2 software platform (http://workbench.sdsc.edu/).
Colony morphology and sporulation analysis.
Ten colonies of confirmed S. coelicolor mutants grown on MS agar plates for 5 days at 30°C were observed using a low-power photomicroscope (Meiji). Representative colonies were photographed with a color digital camera (model CFW01312C) from the Scion Corporation. S. coelicolor spore chains were observed under phase-contrast microscopy at total magnification ×1,000. Aerial mycelium was prepared on glass cover slides and mounted in 50% glycerol (36). Images were captured with an Olympus BX 40 light microscope equipped with an Olympus color digital camera (model DP72) and cellSens software. Micrographs were processed in the software packages ImageJ and Adobe Photoshop.
DNA preparation and manipulation.
Chromosomal DNA from S. coelicolor was isolated using the Wizard genomic DNA purification kit (Promega). Plasmid and cosmid DNA were prepared with the QIAprep miniprep kit (Qiagen). PCR products were prepared for DNA sequencing using the QIAquick PCR purification kit (Qiagen). The DyeEX 2.0 spin kit (Qiagen) was used for the cleanup of all DNA sequencing reactions, and the Applied Biosystems ABI Prism 310 genetic analyzer was used to analyze sequencing reactions.
Transposon mutagenesis and insertion site analysis.
The pJA77 minitransposon system is based on the Tn5 transposon containing the aphI gene (neomycin resistance [Neor]) (4). pJA77 was isolated from E. coli ER2-1 and transformed into wild-type S. coelicolor MT1110 (30) by polyethylene glycol-mediated transformation (41). Following selection of Neor colonies, we screened for Apramycin sensitivity (Apras) to ensure transposition as opposed to plasmid integration via a single crossover. Transformants were also plated on minimal medium, and auxotrophic mutants were eliminated. To establish linkage between the transposon insertion and the mutant phenotype, chromosomal DNA from the insertional mutants was isolated and transformed into MT1110 protoplasts. The Neor transformants were screened visually and by phase-contrast microscopy (4).
To determine the transposon insertion locations, we used inverse PCR and DNA sequencing. Chromosomal DNA from the transposon insertion mutant was digested with Sau3AI to generate short DNA fragments that were ligated into small circular molecules. This DNA served as the template for an inverse PCR (invPCR) reaction utilizing the following transposon-specific primers: Tn5InvF2 (5′-AGGAGACTCGGGCGCCTTCGTCTACCAG) and Tn5InvR3 (5′-CATCATGGCAGAGGCGGAGACGCCGTTC). The PCR products were sequenced. The nested primer Tn5InvSeq4 (5′-GTAGGGGTTTCGGACATTCTGTG) was used to sequence the unknown chromosomal DNA surrounding the transposon insertion, utilizing the known transposon sequence as a priming site. The Basic Local Alignment Search Tool (BLAST) was used to determine the site of transposon insertions. The Streptomyces genome database (http://www.sanger.ac.uk/Projects/S_coelicolor/) was used for gene and protein identification (6).
Confirmation of the transposon insertion site in SCO0928 (rmdA) was accomplished by PCR amplification of the transposon (Tn)-containing fragment using the following primers: SCO0928F (5′-AGGCGGCCCGTAACGGTGCTTGAG), which anneals downstream of the predicted transposon insertion site, and Tn5Inv3R (5′-CATCATGGCAGAGGCGGAGACGCCGTTC), which is transposon specific. The PCR product was sequenced. Insertions of the Tn5062 transposon in SCO0928, SCO0929, and SCO5495 were verified in a similar manner using the following primer pairs: SCO0928 forward (5′-TTCTCCAGGCGTTCGAAGAAGAAG) and SCO0928/SCO0929 reverse 5′-GATGAAACCAAGCCAACCAGGAAG); SCO0929 forward (5′-ACCATGTCGTACCCCTTGAAGAAC) and SCO0928/SCO0929 reverse; SCO5495 forward (5′-CGCCCTGCTCTGGTACATCCAGAC) and SCO5495 reverse (5′-TTGCCGTCGTCCTTGAAGAAGATG).
Mutant construction.
Cosmid DNA mutagenized by in vitro transposon mutagenesis with a Tn5 derivative, Tn5062, was obtained from Paul Dyson (7). ET12567 containing the helper plasmid pUZ8002 was used as a donor in the oriT-mediated conjugal transfer of cosmid DNA from E. coli to S. coelicolor (42). To select for exconjugates and to eliminate E. coli, the plates were overlaid with 25 μg/ml apramycin and 20 μg/ml nalidixic acid, respectively. Apramycin-resistant (Aprar) exconjugates were streaked on MS medium containing kanamycin to screen for kanamycin sensitivity (Kans). Since S. coelicolor cosmids contain both Kanr and Ampr cassettes on the Supercos-1 backbone (44), screening for Aprar Kans colonies allowed the selection of mutant S. coelicolor colonies that had undergone gene disruption via double-crossover recombination.
To construct the rmdA rmdB double mutant, the rmdB::Tn5062 cosmid was conjugated from E. coli ET12567/pUZ8002 into the rmdA mutant, and Kans Aprar Neor colonies were identified and analyzed further. For complementation, a wild-type version of one of the genes (rmdA or rmdB) was introduced into the double mutant strain by interspecies conjugation, and single-crossover integrants of the cosmid were selected.
Protein overexpression and enzymatic assays.
The DNA fragments encoding the PAS9-GGDEF-EAL module of RmdA, the GGDEF-EAL module of RmdB, and full-length SCO1397 were PCR amplified using, respectively, the following primer pairs: SCO0928-F (5′-AATCATATGCATCATCATCATCATCACTTCGAGGGCGCCGCCATAG) and SCO0928-R (5′-ATTGAATTCTCAGCCCGTCGCGTCC); SCO5495-F (5′-AATCATATGCATCATCATCATCATCACCAGTTGCGCGACCCGCTC) and SCO5495-R (5′-ATTGAATTCTCACTCGTCGCTCGCCG); and SCO1397-F (5′-AATCATATGCATCATCATCATCATCACATGACCGGCGGGGTCGC) and SCO1397-R (5′-ATTGAATTCTCAGAGAGCCGTACGACGCC). The amplified PCR fragments were digested with NdeI and EcoRI and cloned into the same restriction sites in pMal-c5x (NEB).
Maltose-binding protein (MBP) fusions were overexpressed in E. coli DH5α and affinity purified on amylose resin according to the specifications of the manufacturer (NEB). Briefly, the overnight cultures expressing MBP fusions were grown to an A600 of 0.6 in LB supplemented with 100 μg/ml ampicillin at 30°C. The cultures were transferred to room temperature, and 0.5 mM (final concentration) isopropyl 1-thio-β-d-galactopyranoside (IPTG) was added. Following a 15-h incubation, cells were harvested by centrifugation and resuspended in amylose column binding buffer (50 mM Tris-HCl, pH 8.0, 350 mM NaCl, 10 mM MgCl2, 0.5 mM EDTA, 10% glycerol). Cells were disrupted using a French pressure cell, and cell debris was removed by centrifugation at 35,000 × g for 45 min at 4°C. Two milliliters (bed volume) of amylose resin (NEB) preequilibrated with the binding buffer was added to the soluble cell extract derived from a 1.5-liter culture and agitated for 1 h at 4°C. The mix was loaded onto a column, and the resin was washed with 200 ml of column binding buffer. Fractions were eluted with 12 ml of binding buffer containing 10 mM maltose. The protein was either used immediately or stored at −80°C in 20% (vol/vol) glycerol (final concentration). Protein concentrations were measured using a Bradford protein assay kit (Bio-Rad) with bovine serum albumin as the protein standard. Proteins were analyzed using SDS-PAGE.
The DGC assays in vitro were performed by measuring the rate of GTP conversion to c-di-GMP as described by Ryjenkov et al. (51), and c-di-GMP PDE activity assays were performed by measuring the rate of c-di-GMP hydrolysis as described by Schmidt et al. (53). The PDE assays were performed at room temperature in PDE assay buffer (50 mM Tris-HCl [pH 8.5], 50 mM NaCl, 0.5 mM EDTA) containing various concentrations of Mg2+ and/or Mn2+. The initial c-di-GMP concentration was 200 μM. The reactions were started by adding 1 μmol purified enzyme. Aliquots were withdrawn at different time points, CaCl2 was added to 10 mM to stop the reaction, and samples were boiled for 5 min. Postreaction processing and nucleotide separation and quantification were accomplished by high-performance liquid chromatography (HPLC) as described earlier (51).
Protein reconstitution with hemin and UV-vis spectroscopy.
Reconstitution of the MBP-RmdA (PAS9-GGDEF-EAL) protein fusion with hemin and flavins in vitro was performed essentially as described earlier (24, 39). A reduced deoxy [Fe(II)] form of the hemoprotein was obtained by reduction with dithionite followed by removal of the reductant using gel filtration on a Sephadex G-25 column. The oxidized [Fe(III); met] form was obtained after protein incubation with an equimolar amount of potassium ferricyanide and subsequent ferricyanide removal by gel filtration. The reduced deoxy forms with CO [Fe(II)-CO] and oxygen [Fe(II)-O2] were obtained by gentle bubbling (for 10 min) through protein solution of CO or air, respectively. Electronic absorption UV-visible (UV-vis) spectra were recorded using a Shimatzu UV-1601PC spectrophotometer.
Intracellular c-di-GMP extraction and measurements.
Intracellular nucleotides were extracted with 40% methanol–40% acetonitrile in 0.1 N formic acid, following the protocol described by Bobrov et al. (8). S. coelicolor was grown for 70 h at 30°C on MS agar overlaid with cellophane disks, roughly corresponding to the time of sporulation in the wild type. Cells were scraped from the cellophane with a sterile scalpel blade, and the wet weights were determined and matched, followed by extraction. c-di-GMP was quantified by liquid chromatography-tandem mass spectrometry carried out at the Mass Spectrometry Core at Michigan State University. The levels of extracted c-di-GMP were determined based on c-di-GMP standards. Average data from three independent measurements are presented.
RESULTS
Identification of the rmdA gene as a regulator of morphology and development in S. coelicolor.
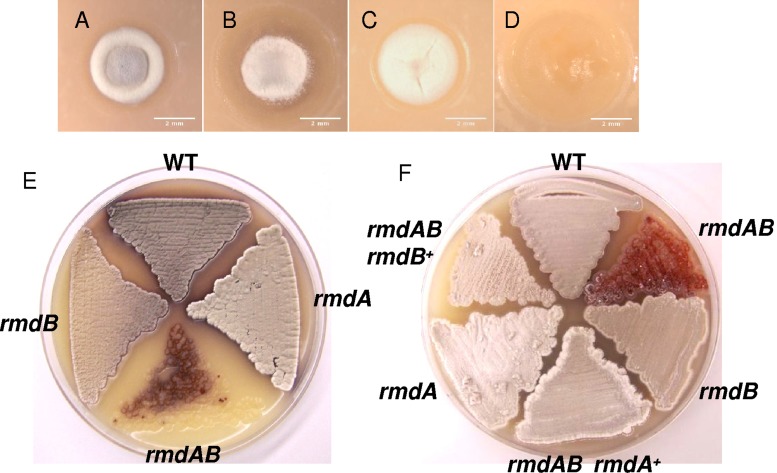
To identify S. coelicolor sporulation mutants, we conducted in vivo Tn5 mutagenesis of the wild-type strain, MT1110, using the protocol described by us earlier (4). Colony morphology of one of the mutants, TH1 (Fig. 1B), was aberrant compared to that of the wild-type strain. While the wild type is characterized by a central gray region of aerial mycelium surrounded by a ring of less-pigmented mycelium (Fig. 1A), TH1 had a white aerial mycelium with little gray pigment. Because gray pigment is present exclusively in spores, low pigmentation of TH1 indicated a potential sporulation defect.
Fig 1.
Effects of inactivation of rmdA and rmdB on S. coelicolor colony morphology and aerial hyphae. (Top) Colony morphology. (A) wild type; (B) TH1 (rmdA::Tn5); (C) TH101 (rmdB::Tn5062); (D) TH106 (rmdA::Tn5 rmdB::Tn5062). Single colonies that were well separated from neighboring colonies were selected for observation using stereomicroscopy. Images were taken on MS media after inocula of dilute spores were grown for 5 days at 30°C, yielding a few colonies per plate. (Bottom) Aerial hyphae. (E) Reduced gray spore pigment production in rmdA and rmdB single mutants and a complete block in aerial hypha formation in the rmdA rmdB double mutant. WT, wild type. (F) Complementation of the rmdA rmdB double mutant with the rmdA or rmdB gene. Strains were grown at 30°C for 5 days on MS medium.
To investigate the nature of this defect, we closely monitored the sporulation process in TH1. In the wild type, mature spore chains are comprised of individual spores that are evenly sized, shaped, and spaced. By day three, the wild type produces long chains with distinct compartments containing spores separated by fully developed septa, which can be observed by phase-contrast microscopy. However, only very rare, short spore chains were visible in TH1 by day three (Fig. 2). On day five, most aerial filaments in TH1 had initiated sporulation, but many spore chains contained incompletely separated spores. However, by day seven, the majority of these spore chains were fully separated (Fig. 2). These results show that the mutation in TH1 resulted in a considerable delay for the onset of sporulation in comparison to the wild-type strain.
Fig 2.
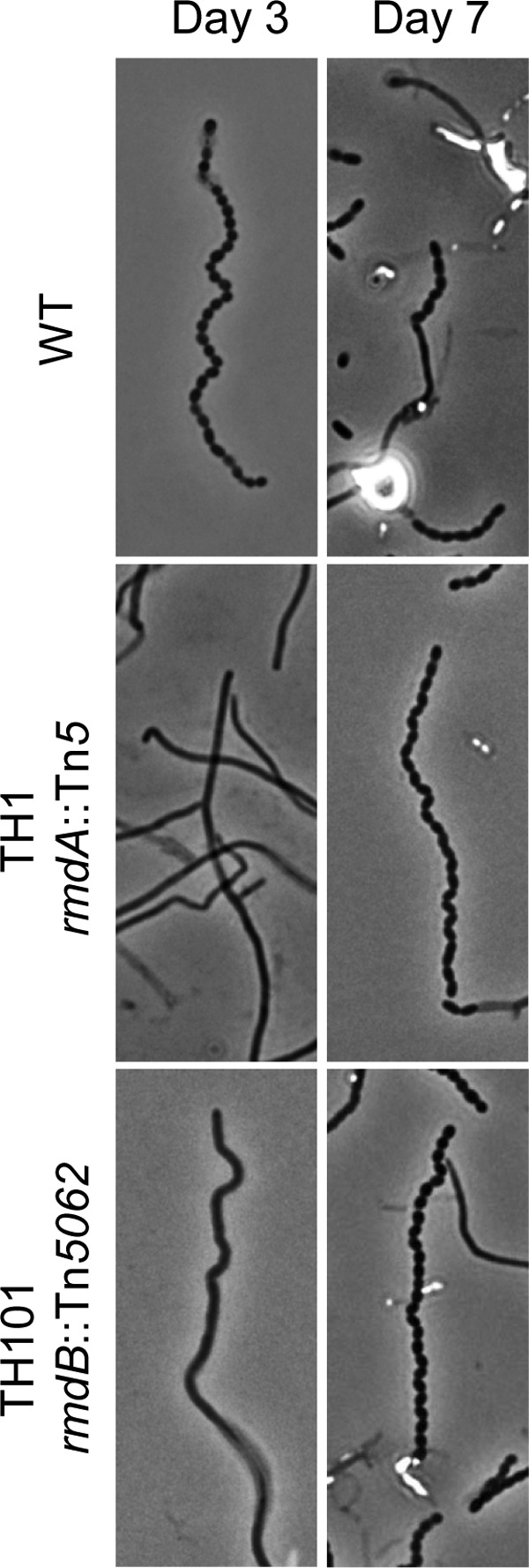
The rmdA and rmdB mutations delay S. coelicolor sporulation. Shown are representative phase-contrast micrographs (×1,000) of S. coelicolor aerial hyphae viewed from impression slides after 3 and 7 days of growth at 30°C on MS agar. The wild-type (WT) strain is characterized by aerial filaments that have undergone division to produce evenly spaced, consistently sized spores separated by complete septa at day 3. The spores begin to disperse by day 7. The rmdA and rmdB mutants exhibit a delay in sporulation, producing primarily undifferentiated aerial hyphae on day three and many long spore chains by day seven.
We determined that the Tn5 insertion in TH1 occurred in codon 581 of SCO0928, hereby designated rmdA (Fig. 3A). To test whether the sporulation defect in TH1 resulted from the Tn5 insertion, we performed linkage analysis. The chromosomal DNA from the TH1 mutant was transformed into the wild type, and transformants expressing the Tn5-encoded Neor marker were selected and assayed further. All seven analyzed transformants displayed a phenotype that was indistinguishable from that of the TH1 mutant (data not shown). To further verify that inactivation of rmdA is responsible for the sporulation defect in TH1, we constructed a new rmdA mutant, TH100, using a cosmid that contains a Tn5062 insertion in codon 120 of rmdA (7). In accord with our expectations, the phenotype of the newly generated rmdA::Tn5062 mutant, TH100, displayed a sporulation phenotype indistinguishable from that of TH1 (data not shown).
Fig 3.
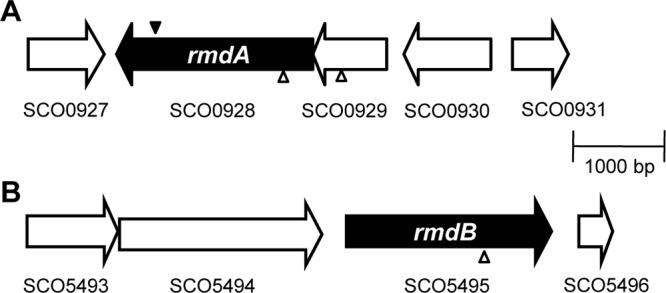
Transposon insertion sites. Genetic maps showing the rmdA (SCO0928) and rmdB (SCO5495) regions. SCO0929 and rmdA form a potential operon. The solid wedge represents the location of the Tn5 minitransposon insertion through which TH1 (rmdA::Tn5) was originally isolated by in vivo mutagenesis. The hollow wedges represent the location of the in vitro Tn5062 transposon insertions in rmdA, SCO0929, and rmdB (7).
rmdA is located downstream of SCO0929 in a putative two-gene operon. SCO0929 encodes a conserved protein of 270 amino acids containing a domain of unknown function, DUF574 (33). To assess a possible role of SCO0929, we constructed a mutant with a Tn5062 transposon mutation in SCO0929. We found that inactivation of SCO929 did not affect sporulation (data not shown), which suggests that rmdA is solely responsible for the developmental delay in TH1 and TH100.
RmdA (SCO0928) functions as a c-di-GMP PDE.
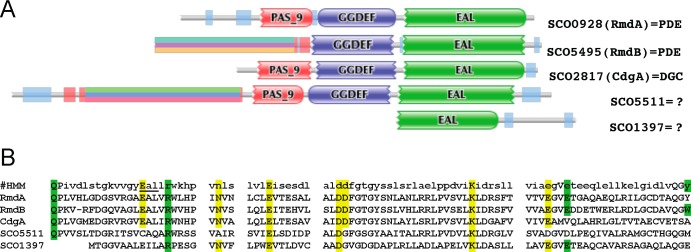
The rmdA gene encodes a 714-amino-acid (aa) protein with the following domain architecture: X-PAS9-GGDEF-EAL (Fig. 4; (21). The N-terminal domain X of approximately 160 aa is present in signaling proteins from numerous species in the genus Streptomyces and other actinomycetes; however, it shows no significant similarity with known protein domains. PAS9 belongs to a large superfamily of PAS domains that often bind small molecules and are involved in signal sensing and/or in protein-protein interactions (58). Sequence analysis of the GGDEF and EAL domains of RmdA revealed that both domains contain residues essential for DGC and PDE activities, respectively, and neither domain contains identifiable deleterious mutations. Since both the GGDEF and EAL domains are potentially enzymatically active, RmdA may possess either one activity or another or may potentially be bifunctional.
Fig 4.
Analysis of S. coelicolor EAL domain proteins. (A) Protein domain architectures derived from the Pfam database (21). (B) Sequence alignment of the most conserved regions of the EAL domains. #HMM, hidden Markov model-derived consensus sequence of the EAL domain. Residues involved in c-di-GMP binding inferred from the X-ray structure of an EAL domain PDE are shown on a green background; residues involved in coordinating catalytic metals are shown on a yellow background (3).
To test for enzymatic activity of RmdA, we expressed rmdA (as an MBP fusion to the PAS9-GGDEF-EAL domain fragment) in two E. coli test strains, MG1655 and MG1655 yhjH (19) (Table 1). MG1655 is a highly motile strain. The inactivation of yhjH, which encodes a dominant c-di-GMP PDE in E. coli, impairs motility in the semisolid agar (19, 50). If RmdA had DGC activity, it would be expected to increase intracellular c-di-GMP levels in strain MG1655 and decrease the diameter of the swimming zone in the semisolid agar. However, no such decrease was detected (not shown). If RmdA had PDE activity, it would be expected to restore swimming of MG1655 yhjH in semisolid agar, and this is what was observed (Fig. 5A).
Fig 5.
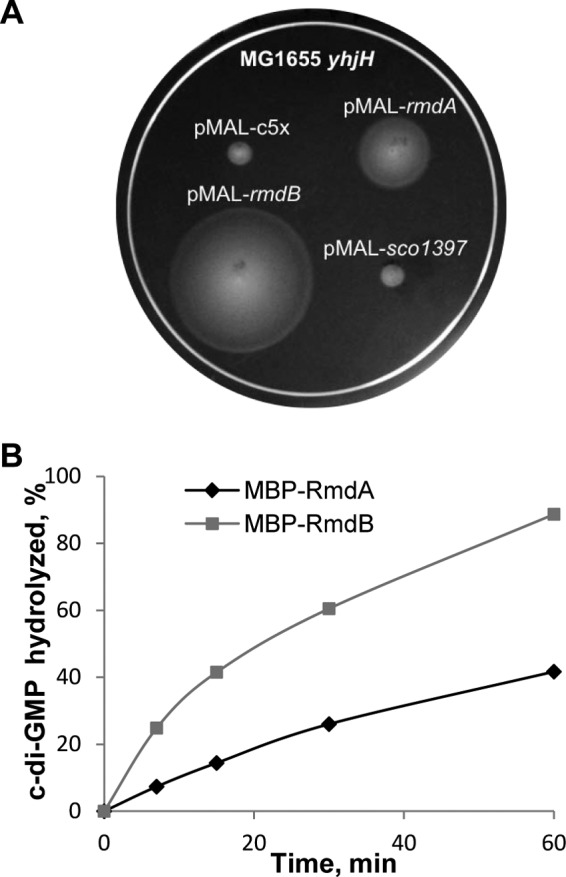
c-di-GMP PDE activity of RmdA and RmdB. (A) Increase in swimming zones in semisolid agar of E. coli strain MG1655 yhjH with MBP-RmdA and MBP-RmdB. Assays were run as described earlier (19). (B) Kinetics of c-di-GMP hydrolysis by MBP-RmdA and MBP-RmdB. The PDE assays shown here were performed in the PDE assay buffer containing 5 mM Mg2+ and 5 mM Mn2+ (MBP-RmdA) or 5 mM Mg2+ (MBP-RmdB) (53).
To verify the predicted PDE function of RmdA, we overexpressed and purified the MBP-RmdA fusion using affinity chromatography. We tested enzymatic activity of the protein in vitro (53). RmdA proved to possess c-di-GMP-specific PDE activity; however, the relative activity in standard, Mg2+-containing buffers was surprisingly low. Addition of Mn2+, which is known to improve catalytic activity of c-di-GMP PDEs (3), enhanced the PDE activity of MBP-RmdA by approximately 2-fold (Fig. 5B).
Based on the PDE activity of RmdA, we expected that the rmdA mutation would increase intracellular levels of c-di-GMP. To test this prediction, we extracted total nucleotides from the wild-type and mutant TH1 grown on MS agar overlaid with cellulose membranes for 70 h at 30°C and analyzed c-di-GMP content via liquid chromatography-tandem mass spectrometry (LC-MS/MS). We found that c-di-GMP levels did not differ significantly between the rmdA mutant and the wild type (Fig. 6), suggesting that only a local pool of c-di-GMP may have been affected. Because the rmdA mutation resulted in no gross alteration in c-di-GMP levels, we conclude that RmdA is specifically involved in regulating S. coelicolor development.
Fig 6.
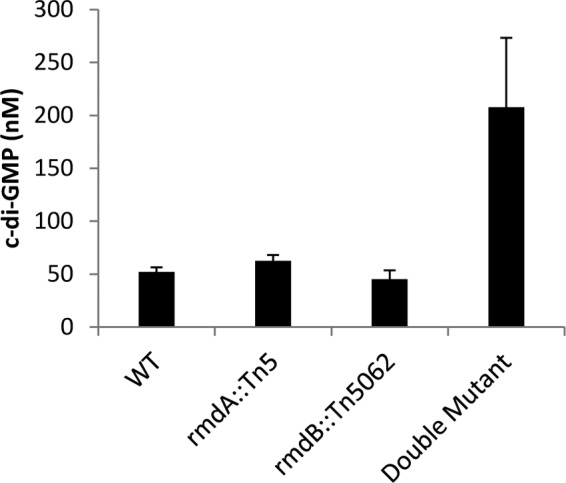
Intracellular c-di-GMP levels in the S. coelicolor PDE mutants. The level of c-di-GMP for the rmdA rmdB double mutant was approximately three times that of the wild type and single mutant strains. c-di-GMP was extracted as described previously (8) from cells grown on cellulose membranes on MS agar for 70 h at 30°C and quantified by LC-MS/MS.
Bioinformatics analysis of putative S. coelicolor c-di-GMP PDEs.
To investigate whether additional c-di-GMP PDEs are involved in controlling c-di-GMP-dependent development in S. coelicolor, we performed a bioinformatics analysis of the EAL and HD-GYP domain proteins encoded in the S. coelicolor genome. In addition to RmdA, four other proteins, SCO1397, SCO2817, SCO5495, and SCO5511, contain EAL domains (21) (Fig. 4), and one, SCO5218, contains an HD-GYP domain.
The sequence of the HD-GYP domain in SCO5218 appears to be consistent with it being enzymatically active. However, our ability to predict activities of this class of proteins based on sequence analysis remains limited (34, 49). In this study, we did not pursue experimental investigation of SCO5218.
Sequence analysis of the EAL domains in SCO1397 and SCO5511 strongly suggested that they are incapable of hydrolyzing c-di-GMP because they miss residues involved in c-di-GMP binding or coordination of essential divalent metals (Fig. 4B) (3, 53). For example, SCO5511 lacks the signature “EAL” motif, while SCO1397 lacks the N-terminal residues of the EAL domain (Fig. 4B). Of the remaining two proteins, SCO2817, designated CdgA, was recently shown to function as a DGC (17). Therefore, among the S. coelicolor EAL domain proteins, only RmdA and SCO5495 may be enzymatically active (Fig. 4B). Below, we show that SCO5495 is a c-di-GMP PDE involved in colony morphology and developmental control, and therefore we designated it RmdB. We used SCO1397, predicted to be enzymatically incompetent in the initial stages of experimental analysis, as a negative control.
RmdA and RmdB appear to be highly conserved among streptomycetes. Orthologs of both proteins are encoded in the genomes of Streptomyces avermitilis, S. venezuelae, S. hygroscopicus, S. griseus, S. flavogriseus, S. bingchenggensis, S. violaceusniger, and S. scabiei. They are also present in other filamentous actinomycetes. Furthermore, the potential rmdA-SCO0929 operon is preserved in streptomycetes and representatives of the suborder Frankineae, e.g., Geodermatophilus obscurus and Nakamurella multipartita. The high degree of conservation suggests that the regulatory mechanism(s) through which RmdA and RmdB are involved in colony morphology and development is likely conserved in filamentous actinomycetes.
RmdA is a hemoprotein.
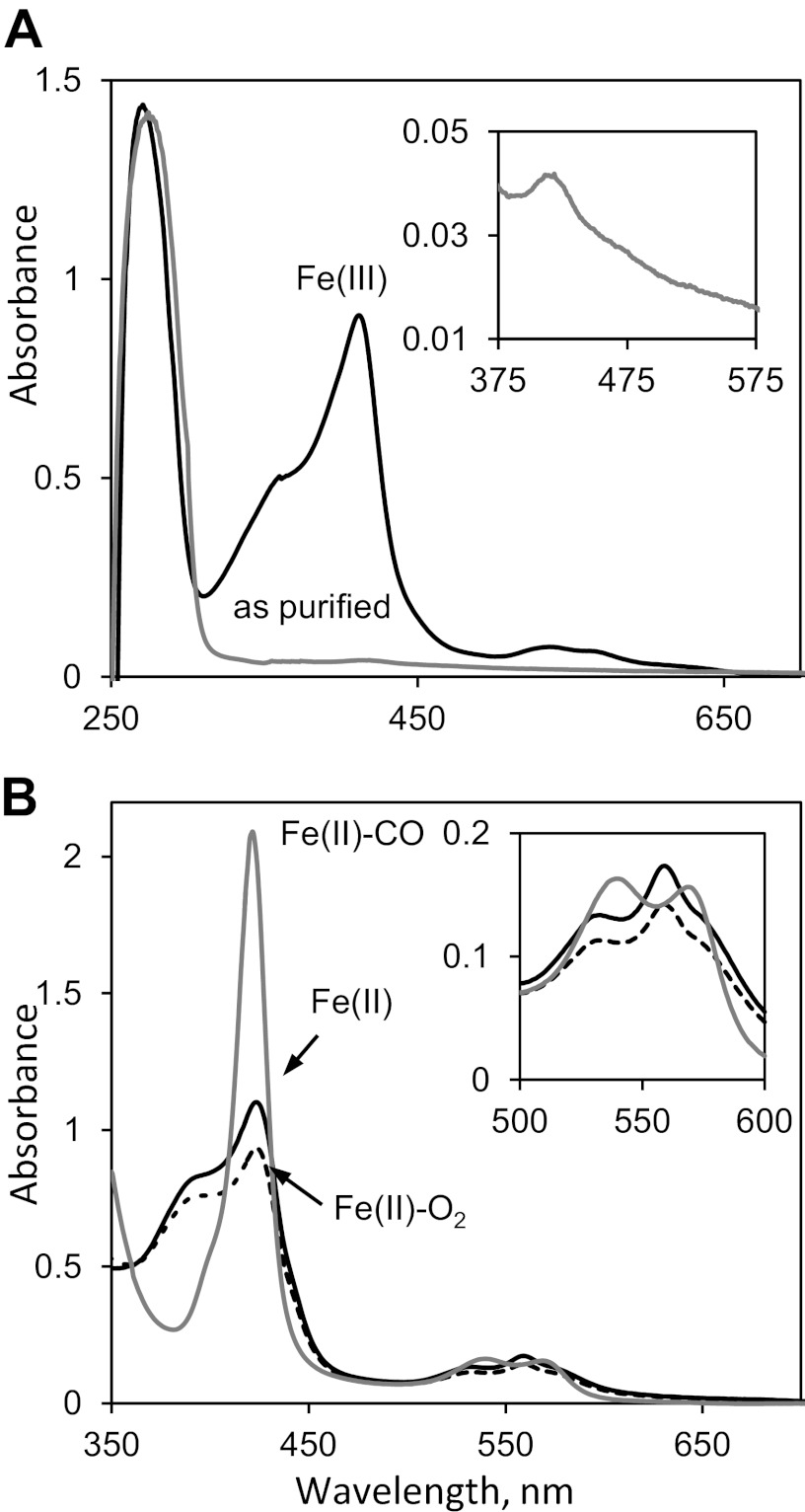
We noticed that in addition to RmdA, two S. coelicolor GGDEF and/or EAL domain proteins, CdgA and SCO5511, contain PAS9 domains (Fig. 4A), which suggests that they may bind a common ligand and respond to the same primary signal. To investigate the nature of the putative ligand bound to the PAS9 domain of RmdA, we reconstituted the MBP-RmdA (PAS9-GGGDEF-EAL) fusion with common PAS domain ligands, FAD, FMN, and hemin (27). MBP-RmdA did not bind FAD or FMN in vitro (not shown), but it did bind hemin with a nearly equimolar stoichiometry (Fig. 7A). Further, upon close inspection of the spectra of the MBP-RmdA protein purified from the E. coli overexpression strain, we noticed that it already contained traces of heme (Fig. 7A, inset).
Fig 7.
Characterization of RmdA as a heme-binding protein. (A) Electronic absorbance spectra of the MBP-RmdA protein. Gray trace, as purified from E. coli; black trace, after in vitro reconstitution with hemin. Inset, expanded version of the spectrum showing a Soret band indicative of traces of heme present in the protein purified from E. coli. (B) Reconstituted with hemin MBP-RmdA protein. Black solid trace, Fe(II), deoxy form; dotted trace, Fe(II), oxy form; gray trace, Fe(II), carbonmonoxy form.
To investigate the specificity of heme binding, we reduced the protein-hemin complex with dithionite under anoxic conditions. We observed a shift in the Soret band (from 412 nm to 424 nm) and sharpening of the α- and β-peaks in the 500- to 600-nm region of the spectrum. These changes are characteristic of the formation of the reduced, Fe(II), deoxy form with hexacoordinated heme iron, where two axial ligands are provided by amino acids of the protein (Fig. 7B). Addition of CO, a gaseous ligand that has high affinity to the majority of hemoproteins, resulted in further sharpening of the Soret band (421.5 nm) and noticeable changes in the α- and β-peaks of heme, suggestive of changes in the heme binding pocket that most likely result from CO replacing a distal (sixth) amino acid ligand of heme (Fig. 7B). Addition of oxygen to the deoxy Fe(II) form also resulted in spectral changes, albeit smaller in scope than those that originated with the addition of CO (Fig. 7B). These results establish that RmdA binds heme specifically and is capable of responding to gaseous ligands, which in turn suggest that conformational changes in the heme-binding pocket might affect the activity of RmdA.
We tested whether PDE activity of the holo-MBP-RmdA protein is affected by CO or oxygen, but we observed no changes using several independently purified protein preparations, nor did we observe changes in PDE activity when the oxidized, Fe(III), met form of the protein was compared to the reduced, Fe(II), deoxy form or the apoprotein was compared to the holoprotein. At present, we don't understand why activity of RmdA appears to be decoupled from the sensory input. Several possibilities exist (see Discussion) and will be explored in the future.
To gain insight into the possibility that CdgA or SCO5511 binds heme, we compared the sequences of their PAS9 domains with that of RmdA. We found these sequences to be highly divergent and lacking conserved residues (e.g., His) that often ligate heme iron. The highest similarity (>30% identity) to the PAS9 domain of RmdA was displayed by the RmdA homologs from Streptomyces species, as well as PAS9 proteins from cyanobacteria and proteobacteria. Therefore, we could not predict whether CdgA or SCO5511 binds heme. It appears that PAS domains have evolved various ways to coordinate the heme ligand. For example, the heme-PAS domains from oxygen-sensing c-di-GMP PDE, E. coli DosP (16, 62), and histidine kinase Bradyrhizobium FixL (25, 29) have low, <16% identity to PAS9 of RmdA.
RmdB (SCO5495) functions as a c-di-GMP PDE.
To investigate the involvement of SCO5495 in S. coelicolor development, we constructed a Tn5062 mutant (Fig. 3B), designated TH101. We observed that colonies of TH101 produced significantly less gray spore pigment than the wild type (Fig. 1C and E). Microscopic analysis revealed that after 3 days of growth, TH101 produced few sporulated aerial filaments compared to the wild type (Fig. 2). After 5 days, the number of spore chains increased, yet most of them displayed a delay in the completion of septation. By day seven, the quantity of spore chains was comparable to that observed for the wild type at 3 days (Fig. 2). These phenotypes were identical to the phenotypes of the rmdA mutants, TH1 and TH100, thus suggesting that RmdB also modulates development in S. coelicolor. The similarity between the rmdA and rmdB phenotypes suggested that RmdB functions as a c-di-GMP PDE.
The 746-aa RmdB protein contains an approximately 300-aa-long N-terminal domain comprising seven transmembrane motifs followed by the GGDEF-EAL domain tandem (Fig. 4A). Neither the GGDEF nor EAL domains of SCO5495 contain readily identifiable residues inconsistent with DGC or PDE activities, respectively. To test for RmdB activity, we cloned its cytoplasmic GGDEF-EAL module downstream of the MBP tag. The effects of this fusion on motility in the semisolid agar were assayed with the E. coli test strains, MG1655 and MG1655 yhjH, and the effect of MBP-RmdB was consistent with it acting as a PDE (Fig. 5A). The MBP-SCO1397 fusion, used as a negative control, showed no PDE activity (Fig. 5A), consistent with the sequence analysis presented above.
We overexpressed and purified the cytoplasmic fragment of RmdB and found that it has PDE activity in vitro. The relative activity of the RmdB fragment was higher than that of RmdA (Fig. 5B), which is consistent with the motility-based assay (Fig. 5A). It is noteworthy that activities of the truncated fusion proteins measured here are only approximations of the in vivo activities of full-length proteins, which are further influenced by their respective sensory domains. The relative contributions of RmdA and RmdB to regulation of development likely depend on environmental conditions.
We measured the effect of the rmdB mutation on intracellular c-di-GMP levels and found no significant differences between TH101 and the wild type (Fig. 6). The lack of perturbations in intracellular c-di-GMP levels is an indication that RmdB is specifically involved in controlling S. coelicolor development.
RmdA and RmdB additively regulate colony morphology, sporulation, and aerial mycelium development.
To investigate the relationship between RmdA and RmdB, we constructed the rmdA rmdB double mutant by inactivating the rmdB gene in strain TH1. The colony morphology of the double mutant, TH106, was strikingly different from those of the wild type and individual rmd mutants in that the double mutant had abnormally large colonies with the bald (bld) phenotype (Fig. 1D), i.e., it was completely blocked in the development of aerial hyphae (Fig. 1E). The introduction into the rmdA rmdB mutant of a cosmid containing the wild-type rmdA or rmdB gene complemented the defect in aerial mycelium formation (Fig. 1F).
We investigated the effect of the double mutant on intracellular c-di-GMP levels and found that these levels were increased by approximately 3-fold compared to those for the wild type (Fig. 6). Taken together, these data demonstrate that two PDEs, RmdA and RmdB, have at least partially overlapping functions in controlling colony morphology, sporulation, and aerial mycelium development in S. coelicolor.
DISCUSSION
S. coelicolor has been a model organism for studying cell division and development in bacteria because mutants in the complex cycle of morphological development can be readily identified both visually and microscopically. Here we relied on decreased gray pigmentation of Tn mutants as an indication of a potential defect in sporulation to identify the rmdA gene. We showed that rmdA encodes a c-di-GMP PDE. Another c-di-GMP PDE identified here, RmdB, is also involved in developmental control and morphological differentiation of S. coelicolor. According to our bioinformatics analysis, RmdA and RmdB are the only EAL domain-containing c-di-GMP PDEs present in S. coelicolor. The membrane-localized SCO5218 containing an HD-GYP domain may potentially function as yet another c-di-GMP PDE in this bacterium. Given the high degree of conservation among RmdA and RmdB homologs, it is likely that c-di-GMP signaling pathways modulate development in various filamentous actinomycetes. This notion emphasizes the need to understand c-di-GMP signaling in this group of bacteria, which includes many species that produce pharmacologically important antibiotics.
Numerous questions regarding the role of c-di-GMP in S. coelicolor remain. One set of questions concerns environmental and intracellular signals that affect expression and activity of the enzymes involved in c-di-GMP signaling. In Streptomyces, development is initiated by nutrient starvation and the production of secondary metabolites like antibiotics (22). How these signals affect c-di-GMP synthesis and degradation remains to be investigated.
In this study, we noticed that RmdA is a hemoprotein and therefore is expected to sense gaseous ligands or redox changes. Interestingly, in several proteobacterial species, including Gluconacetobacter xylinus, E. coli, and Bordetella pertussis (12, 62, 63), the oxygen concentration affects c-di-GMP synthesis or hydrolysis via heme-containing enzymes. However, studies of the role for oxygen in Streptomyces development have been mainly confined to antibiotic production in industrial cultures, not mechanistic studies (2, 37, 64). While RmdA is capable of sensing gaseous ligands, its PDE activity is surprisingly nonresponsive to perturbations. The apparent disconnect between the sensory input and output activity of RmdA may be due to various reasons. It is possible that proper experimental conditions were not found or that the MBP-RmdA fusion was impaired due to the MBP tag or the lack of the N-terminal domain X. It is also possible that RmdA gains responsiveness only in a complex with its cognate DGC. Several c-di-GMP PDEs have now been documented to function in signaling complexes (48, 61). Yet another scenario is that RmdA is a bifunctional GGDEF-EAL protein with a constitutive PDE activity. An inactivation of the EAL domain (either by proteolysis or by interactions with an RmdA partner) may be required to unmask the DGC activity of the GGDEF domain, and the DGC activity may be designed to be regulated via a heme-PAS9 domain. Examples of such unusual regulation in the bifunctional GGDEF-EAL proteins are beginning to emerge (57, 60).
One potential interaction partner of RmdA could be SCO0929, whose gene is located in an apparent operon with rmdA (Fig. 3), and the operon is conserved among filamentous actinomycetes. While in this study we did not observe visible developmental defects of the SCO0929 mutant, additional experimental conditions may be required to uncover its role. Signal sensing and processing in SCO0929-RmdA and the PAS9 domain proteins CdgA and SCO5511 also deserve further investigation.
A second set of questions concerns organization and specificity of c-di-GMP signaling pathways in S. coelicolor. While the issue of whether unique modules comprising DGCs and PDEs signal via dedicated c-di-GMP receptors (effector proteins) to unique targets or whether all DGCs and PDEs contribute to a total intracellular pool remains somewhat controversial, more and more evidence points toward specific cascades as a prevalent regulatory modality. A recent study where each DGC of the tiny bacterium Bdellovibrio was shown to have a specific phenotype that did not overlap with phenotypes controlled by other DGCs (28) provided yet another convincing argument in support of target-specific c-di-GMP signaling modules. We found that either the rmdA or rmdB mutation could cause a pronounced developmental delay, yet neither mutation resulted in significant perturbations in the total intracellular c-di-GMP levels (Fig. 6). This is consistent with the interpretation that a local, as opposed to a total, c-di-GMP pool is important for developmental regulation in S. coelicolor. It further shows a measure of specificity with which the RmdA and RmdB PDEs jointly guard c-di-GMP-dependent targets involved in development.
Recent experiments by the Buttner group showed that overexpression of S. coelicolor DGCs encoded by cdgA or cdgB (SCO4281) block aerial mycelium formation (17, 59), i.e., produce the same bald phenotype as does the rmdA rmdB double mutant. It is likely that RmdA and RmdB counterbalance the effects of CdgA and CdgB. Whether CdgA, CdgB, RmdA, and RmdB are organized in specific signaling modules and, if so, how these modules operate remain to be investigated. It is peculiar that expression of cdgA and cdgB is regulated by BldD, the transcription factor affecting multiple developmentally regulated genes. BldD binding sites are also present upstream of SCO5511 (60), yet another predicted DGC (unpublished data), but they are not present upstream of rmdA or rmdB.
The third set of questions concerns molecular targets of c-di-GMP that result in developmental and colony morphology phenotypes and mechanisms through which c-di-GMP exerts its effects. In order to erect upward into the air and to form aerial hyphae, S. coelicolor filaments must break the surface tension created by the aqueous environment of the vegetative mycelium. Two overlapping mechanisms are involved in overcoming surface tension in S. coelicolor, i.e., chaplin proteins and the SapB protein (10, 15, 18, 31). We hypothesize that these may constitute c-di-GMP targets. However, at present little is known about regulatory pathways affecting synthesis or activities of these proteins.
Another important question that cannot yet be answered concerns identities of the S. coelicolor c-di-GMP receptors/effectors that mediate the effects of c-di-GMP on ultimate targets. The readily recognizable c-di-GMP receptors are represented by PilZ domains (1, 50), I sites in the enzymatically inactive GGDEF domains (11), or enzymatically inactive EAL domains (40). c-di-GMP receptors that are not based on PilZ, EAL, or GGDEF domains are difficult to recognize because their c-di-GMP-binding patterns are not readily identifiable (38). The PilZ domains are not encoded in the Streptomyces genomes. Among the GGDEF domain proteins, only CdgB and SCO5511 contain I sites (60), and both function as a DGC (60; unpublished data). SCO1397 is the only enzymatically inactive EAL domain protein that is not a DGC. While we cannot completely exclude the possibility that it binds c-di-GMP, this is unlikely. Overexpressed SCO1397 did not improve motility of MG1655 yhjH in the semisolid agar (Fig. 5A), in contrast to the case with other c-di-GMP receptors that serve as c-di-GMP sinks and decrease intracellular c-di-GMP levels (29). It therefore appears that identification of c-di-GMP receptors in streptomycetes will present a formidable challenge.
Finally, we would like to note that while the importance of the c-di-GMP signaling pathways in lifestyle changes in the proteobacteria has been well appreciated and the mechanisms of regulation are being elucidated (27, 39, 46), the studies of c-di-GMP signaling pathways in Gram-positive bacteria are still in their infancy. Our study, along with those of Buttner and colleagues (17, 59), reveals an apparently conserved function of c-di-GMP signaling in modulation of development in actinomycetes. This extends the realm of processes controlled by c-di-GMP and suggests that future studies in the Gram-positive bacteria may result in new, yet-unanticipated processes controlled by this second messenger, new kinds of c-di-GMP receptors, and new regulatory mechanisms.
ACKNOWLEDGMENTS
We are indebted to Paul Dyson for providing the Tn5062 mutagenized cosmids used for strain construction and to A. Daniel Jones, Lijun Chen, and Bev Chamberlin for c-di-GMP quantification via LC-MS.
Funding for this study was provided by the William J. von Liebig Foundation (to J.A.B.), NSF grant MCB 1052575 (to M.G.), and a Sigma Xi Grant-in-Aid of Research, administered by the National Academy of Sciences (to R.C.J.). We are grateful to the von Liebig Foundation for fellowships (to T.D.H. and N.T.K.), to the American Society for Microbiology for Undergraduate Research Fellowships (to T.D.H., R.C.J., and N.T.K.), and for Merck/AAAS Undergraduate Science Research Program funding (to J.A.B. and R.M.G.).
Footnotes
Published ahead of print 29 June 2012
REFERENCES
- 1. Amikam D, Galperin MY. 2006. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22:3–6 [DOI] [PubMed] [Google Scholar]
- 2. Bankar SB, Singhal RS. 2011. Improved poly-epsilon-lysine biosynthesis using Streptomyces noursei NRRL 5126 by controlling dissolved oxygen during fermentation. J. Microbiol. Biotechnol. 21:652–658 [PubMed] [Google Scholar]
- 3. Barends TR, et al. 2009. Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase. Nature 459:1015–1018 [DOI] [PubMed] [Google Scholar]
- 4. Bennett JA. 2007. Molecular genetic analysis of division and development in Streptomyces coelicolor. Ph.D. dissertation Duquesne University, Pittsburgh, PA [Google Scholar]
- 5. Bennett JA, McCormick JR. 2001. Two new loci affecting cell division identified as suppressors of an ftsQ-null mutation in Streptomyces coelicolor A3(2). FEMS Microbiol. Lett. 202:251–256 [DOI] [PubMed] [Google Scholar]
- 6. Bentley SD, et al. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147 [DOI] [PubMed] [Google Scholar]
- 7. Bishop A, Fielding S, Dyson P, Herron P. 2004. Systematic insertional mutagenesis of a streptomycete genome: a link between osmoadaptation and antibiotic production. Genome Res. 14:893–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bobrov AG, et al. 2011. Systematic analysis of cyclic di-GMP signalling enzymes and their role in biofilm formation and virulence in Yersinia pestis. Mol. Microbiol. 79:533–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bordeleau E, Fortier LC, Malouin F, Burrus V. 2011. c-di-GMP turn-over in Clostridium difficile is controlled by a plethora of diguanylate cyclases and phosphodiesterases. PLoS Genet. 7:e1002039 doi:10.1371/journal.pgen.1002039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Capstick DS, Willey JM, Buttner MJ, Elliot MA. 2007. SapB and the chaplins: connections between morphogenetic proteins in Streptomyces coelicolor. Mol. Microbiol. 64:602–613 [DOI] [PubMed] [Google Scholar]
- 11. Chan C, et al. 2004. Structural basis of activity and allosteric control of diguanylate cyclase. Proc. Natl. Acad. Sci. U. S. A. 101:17084–17089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chang AL, et al. 2001. Phosphodiesterase A1, a regulator of cellulose synthesis in Acetobacter xylinum, is a heme-based sensor. Biochemistry 40:3420–3426 [DOI] [PubMed] [Google Scholar]
- 13. Chater KF. 2000. Developmental decisions during sporulation in the aerial mycelium in Streptomyces, p 33–48 In Brun YV, Shimkets LJ. (ed), Prokaryotic development. ASM Press, Washington, DC [Google Scholar]
- 14. Christen M, Christen B, Folcher M, Schauerte A, Jenal U. 2005. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J. Biol. Chem. 280:30829–30837 [DOI] [PubMed] [Google Scholar]
- 15. Claessen D, et al. 2003. A novel class of secreted hydrophobic proteins is involved in aerial hyphae formation in Streptomyces coelicolor by forming amyloid-like fibrils. Genes Dev. 17:1714–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Delgado-Nixon VM, Gonzalez G, Gilles-Gonzalez MA. 2000. Dos, a heme-binding PAS protein from Escherichia coli, is a direct oxygen sensor. Biochemistry 39:2685–2691 [DOI] [PubMed] [Google Scholar]
- 17. den Hengst CD, et al. 2010. Genes essential for morphological development and antibiotic production in Streptomyces coelicolor are targets of BldD during vegetative growth. Mol. Microbiol. 78:361–379 [DOI] [PubMed] [Google Scholar]
- 18. Elliot MA, et al. 2003. The chaplins: a family of hydrophobic cell-surface proteins involved in aerial mycelium formation in Streptomyces coelicolor. Genes Dev. 17:1727–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fang X, Gomelsky M. 2010. A post-translational, c-di-GMP-dependent mechanism regulating flagellar motility. Mol. Microbiol. 76:1295–1305 [DOI] [PubMed] [Google Scholar]
- 20. Ferreira RB, Antunes LC, Greenberg EP, McCarter LL. 2008. Vibrio parahaemolyticus ScrC modulates cyclic dimeric GMP regulation of gene expression relevant to growth on surfaces. J. Bacteriol. 190:851–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Finn RD, et al. 2010. The Pfam protein families database. Nucleic Acids Res. 38:D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Flardh K, Buttner MJ. 2009. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat. Rev. Microbiol. 7:36–49 [DOI] [PubMed] [Google Scholar]
- 23. Galperin MY, Natale DA, Aravind L, Koonin EV. 1999. A specialized version of the HD hydrolase domain implicated in signal transduction. J. Mol. Microbiol. Biotechnol. 1:303–305 [PMC free article] [PubMed] [Google Scholar]
- 24. Gomelsky M, Kaplan S. 1998. AppA, a redox regulator of photosystem formation in Rhodobacter sphaeroides 2.4.1, is a flavoprotein. Identification of a novel fad binding domain. J. Biol. Chem. 273:35319–35325 [DOI] [PubMed] [Google Scholar]
- 25. Gong W, et al. 1998. Structure of a biological oxygen sensor: a new mechanism for heme-driven signal transduction. Proc. Natl. Acad. Sci. U. S. A. 95:15177–15182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7:263–273 [DOI] [PubMed] [Google Scholar]
- 27. Henry JT, Crosson S. 2011. Ligand-binding PAS domains in a genomic, cellular, and structural context. Annu. Rev. Microbiol. 65:261–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hobley L, et al. 2012. Discrete cyclic di-GMP-dependent control of bacterial predation versus axenic growth in Bdellovibrio bacteriovorus. PLoS Pathog. 8:e1002493 doi:10.1371/journal.ppat.1002493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Key J, Moffat K. 2005. Crystal structures of deoxy and CO-bound bjFixLH reveal details of ligand recognition and signaling. Biochemistry 44:4627–4635 [DOI] [PubMed] [Google Scholar]
- 30. Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. 2000. Practical Streptomyces genetics. The John Innes Foundation, Colney, United Kingdom [Google Scholar]
- 31. Kodani S, et al. 2004. The SapB morphogen is a lantibiotic-like peptide derived from the product of the developmental gene ramS in Streptomyces coelicolor. Proc. Natl. Acad. Sci. U. S. A. 101:11448–11453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kumar M, Chatterji D. 2008. Cyclic di-GMP: a second messenger required for long-term survival, but not for biofilm formation, in Mycobacterium smegmatis. Microbiology 154:2942–2955 [DOI] [PubMed] [Google Scholar]
- 33. Letunic I, Doerks T, Bork P. 2012. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 40:D302–D305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lovering AL, Capeness MJ, Lambert C, Hobley L, Sockett RE. 2011. The structure of an unconventional HD-GYP protein from Bdellovibrio reveals the roles of conserved residues in this class of cyclic-di-GMP phosphodiesterases. mBio 2(5):e00163-11 doi:10.1128/mBio. 00163-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. MacNeil DJ. 1988. Characterization of a unique methyl-specific restriction system in Streptomyces avermitilis. J. Bacteriol. 170:5607–5612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McCormick JR, Losick R. 1996. Cell division gene ftsQ is required for efficient sporulation but not growth and viability in Streptomyces coelicolor A3(2). J. Bacteriol. 178:5295–5301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mehmood N, et al. 2011. Oxygen supply controls the onset of pristinamycins production by Streptomyces pristinaespiralis in shaking flasks. Biotechnol. Bioeng. 108:2151–2161 [DOI] [PubMed] [Google Scholar]
- 38. Mills E, Pultz IS, Kulasekara HD, Miller SI. 2011. The bacterial second messenger c-di-GMP: mechanisms of signalling. Cell. Microbiol. 13:1122–1129 [DOI] [PubMed] [Google Scholar]
- 39. Moskvin OV, Kaplan S, Gilles-Gonzalez MA, Gomelsky M. 2007. Novel heme-based oxygen sensor with a revealing evolutionary history. J. Biol. Chem. 282:28740–28748 [DOI] [PubMed] [Google Scholar]
- 40. Newell PD, Monds RD, O'Toole GA. 2009. LapD is a bis-(3′,5′)-cyclic dimeric GMP-binding protein that regulates surface attachment by Pseudomonas fluorescens Pf0-1. Proc. Natl. Acad. Sci. U. S. A. 106:3461–3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Oh SH, Chater KF. 1997. Denaturation of circular or linear DNA facilitates targeted integrative transformation of Streptomyces coelicolor A3(2): possible relevance to other organisms. J. Bacteriol. 179:122–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Paget MS, Chamberlin L, Atrih A, Foster SJ, Buttner MJ. 1999. Evidence that the extracytoplasmic function sigma factor sigmaE is required for normal cell wall structure in Streptomyces coelicolor A3(2). J. Bacteriol. 181:204–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Paul R, et al. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 18:715–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Redenbach M, et al. 1996. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol. Microbiol. 21:77–96 [DOI] [PubMed] [Google Scholar]
- 45. Romling U, Simm R. 2009. Prevailing concepts of c-di-GMP signaling. Contrib. Microbiol. 16:161–181 [DOI] [PubMed] [Google Scholar]
- 46. Ross P, et al. 1990. The cyclic diguanylic acid regulatory system of cellulose synthesis in Acetobacter xylinum. Chemical synthesis and biological activity of cyclic nucleotide dimer, trimer, and phosphothioate derivatives. J. Biol. Chem. 265:18933–18943 [PubMed] [Google Scholar]
- 47. Ross P, et al. 1987. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325:279–281 [DOI] [PubMed] [Google Scholar]
- 48. Ryan RP, Dow JM. 2010. Intermolecular interactions between HD-GYP and GGDEF domain proteins mediate virulence-related signal transduction in Xanthomonas campestris. Virulence 1:404–408 [DOI] [PubMed] [Google Scholar]
- 49. Ryan RP, et al. 2006. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc. Natl. Acad. Sci. U. S. A. 103:6712–6717 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50. Ryjenkov DA, Simm R, Romling U, Gomelsky M. 2006. The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J. Biol. Chem. 281:30310–30314 [DOI] [PubMed] [Google Scholar]
- 51. Ryjenkov DA, Tarutina M, Moskvin OV, Gomelsky M. 2005. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 187:1792–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schirmer T, Jenal U. 2009. Structural and mechanistic determinants of c-di-GMP signalling. Nat. Rev. Microbiol. 7:724–735 [DOI] [PubMed] [Google Scholar]
- 53. Schmidt AJ, Ryjenkov DA, Gomelsky M. 2005. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J. Bacteriol. 187:4774–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tal R, et al. 1998. Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: genetic organization and occurrence of conserved domains in isoenzymes. J. Bacteriol. 180:4416–4425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tamayo R, Pratt JT, Camilli A. 2007. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu. Rev. Microbiol. 61:131–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tamayo R, Tischler AD, Camilli A. 2005. The EAL domain protein VieA is a cyclic diguanylate phosphodiesterase. J. Biol. Chem. 280:33324–33330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tarutina M, Ryjenkov DA, Gomelsky M. 2006. An unorthodox bacteriophytochrome from Rhodobacter sphaeroides involved in turnover of the second messenger c-di-GMP. J. Biol. Chem. 281:34751–34758 [DOI] [PubMed] [Google Scholar]
- 58. Taylor BL, Zhulin IB. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tran NT, Den Hengst CD, Gomez-Escribano JP, Buttner MJ. 2011. Identification and characterization of CdgB, a diguanylate cyclase involved in developmental processes in Streptomyces coelicolor. J. Bacteriol. 193:3100–3108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Trimble MJ, McCarter LL. 2011. Bis-(3′-5′)-cyclic dimeric GMP-linked quorum sensing controls swarming in Vibrio parahaemolyticus. Proc. Natl. Acad. Sci. U. S. A. 108:18079–18084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tuckerman JR, Gonzalez G, Gilles-Gonzalez MA. 2011. Cyclic di-GMP activation of polynucleotide phosphorylase signal-dependent RNA processing. J. Mol. Biol. 407:633–639 [DOI] [PubMed] [Google Scholar]
- 62. Tuckerman JR, et al. 2009. An oxygen-sensing diguanylate cyclase and phosphodiesterase couple for c-di-GMP control. Biochemistry 48:9764–9774 [DOI] [PubMed] [Google Scholar]
- 63. Wan X, et al. 2009. Globins synthesize the second messenger bis-(3′-5′)-cyclic diguanosine monophosphate in bacteria. J. Mol. Biol. 388:262–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wei ZH, Bai L, Deng Z, Zhong JJ. 2011. Enhanced production of validamycin A by H2O2-induced reactive oxygen species in fermentation of Streptomyces hygroscopicus 5008. Bioresour. Technol. 102:1783–1787 [DOI] [PubMed] [Google Scholar]
- 65. Wolf A, Visick K. (ed). 2010. The second messenger cyclic di-GMP, p 356 ASM Press, Washington, DC [Google Scholar]