Abstract
PfmR is one of four TetR family transcriptional regulators found in the extremely thermophilic bacterium, Thermus thermophilus HB8. We identified three promoters with strong negative regulation by PfmR, both in vivo and in vitro. PfmR binds pseudopalindromic sequences, with the consensus sequence of 5′-TACCGACCGNTNGGTN-3′ surrounding the promoters. According to the amino acid sequence and three-dimensional structure analyses of the PfmR-regulated gene products, they are predicted to be involved in phenylacetic acid and fatty acid metabolism. In vitro analyses revealed that PfmR weakly cross-regulated with the TetR family repressor T. thermophilus PaaR, which controls the expression of the paa gene cluster putatively involved in phenylacetic acid degradation but not with another functionally identified TetR family repressor, T. thermophilus FadR, which is involved in fatty acid degradation. The X-ray crystal structure of the N-terminal DNA-binding domain of PfmR and the nucleotide sequence of the predicted PfmR-binding site are quite similar to those of the TetR family repressor QacR from Staphylococcus aureus. Similar to QacR, two PfmR dimers bound per target DNA. The bases recognized by QacR within the QacR-binding site are conserved in the predicted PfmR-binding site, and they were important for PfmR to recognize the binding site and properly assemble on it. The center of the PfmR molecule contains a tunnel-like pocket, which may be the ligand-binding site of this regulator.
INTRODUCTION
TetR family transcriptional regulators are one of the major families of bacterial transcriptional regulators and are widely distributed among bacteria (23, 30, 32). This family protein has an N-terminal helix-turn-helix (HTH) DNA-binding domain, which exhibits high sequence similarity among the family members (30, 42). The TetR family protein is composed of approximately 10 α-helices per monomer and forms a dimer (42). The TetR family regulators are mostly repressors, and, interestingly, they control various genes with products involved in multidrug resistance, as well as enzymes in catabolic pathways, antibiotic biosynthesis, osmotic stress, and pathogenicity (30). The TetR family proteins bind their operators, composed of ∼10- to ∼30-bp palindromic sequences, to repress the target genes and are released from the DNA when the regulators bind their cognate ligands (42). For example, the Escherichia coli TetR protein dimer binds its operator, composed of a 15-bp palindromic sequence, upstream of the tetracycline transporter gene tetA, and tetracycline binds this protein to derepress the target gene expression (12, 14, 25, 26). In the case of the QacR protein from Staphylococcus aureus, two dimers bind the operator of the multidrug transporter gene qacA, which is composed of a 28-bp palindromic sequence (9). Each dimer binds one half of the operator; thus, the monomers bind proximally and distally relative to the center of the palindromic sequence (10, 34). The ligands of QacR have been identified and include rhodamine 6G and malachite green (35). The ActR protein from Streptomyces coelicolor, which regulates the expression of the ActA efflux pump, uses actinorhodin or various actinorhodin biosynthetic intermediates as ligands (41). Simocyclinone is a ligand of the SimR protein from Streptomyces antibioticus and controls the expression of the efflux pump gene simX (18). Two mechanisms of derepression by the TetR family repressors have been proposed. One involves the ligands binding around the center of the repressor molecules, causing the proteins to undergo a conformational change to increase the distance between the DNA recognition helices and thereby dissociate from DNA (26, 35, 41). The other involves the ligand-binding capture of one of the apo-repressors in a conformation that is not compatible with DNA binding rather than inducing a conformational change (18, 31).
Thermus thermophilus HB8, which belongs to the phylum Deinococcus-Thermus, is an extremely thermophilic bacterium isolated from the water at a Japanese hot spring and grows at an optimum temperature range of 65 to 72°C (27). Its genome is composed of the 1.85-Mbp chromosomal DNA, the 0.26-Mbp plasmid pTT27, and the 9.32-kbp plasmid pTT8, encoding 1,973, 251, and 14 open reading frames (ORFs), respectively (NCBI accession numbers NC_006461, NC_006462, and NC_006463, respectively). Recently, a third plasmid, pVV8, encoding 91 ORFs was discovered, but a laboratory strain does not harbor this plasmid (24). With its relatively small genome, T. thermophilus HB8 is considered to represent a minimal model of life, and structural and functional genomics studies have been performed on this strain (http://www.thermus.org/e_index.htm). Based on the genome sequence analysis, this strain is predicted to have approximately 70 transcriptional regulators, but only a few of them have been characterized. T. thermophilus HB8 encodes four TetR family proteins, TTHA0101 (FadR), TTHA0167, TTHA0973 (PaaR), and TTHB023 (NCBI accession numbers YP_143367, YP_143433, YP_144239, and YP_145262, respectively), which share 44 to 57% amino acid sequence similarity with one another. FadR negatively regulates the expression of 21 genes under nine promoters, including those involved in fatty acid (FA) degradation (1). FadR binds a medium-to-long straight chain (C10 to C18) fatty acyl-coenzyme A (CoA) molecule as a ligand to derepress the target genes (1). PaaR negatively regulates the expression of two operons encoding several putative paa genes for phenylacetic acid (PAA) degradation (33). Phenylacetyl (PA)-CoA is a ligand of PaaR and was effective for transcriptional derepression (33). In this study, we performed functional and structural analyses of the TTHB023 protein and discussed its similarities with several other TetR family proteins.
MATERIALS AND METHODS
Purification of recombinant PfmR proteins.
The T. thermophilus pfmR (TTHB023) gene was amplified by genomic PCR using the primers P01 and P02 (see Table S1 in the supplemental material), and the amplified fragment was cloned under the control of the T7 promoter (NdeI-BamHI sites) in the E. coli expression vector pET-21a(+) (Merck), to construct pET-ttPfmR.
E. coli BL21(DE3) cells (Merck) harboring pET-ttPfmR were cultured at 37°C in 6 liters of Luria-Bertani (LB) broth containing supplement A [0.05% glucose, 0.2% lactose monohydrate, 0.5% glycerol, 50 mM Na2HPO4, 50 mM KH2PO4, 25 mM (NH4)2SO4, 1 mM MgSO4] and 50 μg of ampicillin ml−1 for 16 h. The cells (18 g) were resuspended in 60 ml of 20 mM Tris-HCl (pH 8.0) supplemented with 50 mM NaCl and 5 mM β-mercaptoethanol and were disrupted by sonication in ice water. The same volume of buffer, preheated at 70°C, was added to the cell lysate. This mixture was incubated for 10 min at 70°C and then ultracentrifuged (200,000 × g) for 1 h at 4°C. The supernatant was applied to a Resource Iso column (GE Healthcare), preequilibrated with 50 mM sodium phosphate buffer (pH 7.0) containing 1.5 M (NH4)2SO4. The flowthrough fraction was collected and desalted by fractionation on a HiPrep 26/10 desalting column (GE Healthcare), preequilibrated with 20 mM Tris-HCl (pH 8.0). The sample was then applied to a Resource Q column (GE Healthcare), preequilibrated with the same buffer, and the bound protein was eluted with a linear gradient of 0 to 0.5 M NaCl. The target fractions were collected and applied to a HiLoad 16/60 Superdex 75 prep grade (GE Healthcare) column, preequilibrated with 20 mM Tris-HCl (pH 8.0) containing 0.15 M NaCl. The target fractions were collected and concentrated with a Vivaspin 20 concentrator (5,000-molecular-weight cutoff; Sartorius).
Selenomethionine (SeMet)-containing recombinant PfmR (Se-PfmR) was generated using the methionine auxotroph E. coli B834(DE3) strain (Merck) as the host. The recombinant strain was grown in LeMaster medium (19) containing 50 μg of SeMet ml−1, supplement A (see above), and 50 μg of ampicillin ml−1, for 16 h. Se-PfmR was purified in basically the same manner as the native protein (see above).
Disruption of the T. thermophilus pfmR gene.
The upstream region (positions 14246 to 13724 of pTT27) of the pfmR gene, followed by the thermostable kanamycin resistance marker gene and the downstream region (positions 13193 to 12674 of pTT27) of the pfmR gene, was inserted into pUC19 (HindIII-EcoRI sites) to construct the plasmid pUC-ΔpfmR. The plasmid was transformed into the T. thermophilus HB8 strain, and the kanamycin-resistant clones were isolated as disruptants of the pfmR gene (ΔpfmR), as described previously (11). Genomic PCR was performed to confirm the replacement of the pfmR gene by the kanamycin resistance marker gene by using primers corresponding to the kanamycin resistance marker gene and the regions upstream or downstream of the pfmR gene and monitoring the expected lengths of the DNA fragments amplified by the primers P07/P10, P07/P12, P08/P09, and P08/P11 (see Table S1 in the supplemental material). We also confirmed by genomic PCR that a DNA fragment derived from the pfmR gene, which was amplified from the genome of the wild type, was not amplified from that of the mutant, by using primers P13/P14 (see Table S1).
DNA microarray.
The T. thermophilus HB8 strain was cultured at 70°C for 6 h in 1 liter of TT medium, as described previously (2). The crude RNA was extracted from each cell, and after the cDNA was synthesized, it was fragmented and labeled with biotin-dideoxy UTP, as described previously (2). The 3′-terminally labeled cDNA was hybridized to a TTHB8401a520105F GeneChip (Affymetrix), and then the array was washed, stained, and scanned as described previously (2). The raw intensities for the seven independently cultured wild-type and three ΔpfmR strains were each summarized to 2,266 ORFs, including 38 ORFs out of 91 on pVV8, using the GeneChip Operating Software, version 1.2 (Affymetrix). The data sets were then normalized through the following steps, using the Subio Platform (Subio), including the shifting of low signals less than 1.0 to 1.0, the log2-based transformation of the data, and global normalization (normalized to the 75th percentile [third quartile]). We excluded 142 genes with detection levels labeled as absent (29) in both the wild-type and ΔpfmR strains. The remaining data of 2,124 ORFs were used for the following analysis. The t test P values of the observed differences in the normalized intensities between the wild-type and ΔpfmR strains were calculated using the Subio Platform, and then from these values their false-discovery rates (q values) (37) were calculated using the R program (http://www.R-project.org).
BIAcore biosensor assay.
A DNA fragment (0.1 mM), biotinylated at the 5′ end of one strand, was diluted to 50 nM in 10 mM HEPES-NaOH (pH 7.4) buffer containing 0.5 M NaCl, 3 mM EDTA, and 0.005% surfactant P20, and then applied to a streptavidin-coated (SA) biosensor chip (GE Healthcare), as described previously (33). All experiments for measuring the DNA and protein interactions were performed at 25°C using buffer A (10 mM HEPES-NaOH, pH 7.4, 0.3 M NaCl, 3 mM EDTA, 0.005% surfactant P20). The protein was diluted with the same buffer and then injected over the DNA surface at a flow rate of 20 μl min−1. Sensorgrams were recorded and normalized to a baseline of 0 resonance units (RU). An equivalent volume of each protein dilution was also injected over a nontreated surface to determine the bulk refractive index background. At the end of each cycle, the bound protein was removed by injecting 80 μl of 3 M NaCl to regenerate the chip. The association and dissociation rate constants (kon and koff, respectively) and the dissociation constant (Kd) values were determined by 1:1 Langmuir local fitting with the BIAevaluation, version 3.0, software (GE Healthcare). For the low-affinity binding, the binding data were fit using general fit-steady-state affinity, assuming a 1:1 binding interaction, with the BIAevaluation, version 3.0, software (GE Healthcare).
In vitro transcription assay. (i) Preparation of templates.
The construction of the plasmids containing the upstream regions of the TTHA0750, TTHA0987, and TTHB023 genes was performed in basically the same manner as described previously (36), using the oligonucleotides P15/P16, P17/P18, and P19/P20, respectively (see Table S1 in the supplemental material). Using each plasmid as the template, PCR was performed with the primers P21 and P22 (see Table S1) to prepare the template DNA for the transcription assay.
(ii) Runoff transcription.
Assays were performed in 15-μl reaction mixtures, in the absence or presence of the transcription factor, in basically the same manner as described previously (36). The template DNA was preincubated with or without the transcription factor at 55°C for 5 min. T. thermophilus RNA polymerase (RNAP)-σA holoenzyme was added, and the mixture was further incubated for 5 min. Transcription was initiated by the addition of 1.5 μCi of [α-32P]CTP and unlabeled ribonucleotide triphosphates. After incubation for 10 min, the reaction was stopped, and the sample was fractionated on a 10% polyacrylamide gel containing 8 M urea and analyzed by autoradiography.
Identification of the transcriptional start site.
Total RNA, isolated from wild-type T. thermophilus HB8 cells cultured at 70°C for 6 h in rich medium, was treated with DNase I, followed by ethanol precipitation, as described previously (36). Rapid amplification of cDNA ends (5′ RACE) was performed with a 5′ Full RACE Core Set (TaKaRa Bio), as described previously (33). The first-strand cDNA was synthesized with the 5′ phosphorylated primers P23, P24, and P25 for the TTHA0750, TTHA0987, and TTHB023 genes, respectively (see Table S1 in the supplemental material). The RNA was digested by RNase H, and the single-stranded cDNA was ligated with T4 RNA ligase to construct DNA concatemers. PCR was performed using the DNA concatemers as templates and P26/P27, P28/P29, and P30/P31 (see Table S1 in the supplemental material) as primers for the TTHA0750, TTHA0987, and TTHB023 genes, respectively. The second PCR was performed using the primers P32/P33, P34/P35, and P36/P37 (see Table S1) for the TTHA0750, TTHA0987, and TTHB023 genes, respectively. The amplified DNA fragments were cloned into the plasmid pT7Blue (Merck), and the nucleotide sequences of seven clones for each gene were analyzed.
RT-PCR.
Total RNA isolated from the wild-type T. thermophilus HB8 strain cultured for 11.3 h in rich medium was treated with DNase I, followed by ethanol precipitation, as described previously (36). Using the RNA (1 μg) as the template, reverse transcription (RT) was performed using a PrimeScript RT-PCR kit (TaKaRa Bio), as described previously (33). Using the reaction mixture (1 μl) as the template, PCR was performed in the presence of 0.2 μM concentrations of each primer (see Table S1 in the supplemental material) in a 25-μl reaction mixture. Primers P38 and P39, which correspond to the pTT27 genome positions 13209 to 13190 and 10206 to 10187, respectively, were used to detect the operon composed of the genes TTHB023 to TTHB018. The PCR analysis involved 30 cycles at 96°C for 30 s, 58°C for 30 s, and 72°C for 3.5 min in GC buffer II (TaKaRa Bio). To confirm the absence of genomic DNA contamination of the total RNA fraction used as the template, PCR was performed with no RT as a control.
Crystallization and X-ray crystal structure analysis of PfmR.
Crystallization of PfmR was performed by the sitting-drop vapor diffusion method by mixing 0.7 μl of the recombinant Se-PfmR protein (9.7 mg ml−1), obtained as described above, with an equal volume of reservoir solution containing 0.1 M phosphate-citrate (pH 4.2) and 40% (vol/vol) polyethylene glycol (PEG) 3000, followed by equilibration against 0.1 ml of the reservoir solution at 293 K. Crystals grew within 80 days to maximum dimensions of 150 by 100 by 10 μm.
A crystal was mounted on a cryo-loop and flash-cooled in a nitrogen gas stream at 100 K. Single anomalous dispersion (SAD) data were collected at a wavelength of 0.9791 Å with a MAR MX-225 charge-coupled-device (CCD) detector (Rayonix LLC), using the RIKEN Structural Genomics Beamline II Synchrotron (BL26B2) (40) at SPring-8 (Hyogo, Japan) (proposal number 20110005). The oscillation angle was 1°, the exposure time was 4 s per frame, and the camera distance was 200 mm. The collected data were processed with the HKL2000 program suite (28). Selenium sites were determined with the SOLVE program (38), and the resulting phases were improved with the RESOLVE program (38). The initial model was built with the Buccaneer program (6) in the CCP4 suite (5), and further manual model building was performed using Coot (7). Simulated annealing, energy minimization, and B factor refinement were performed using the CNS program package (4). Cycles of manual modeling and CNS refinement were performed, and 10% of the total reflections were randomly chosen for the Rfree sets. The quality of the structure was validated using the ADIT! Validation Server in PDBj (http://pdbdep.protein.osaka-u.ac.jp/validate/en/) and the MolProbity server (http://molprobity.biochem.duke.edu/). The data collection and refinement statistics are presented in Table 1.
Table 1.
X-ray data collection and refinement statistics
| Parameter | Valuea |
|---|---|
| Data collection | |
| Source | BL26B2 (SPring-8) |
| Wavelength (Å) | 0.9791 |
| Resolution (Å) | 50–2.27 |
| Space group | P21 |
| No. of molecules in an asymmetric unit | 4 |
| Unit cell parameters | |
| a, b, c (Å) | 49.7, 57.6, 133.4 |
| α, β, γ (°) | 90, 94, 90 |
| No. of measured reflections | 244,272 |
| No. of unique reflections | 35,016 |
| Completeness (%) | 99.8 (100) |
| Redundancy | 7.0 (6.8) |
| I/σ(I) | 18.6 (7.2) |
| Rmerge (%)b | 8.1 (26.3) |
| Phasing | |
| No. of Se atoms used | 18 |
| Figure of merit | 0.34 |
| Figure of merit after density modification | 0.58 |
| Refinement | |
| Resolution range (Å) | 28.8-2.27 |
| Rwork (%)c | 21.2 |
| Rfree (%)d | 26.9 |
| No. of protein atoms | 5,984 |
| No. of water atoms | 150 |
| Wilson B factor (Å2) | 38.4 |
| Avg B factor (Å2) | |
| Protein | 47.7 |
| Water | 53.1 |
| RMSD bond lengths (Å) | 0.007 |
| RMSD bond angles (°) | 1.1 |
| Ramachandran analysise | |
| Favored (%) | 98.1 |
| Outliers (%) | 0.0 |
Values in parentheses are for the highest-resolution shell.
Rmerge = ΣhΣi|Ih,i − <Ih>|/ΣhΣiIh,i, where Ih,i is the ith measured diffraction intensity of reflection h and <Ih> is the mean intensity of reflection h.
Rwork is the R-factor, calculated as Σ||Fo| − |Fc||/Σ|Fo|, where Fo and Fc are the observed and calculated structure factors, respectively.
Rfree is the R-factor calculated using 10% of the data that were excluded from the refinement.
Calculated by MolProbity.
DLS photometry.
The dynamic light scattering (DLS) of a solution (15 μl) containing 50 μM PfmR dimer, 10 mM sodium phosphate (pH 7.0), and 0.3 M NaCl, in the absence or presence of 25 μM DNA, was measured using a DynaPro-801 at 35°C (Protein Solutions). Data were analyzed using the instrument control software package Dynamics, version 5.26 (Protein Solutions). The data filter of the baseline limit was set to 1 ± 0.001 before regularization. At least 18 readings were recorded during each measurement. The Gaussian monomodal mode was used for the analysis. The experiment was conducted three times for each sample, and the molecular mass and polydispersity values were expressed as means ± standard deviations (SD).
Other methods.
The culture conditions for T. thermophilus HB8 in minimal medium were described previously (1, 2). The protein concentration was determined by measuring the absorbance at 280 nm (15). The N-terminal sequence analysis of the protein was performed with a protein sequencer (Procise HT; Applied Biosystems). To estimate the molecular mass of T. thermophilus PfmR, gel filtration chromatography was performed with a Superdex 75 HR 10/30 column (GE Healthcare). BLAST and conserved-domain database (CDD) searches were performed on the http://blast.ncbi.nlm.nih.gov/Blast.cgi and http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml websites, respectively. A nucleotide sequence motif search was performed with the GENETYX program, version 8.0 (GENETYX).
Accession numbers.
The microarray data discussed in this study have been deposited in the NCBI Gene Expression Omnibus (GEO [http://www.ncbi.nlm.nih.gov/geo/]) under the accession number GSE36912. The atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB) under the accession number 3VPR.
RESULTS
Initial characterization of the TTHB023 (PfmR) protein.
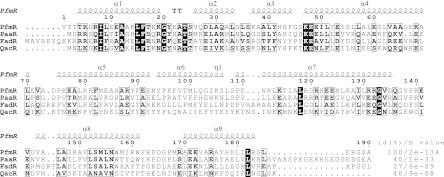
The TTHB023 ORF encodes 191 amino acid residues, with a predicted molecular mass of 21.7 kDa. Based on the results of a CDD (21) search, the protein has the conserved domain of the TetR family transcriptional regulators, comprising residues Ile9 to Ile55 (Ile8 to Ile54 in Fig. 1) with an E value of 6.22e−16 for the consensus sequence (pfam00440). According to a BLAST search, the homologous proteins most closely related to the TTHB023 protein exist in several Thermus species, with E values of 4e−126 to 2e−95. The TTHB023 protein shares homology with the TetR family regulators T. thermophilus PaaR and FadR, with E values of 1e−33 and 6e−09, respectively (Fig. 1). We found that the TTHB023 protein is a transcriptional regulator of functionally uncharacterized genes predicted to be involved in FA and PAA metabolism (see below); therefore, we named this protein T. thermophilus PfmR (phenylacetic acid and fatty acid metabolism regulator).
Fig 1.
Sequence alignment of T. thermophilus PfmR with representative homologous proteins. Strictly conserved residues are represented by white letters on a black background, and similar residues are depicted by boxed bold letters. PaaR, T. thermophilus PaaR (33); FadR, T. thermophilus FadR (1); QacR, S. aureus QacR (9). The sequences were aligned using Clustal W2 (16). The secondary structure of PfmR (chain A) was predicted with the DSSP program (13), and the figure was generated with ESPript, version 2.2 (8). α, η, and T represent the α-helix, 310-helix, and turn, respectively. The percent identities [id(%)] and the E values relative to PfmR, determined by BLAST, are indicated on the right.
PfmR was overexpressed in E. coli, and the recombinant protein was purified from the cell lysate to >95%, based on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (see Fig. S1 in the supplemental material). The N-terminal amino acid sequence of the purified protein was Val-Thr-Thr-Thr-Arg, indicating that the N-terminal methionine residue had been removed (data not shown). The DLS measurement of PfmR yielded a molecular mass of 47.8 ± 2.2 kDa, with a polydispersity value of 16.0% ± 3.8%. The molecular mass of PfmR, as estimated by gel filtration chromatography, was ∼34.0 kDa (see Fig. 7A, panel a, and B). These results suggested that PfmR exists as a homodimer in solution.
Fig 7.
Gel filtration analyses of T. thermophilus PfmR and the PfmR-DNA complex. (A) Elution profiles of the samples (26 μl) comprising 50 μM PfmR dimer (a), 50 μM PfmR dimer plus a 25 μM DNA fragment derived from the upstream region of the TTHB023 gene (see the text) (b), 50 μM PfmR dimer plus a 12.5 μM DNA fragment derived from the upstream region of the TTHB023 gene (c), 50 μM PfmR dimer plus a 25 μM DNA fragment derived from the upstream region of the TTHA0890 gene (see the text) (d), 50 μM PfmR dimer plus a 25 μM DNA fragment derived from the upstream region of the TTHB023 gene containing the A6C/T6′G mutations (see the text) (e), and 50 μM PfmR dimer plus a 25 μM DNA fragment derived from the upstream region of the TTHB023 gene containing the C8A/G8′T mutations (see the text) (f). The elution buffer was 10 mM sodium phosphate (pH 7.0) and 0.3 M NaCl. Relative absorbances at 220 nm (solid line) and 260 nm (dashed line) are indicated. The elution volume at the top of each peak is indicated. (B) Molecular masses of PfmR (open triangle) and PfmR with the DNA fragment derived from the upstream region of the TTHB023 gene (closed triangle). The partition coefficient (Kav) value was calculated as Kav = (elution volume − void volume)/(geometric column volume − void volume). As standards, the Kav values of conalbumin (75 kDa), ovalbumin (44 kDa), carbonic anhydrase (29 kDa), RNase A (13.7 kDa), and aprotinin (6.5 kDa) (open circles) were plotted.
Screening of T. thermophilus PfmR-regulated genes.
Using GeneChip technology, we observed that the expression profile of the pfmR mRNA did not vary significantly during cultivation in rich medium at 70°C (A600 of 0.1 to 5.0) (see Fig. S2 in the supplemental material). In order to investigate the effects of PfmR in vivo, we disrupted the pfmR gene and compared the growth of this strain with that of the wild type. The ΔpfmR strain was viable, with almost the same growth characteristics as those of the wild type in both rich medium (see Fig. S2) and synthetic medium containing 2% sucrose or 0.025% palmitic acid sodium salt as the sole carbon source (data not shown), indicating that the gene is not essential in this strain for growth in these media.
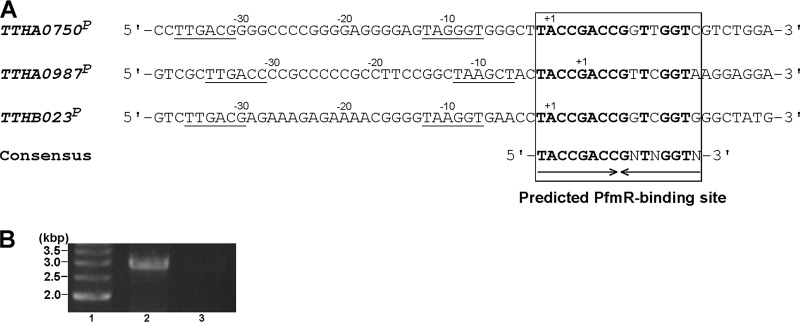
To identify the genes regulated by PfmR, wild-type and ΔpfmR strains were cultured in rich medium, and the expression of each mRNA was analyzed on a GeneChip. The expression level of the pfmR mRNA in the ΔpfmR strain, relative to that in the wild type, was 0.003 (q value of 0.05), indicating that the pfmR gene was disrupted. From the 2,124 genes analyzed, we selected the genes that showed altered expression in the ΔpfmR strain, with q values of ≤0.05. In total, 30 genes were selected as candidate genes regulated by PfmR (see Table S2 in the supplemental material). Among the genes, 12 were upregulated in the ΔpfmR strain (see Table S2). We analyzed the nucleotide sequences upstream of the 30 genes with expression levels that were altered in the ΔpfmR strain. We found that the upstream portions of the TTHA0750, TTHA0987, and TTHB023 (pfmR) genes included similar pseudopalindromic sequences: 5′-TACCGACCGGTTGGTC-3′, 5′-TACCGACCGTTCGGTA-3′, and 5′-TACCGACCGGTCGGTG-3′, respectively (Fig. 2A). The expression levels of the TTHA0750 and TTHA0987 genes in the ΔpfmR strain were increased 3.568-fold (q = 0.05) and 3.851-fold (q = 0.05), respectively, relative to those in the wild-type strain (see Table S2). These results suggested that PfmR binds the aforementioned sequences and negatively regulates these genes. In this study, 20 genes among the 38 pVV8-derived genes on the GeneChip were not used for the expression analysis because their detection levels were labeled as absent in both the wild-type and ΔpfmR strains. The detection levels of the remaining 18 genes were labeled as present in the wild-type strain but absent in the ΔpfmR strain. Moreover, the TTHV085 and TTHV086 genes on pVV8 were not amplified from the genome of the ΔpfmR strain (data not shown). The reason why the 12 genes on pVV8 were selected as significantly downregulated genes in the ΔpfmR strain (see Table S2) might be that this strain does not harbor pVV8. The altered expression of the other genes listed in Table S2, which do not have potential PfmR-binding sites, might be attributed to the side effects of either the pfmR gene deletion and/or the lack of pVV8.
Fig 2.
(A) Nucleotide sequence alignment of the promoters of the TTHA0750, TTHA0987, and TTHB023 genes regulated by T. thermophilus PfmR. The predicted PfmR-binding sites and the consensus sequence are indicated. The conserved bases in the PfmR binding sites are indicated by bold letters. Possible −10 and −35 hexamer sequences of the promoters are underlined. The transcription start site of each gene (+1), as determined by a 5′ RACE experiment, is indicated. (B) RT-PCR analysis to confirm the operon composed of the genes TTHB023 to TTHB018 (lane 2). As a control, PCR was also performed with no RT, using the same primers (lane 3). The samples were fractionated on a 1% agarose gel, which was stained with ethidium bromide and photographed. Lane 1, 500-bp DNA ladder markers.
Identification of the target genes of T. thermophilus PfmR.
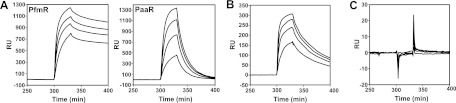
We confirmed the ability of PfmR to bind a double-stranded DNA (dsDNA) with the aforementioned pseudopalindromic sequence by means of a BIAcore analysis. The dsDNA fragment containing the upstream region of the TTHB023 gene (5′-TGAACCTACCGACCGGTCGGTGGGCTAT-3′) (the predicted PfmR-binding sequence is underlined) was immobilized on the streptavidin surface of a sensor chip through biotin conjugation to the 5′ end of one of the strands, and then PfmR was injected over the DNA surface at 25°C. We found that PfmR bound the DNA, with kon, koff, and Kd values of (5.6 ± 1.1) × 105 M−1 s−1, (4.3 ± 0.3) × 10−3 s−1, and 7.9 ± 1.4 nM, respectively (Fig. 3A and Table 2). PfmR did not bind the upstream region of the TTHA0890 gene, which contains a binding site for T. thermophilus FadR, another TetR family regulator (1) (Fig. 3C and Table 2). These results indicated that PfmR specifically binds the upstream region of the TTHB023 gene, containing the predicted PfmR-binding site, under these experimental conditions.
Fig 3.
BIAcore biosensor analyses of the interactions between the T. thermophilus TetR family transcriptional regulators and DNA. (A) A dsDNA fragment corresponding to the upstream region of the TTHB023 gene (see the text), which contains the predicted PfmR-binding site, was immobilized on the sensor chip, and then the PfmR or PaaR protein was injected over the DNA surface at concentrations of 0.4, 0.3, 0.2, and 0.1 μM dimer in buffer A. (B) A dsDNA fragment corresponding to the upstream region of the TTHA0963 gene (33), which contains the PaaR-binding site, was immobilized on the sensor chip, and then the PfmR protein was injected over the DNA surface, in the same manner as described for panel A. (C) A dsDNA fragment corresponding to the upstream region of the TTHA0890 gene (1), which contains the FadR-binding site, was immobilized on the sensor chip, and then the PfmR protein was injected over the DNA surface, in the same manner as described for panel A.
Table 2.
Cross-promoter recognition among three TetR family regulators from T. thermophilus HB8a
| Protein | PTTHB023 (PfmR-binding site) |
PTTHA0963 (PaaR-binding site) |
PTTHA0890 (FadR-binding site) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| kon (M−1s−1) | koff (s−1) | Kd (nM) | kon (M−1s−1) | koff (s−1) | Kd (nM) | kon (M−1s−1) | koff (s−1) | Kd (nM) | |
| PfmR | (5.6 ± 1.1) × 105 | (4.3 ± 0.3) × 10−3 | 7.9 ± 1.4 | (7.2 ± 0.9) × 105 | (1.7 ± 0.0) × 10−2 | (2.4 ± 0.3) × 101 | ND | ND | ND |
| PaaRb | (2.6 ± 0.4) × 105 | (6.0 ± 0.2) × 10−2 | (2.3 ± 0.1) × 102 | (9.3 ± 1.0) × 105 | (1.0 ± 0.1) × 10−3 | 1.1 ± 0.1 | ND | ND | ND |
| FadRc | ND | ND | (8.3 ± 1.3) × 104 | ND | ND | (7.2 ± 1.4) × 102 | (9.4 ± 1.2) × 105 | 0.1 ± 0.1 | (9.0 ± 2.2) × 101 |
The kinetic constants were determined using a BIAcore system, as described in the Materials and Methods and the legends to Fig. 3 and Fig. S3 in the supplemental material. Each value is the mean ± SD of the injection series. ND, not detected.
The values for PTTHA0963 are from reference 33.
The values for PTTHA0890 are from reference 1.
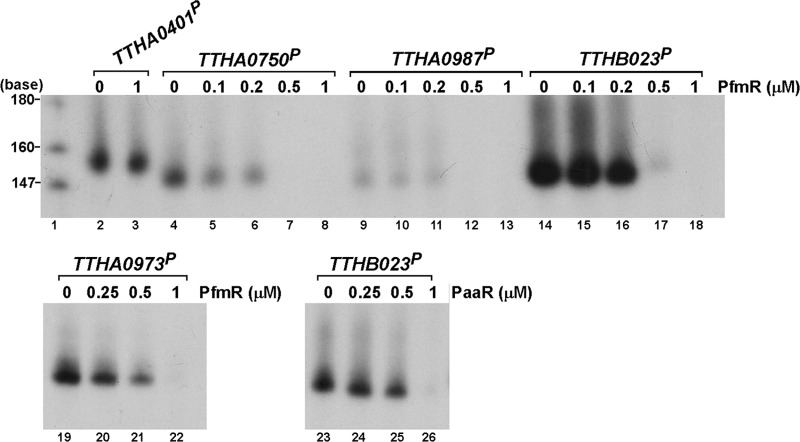
Next, we investigated the effects of PfmR on transcription in vitro. DNA fragments containing the predicted PfmR-binding sites upstream of the TTHA0750, TTHA0987, and TTHB023 genes (Fig. 2A) were constructed and used as templates. All of the obtained clones contained the upstream regions of the TTHA0750 and TTHA0987 genes but lacked a C base at a position between −24 and −27 and between −23 and −27, respectively. We used these templates for the following in vitro transcription experiments because the correct DNA fragments could not be obtained, perhaps due to instability in the host strain. We found that all of the templates were transcribed by T. thermophilus RNAP although the efficiencies differed, depending on the template (Fig. 4). The transcription reaction was repressed in the presence of PfmR in each case. PfmR did not affect the transcription of the TTHA0401 gene, which is regulated by FadR (1), indicating that PfmR specifically represses the transcription of the DNAs containing the predicted PfmR-binding site under these experimental conditions. The transcriptional start sites of the PfmR-regulated genes in vivo were determined by means of 5′ RACE. They were identified within the probable PfmR-binding sites (Fig. 2A). The potential promoter sequences were found in close proximity to the predicted PfmR-binding sites (Fig. 2A). These results agree well with those from the in vitro transcription assays and support the proposed function of PfmR as a transcriptional repressor of the aforementioned genes. To find other possible PfmR-binding sites, we searched for potential binding sites in the whole genome of T. thermophilus HB8 by using the consensus sequence (5′-TACCGACCGNTNGGTN-3′) as a query. However, no other sequences besides the three described above were found.
Fig 4.
Effects of T. thermophilus TetR family transcriptional regulators on transcription in vitro. Runoff transcription assays were performed with templates containing the upstream sequences of the genes regulated by PfmR (PTTHA0750, PTTHA0987, and PTTHB023), FadR (PTTHA0401) (1), and PaaR (PTTHA0973) (33) in the absence or presence of PfmR or PaaR. After the reaction, the samples were fractionated on the polyacrylamide gel, followed by autoradiography. Lane 1, [α-32P]dCTP-labeled MspI fragments of pBR322.
According to the genome analysis, the genes TTHB023 to TTHB018 form an operon (NCBI accession number NC_006462). We confirmed the operon structure by RT-PCR analysis (Fig. 2B). Thus, the eight genes summarized in Table 3 may be negatively regulated by PfmR. Since the transcripts of these PfmR-regulated genes were observed in T. thermophilus HB8 cultivated in rich medium, the genes may be slightly expressed under noninducing conditions. To determine the cellular role of PfmR, we predicted the functions of the PfmR-regulated gene products by investigating their amino acid sequences and structural features because they are either biochemically or biophysically uncharacterized (Table 3). The PfmR-regulated gene products, except for TTHB021 and TTHB023, are predicted to be enzymes that are possibly involved in FA biosynthesis, FA degradation, and PAA degradation.
Table 3.
Features of the T. thermophilus PfmR-regulated gene productsa
| Gene | Annotation for product | Domain | E value to the domain | Representative homolog (E value) | Possible cellular role | Reference |
|---|---|---|---|---|---|---|
| TTHA0750 | 3-oxoacyl-acyl carrier protein reductase | Beta-keto acyl carrier protein reductase (cd05333) | 1.01e−98 | Escherichia coli FabG (2e−55) | FA synthesis | PDB 1ULS |
| TTHA0987 | Beta-ketoadipyl CoA thiolase | Thiolase (cd00751) | 0 | Escherichia coli PaaE (7e−150) | PAA degradation | PDB 1ULQ |
| Beta-ketoadipyl CoA thiolase, validated (PRK09050) | 0 | |||||
| Putative acyltransferase, provisional (PRK05790) | 0 | Escherichia coli FadA (5e−103) | FA degradation | |||
| 3-Oxoadipyl-CoA thiolase (TIGR02430) | 0 | |||||
| Acetyl-CoA acetyltransferases (TIGR01930) | 0 | |||||
| Beta-ketoadipyl CoA thiolase, provisional (PRK13359) | 0 | |||||
| TTHB018 | Hypothetical protein | PaaI (PaaD) thioesterase (cd03443) | 5.35e−20 | Pseudomonas putida PaaD (6−09) | PAA degradation | |
| TTHB019 | MaoC (monoamine oxidase C)-related acyl dehydrogenase | MaoC-like (cd03446) | 3.89e−70 | Pseudomonas putida PaaN (3e−25) | PAA degradation | |
| TTHB020 | 3-oxoacyl-acyl carrier protein reductase | Beta-keto acyl carrier protein reductase (cd05333) | 1.22e−61 | Escherichia coli FabG (3e−33) | FA synthesis | PDB 2A4K |
| TTHB021 | Hypothetical protein | Beta-lactamase (pfam00144) | 1.67e−05 | Not detected | Unknown | |
| TTHB022 | Putative acyl-CoA dehydrogenase | SCAD-SBCAD (cd01158)b | 1.23e−150 | Bacillus subtilis AcdA (1e−98) | FA degradation | |
| TTHB023 | PfmR | Bacterial regulatory proteins, TetR family (pfam00440) | 6.22e−16 | Thermus thermophilus PaaR (1e−33) | Transcriptional regulation | This study |
A BLAST search was performed for each gene product, and the representative conserved domain, the E value to the domain, and the representative homolog with its E value are shown.
SCAD, short chain acyl-CoA dehydrogenase; SBCAD, short/branched chain acyl-CoA dehydrogenase.
Cross-regulation of T. thermophilus PfmR with PaaR.
T. thermophilus PaaR negatively regulates the expression of the putative paa genes (33). We found that PfmR interacted with the PaaR-binding sequence with a Kd value of ∼24 nM although this value is ∼3-fold higher than that for the interaction with the proper PfmR-binding sequence (∼7.9 nM) (Fig. 3A and B and Table 2). Conversely, PaaR interacted with the PfmR-binding sequence with a Kd value of ∼230 nM, which is 2 orders of magnitude higher than that for the interaction with the proper PaaR-binding sequence (∼1.1 nM) (Fig. 3A and Table 2). We also investigated the cross-regulation by means of a transcription assay in vitro (Fig. 4). The transcription of the DNA fragment containing the predicted PaaR-binding site (the TTHA0973 promoter [PTTHA0973]) was repressed by PfmR although the effect was weak compared to those of fragments containing the predicted PfmR-binding sites (PTTHA0750, PTTHA0987, and PTTHB023). Transcription of the DNA fragment containing the predicted PfmR-binding site (PTTHB023) was repressed by PaaR. These results were consistent with those of the aforementioned BIAcore analysis and indicated that PfmR cross-regulates with PaaR, which is supported by the observation that the nucleotide sequence of the predicted PfmR-binding site is about 44% identical to that of the PaaR-binding sites (Fig. 5). On the other hand, according to the DNA microarray analysis, the altered expression of the putative paa gene cluster (TTHA0963 to TTHA0973), which may be the main target of PaaR (33), was statistically insignificant; i.e., the expression level in the strain relative to that in the wild type was 0.383 to 0.750 (q values of 0.30 to 0.57). Moreover, no PfmR-regulated genes were obtained by the genomic SELEX experiment for the selection of DNA fragments containing the PaaR-binding sites (33). Thus, the cross-regulation of PfmR with PaaR might be weak in vivo. Cross-regulation of FadR with PfmR or PaaR was not observed in either the BIAcore analysis (Fig. 3 and Table 2; see also Fig. S3 in the supplemental material) or the transcription assay (Fig. 4; see also Fig. S4 in the supplemental material). These results are supported by the observation that the nucleotide sequence of the predicted FadR-binding site is unlike the sequences of the PfmR- and PaaR-binding sites (1).
Fig 5.
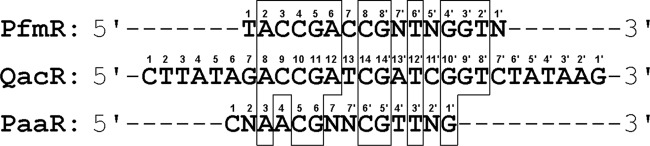
Sequence alignment of the predicted binding site of T. thermophilus PfmR with binding sites of other TetR family regulators. QacR, S. aureus QacR (34); PaaR, predicted T. thermophilus PaaR (33). One half-site is numbered 1 to 8, 1 to 14, or 1 to 7 (reading from the left) and the other is 1′ to 8′, 1′ to 14′, or 1′ to 7′ (reading from the right). Identical bases are boxed. N represents G, A, T, or C.
PaaR and FadR bind PA-CoA and medium-to-long (C10 to C18) straight chain fatty acyl-CoA as ligands, respectively, to derepress their target genes (1, 33); however, these compounds had no effects on the activity of PfmR (data not shown). In addition, T. thermophilus PfmR did not bind malonyl-CoA and acetyl-CoA (data not shown).
Three-dimensional structure of T. thermophilus PfmR.
The crystal structure of Se-PfmR, produced in E. coli, was determined by the SAD method and was refined to 2.27-Å resolution, with crystallographic Rwork and Rfree factors of 21.2 and 26.9%, respectively (Table 1). The asymmetric unit of the crystal comprised two homodimers (chains A-B and C-D) of PfmR. The overall structure of PfmR is shown in Fig. 6A. The final model exhibits the three-dimensional structure of a typical TetR family protein containing nine α-helices (Fig. 1 and 6A) (42). Note that there are several disordered regions that are not included in the model: Ala190 for chain A; Val1, Thr2, and Ala190 for chain B; Val1 to Arg5 and Gly188 to Ala190 for chain C; and Asp165 to Pro167, Ser189, and Ala190 for chain D. Each monomer is essentially identical, with a root mean square deviation (RMDS) of ∼0.74 Å in the superposition of the corresponding Cα atoms of residues 9 to 186. The PfmR structure was compared with the structures in the PDB database using the PDBeFold server (http://www.ebi.ac.uk/msd-srv/ssm/). The closest structures were the probable transcriptional regulator from Rhodococcus jostii RHA1 (PDB code 3HIM; Z = 9.7; RMSD, 1.4 Å; number of matched residues, 175; sequence identity [IDE], 28%), the HTH-type transcriptional repressor KstR2 from R. jostii RHA1 (PDB code 2IBD; Z = 7.9; RMSD, 2.4 Å; number of matched residues, 183; IDE, 30%), Bacillus subtilis FadR in complex with lauroyl-CoA (PDB code 1VI0; Z = 6.6; RMSD, 2.4 Å; number of matched residues, 180; IDE, 19%), and a Staphylococcus QacR mutant (Glu58Gln) in complex with berberine (PDB code 3BTI; Z = 5.5; RMSD, 2.3 Å; number of matched residues, 171; IDE, 20%).
Fig 6.
X-ray crystal structure of T. thermophilus PfmR. (A) Ribbon diagram of the PfmR dimer chains A (red) and B (gray). (B) Molecular surface representation of the PfmR dimer. Red and blue surfaces represent negative and positive electrostatic potentials (−5 kBT and +5 kBT, where kB is the Boltzmann constant and T is the temperature), respectively. The electrostatic potentials were calculated using the Adaptive Poisson-Boltzmann Solver (APBS) (3) with the PyMol APBS tools. (C) The N-terminal HTH DNA-binding domain of the S. aureus QacR proximal monomer (chain A) in complex with DNA (PDB code 1JT0) (34). DNA strands are shown in yellow and orange. Oxygen, nitrogen, and phosphate atoms are shown in red, blue, and light blue, respectively. The protein domain is gray, and the DNA-binding residues are indicated by stick models. (D) The N-terminal HTH domain of the PfmR monomer (chain A) superimposed on the corresponding domain of the QacR monomer in complex with DNA is shown, as described for panel C. The putative DNA-binding residues are indicated by stick models. (E) Stereo view around the center of the PfmR molecule. Chains A and B are shown in red and gray, respectively. The putative ligand-binding tunnel-like pocket is indicated by a mesh. The residues comprising the pocket are depicted by stick models. (F) Schematic model structures of the residues comprising the tunnel-like pocket of PfmR. The residues in parentheses are from chain A. The other residues are from chain B. These figures were drawn using the Pymol program (http://www.pymol.org/).
The N-terminal domain (α1 to α3) of PfmR, containing a typical HTH motif (α2 to α3) with a positively charged surface (Fig. 6B), may be the DNA-binding domain, as in the cases of other TetR family proteins (42). The structure of the domain (chain A, Val26 to Phe44) resembles that of the DNA-binding domain (chain A, Thr25 to Phe43) in the DNA-binding form of the proximal monomer of QacR (PDB code 1JT0) (34), with an RMSD of 0.43 Å (Fig. 6C and D). According to the model of the PfmR-DNA complex, four of the seven QacR residues (Thr25, Ser34, Ser35, Tyr40, Tyr41, His42, and Lys46), which hydrogen bond with the backbone of the major groove of DNA in the QacR-DNA complex, are conserved in PfmR (Ser36, Tyr41, His43, and Lys47). Furthermore, the Lys36 and Tyr40 residues of QacR, which contact the bases of the G14′ and T12′ nucleotides within the QacR-binding site, respectively (Fig. 5) (34), are conserved in PfmR (Lys37 and Tyr41). The distance between the two Cα atoms of the amino acid residues located in the center of the α3 helices of PfmR (Tyr41), which may interact with the major groove of the DNA, is ∼42.6 Å. This distance is greater than that of the DNA-binding form (37 Å) and shorter than the DNA-unbound form (48 Å) of QacR (35). Interestingly, the nucleotide sequence of the predicted PfmR-binding site is about 69% identical to that of the central 16 bases of the QacR-binding site, and the G14′ and T12′ bases of the QacR-binding site are conserved in the predicted PfmR-binding site (G8′ and T6′) (Fig. 5).
Many characterized TetR family regulators bind small molecules as ligands in similarly positioned pockets near the center of the molecules (42). In the PfmR structure, a similar pocket exists at the center of each monomer molecule (Fig. 6E). The pocket is composed of aromatic residues (Phe and Trp), hydrophobic residues (Met and Leu), and polar residues (His, Asn, and Arg), including three residues derived from another monomer, and these residues are exposed within the pocket (Fig. 6E and F). These are the common features of the ligand-binding pockets of the characterized TetR family regulators (42). Excess electron density was not observed in the pocket of PfmR.
DNA-binding by T. thermophilus PfmR.
According to the X-ray crystal structural analysis of PfmR, the DNA-binding mechanism of PfmR was predicted to be similar to that of QacR. In the case of QacR, two dimers bind the operator (10). We confirmed the stoichiometry of DNA binding by PfmR. DLS measurements of the sample containing PfmR and the 28-bp DNA fragment, derived from the upstream region of the TTHB023 gene and containing the predicted PfmR-binding site (see above), yielded a molecular mass of 109.5 ± 2.0 kDa (polydispersity value of 15.9% ± 1.9%), which is close to that of the two PfmR dimers plus one DNA fragment (103.7 kDa). The molecular mass of the sample was confirmed by gel filtration chromatography, and the main peak was observed at the elution volume of 9.2 ml, corresponding to the molecular mass of ∼85.7 kDa (Fig. 7A, panel b, and B). Since the peak of the DNA fragment at the elution volume of 10.8 ml (see Fig. S5 in the supplemental material) was not observed in the elution profile, the ∼85.7-kDa peak is probably the PfmR-DNA complex. When the ratio of PfmR to DNA was increased, another peak was observed at the elution volume of 11.1 ml (∼34.0 kDa), which probably corresponds to the excess free PfmR dimer (Fig. 7A, panels a and c). Since the molecular mass of the PfmR dimer estimated by the gel filtration chromatography was smaller than the calculated value (43.4 kDa), the peak at the elution volume of 9.2 ml may correspond to two PfmR dimers plus one DNA fragment. In the elution profile of PfmR with the DNA fragment derived from the upstream region of TTHA0890 gene, which PfmR did not bind in the BIAcore analysis (Fig. 3C and Table 2), the main peak was observed at the fraction of one PfmR dimer (Fig. 7A, panel d). These results suggested that two PfmR dimers bind per target DNA, which is the same DNA binding stoichiometry as that of QacR. In order to confirm the importance of the two conserved bases (G8′ and T6′) in the predicted PfmR-binding site, we investigated the effects of mutations at these positions on the DNA binding of PfmR. Based on the upstream sequence of the TTHB023 gene, two DNA sequences were designed containing the mutations of A6C/T6′G and C8A/G8′T within the predicted PfmR-binding site, i.e., 5′-TGAACCTACCGCCCGGGCGGTGGGCTAT-3′ and 5′-TGAACCTACCGACATGTCGGTGGGCTAT-3′ (mutated bases are underlined), respectively, and PfmR binding was investigated by means of gel filtration chromatography. When PfmR was mixed with the A6C/T6′G mutant DNA fragment, peaks were observed at the elution volumes of 9.6 ml and 10.8 ml (Fig. 7A, panel e). Since the DNA fragment was eluted at the elution volume of 10.8 ml (see Fig. S5 in the supplemental material), the latter peak may be derived from PfmR and the DNA fragment although it is unclear whether the complex is actually formed. The peak at 9.6 ml was slightly different from that of two PfmR dimers plus DNA and might reflect improper PfmR assembly on the DNA. In the assay of PfmR with the C8A/G8′T mutant DNA fragment, the main peak was one PfmR dimer, and the peak of two PfmR dimers was not observed (Fig. 7A, panel f). The DNA fragment peak that eluted at the elution volume of 10.8 ml (see Fig. S5) might have shifted to the elution volume of 10.5 ml due to binding only one PfmR dimer (Fig. 7A, panel f). These results suggested that the T6′ and G8′ bases are important for PfmR to recognize the PfmR binding-site and properly assemble on it, in a similar manner to the recognition of the conserved bases (G14′ and T12′) within the QacR-binding site by QacR.
DISCUSSION
In this study, we found that one of the four TetR family transcriptional regulators of T. thermophilus HB8, which we named T. thermophilus PfmR, is a strong repressor of eight genes (under three promoters), which are predicted to be involved in PAA degradation and FA degradation and biosynthesis. One of the merits of the coregulation of both PAA and FA metabolism might be that the acetyl-CoA molecules produced by FA and/or PAA degradation (20, 39) can be immediately used for FA biosynthesis. In T. thermophilus HB8, the TetR family regulator PaaR negatively regulates 11 putative paa genes (under two promoters) (33). Since PfmR weakly cross-regulated with PaaR in vitro, the PAA degradation might be controlled by the two regulators although supporting in vivo evidence has not been obtained. In addition to the PfmR-regulated gene products, T. thermophilus HB8 has other proteins involved in FA degradation, including another FadA paralog (TTHA0891), with expression controlled by the TetR family regulator FadR (1), with which PfmR did not cross-regulate. Moreover, this strain also has other proteins for FA biosynthesis, including another FabG paralog (TTHA0415), as well as TTHA0304 (FabI), TTHA0413 to TTHA0417 [FabB(F), AcpP, FabG, FabD, and FabH], TTHA1123 (AccC), TTHA1124 (AccB), TTHA1767 (AccA), and TTHA1768 (AccD) (NCBI accession number NC_006461). Since the predicted PfmR-binding sites were not found upstream of the genes involved in FA synthesis and since their expression levels were not significantly altered in the ΔpfmR strain (expression levels relative to those in the wild-type strain were 0.901 to 1.031 [q values of 0.47 to 0.67]), these genes may not be regulated by PfmR. Therefore, if the PfmR-regulated gene products are involved in FA metabolism, then they might function in a bypass of the main metabolic pathways or play some accessory roles for the main pathways. The ΔpfmR strain probably lacked the pVV8 plasmid. We found that this plasmid was also unstable in the wild-type strain (data not shown), and a wild-type strain does not harbor it (24). Thus, it is unclear whether PfmR is actually involved in the maintenance of pVV8.
The X-ray crystal structure of PfmR revealed that it adopts the typical three-dimensional structure of the TetR family proteins, with the putative N-terminal DNA-recognition helices α3 and α3′. The DNA-binding domains of the TetR family regulators are flexible, and the distance between the DNA-recognition helices, α3 and α3′, of the apo-form is not always compatible with the binding to two consecutive major grooves of DNA; therefore, the DNA is captured when the conformation is suitable for DNA binding (17, 42). The PfmR structure may be the ligand-free form because excess electron density was not observed in the putative ligand-binding pocket (see below). If the ligand-free form of PfmR is also flexible, then the structure determined in this study represents one of several possible conformations. In the case of QacR, two dimer molecules recognize a long 28-bp operator (34). The distal monomer of the QacR dimer binds around A4 to C10 (or G10′ to T4′), and the other (proximal) monomer binds G14′ to T8′ (or A8 to C14) (Fig. 5) (34). Several DNA recognition amino acid residues of the proximal QacR monomer are conserved in PfmR, and the nucleotide sequence of the binding site for the proximal QacR monomer is quite similar to that of the predicted PfmR-binding site (Fig. 5). In fact, two PfmR dimers bound per target DNA, which is the same DNA-binding stoichiometry as that of QacR. Furthermore, the G8′ and T6′ bases within the predicted PfmR-binding site were important for PfmR to recognize the binding site and properly assemble on it. The two bases are conserved within the QacR-binding site (G14′ and T12′), and they are recognized by Lys36 and Tyr40, respectively, which are also conserved in PfmR (Lys37 and Tyr41). Thus, the DNA-binding mechanism of PfmR may be similar to that of QacR. If Lys37 and Tyr41 of PfmR recognize the G8′ and T6′ bases, then the conformation of the DNA-binding site in the DNA-binding form may differ from that determined in this study because in the model of the PfmR-DNA complex, the two residues are distal from the bases, compared to those in the structure of the QacR-DNA complex (Fig. 6C and D). If the proximal PfmR monomer binds DNA in a similar manner to the proximal QacR monomer, then the distal PfmR monomer may not specifically recognize the bases unless a large conformational change or subunit rotation occurs because the nucleotide sequence possibly recognized by the distal monomer is not conserved, unlike the case of the QacR-binding site (Fig. 5). According to one of the two proposed structural mechanisms of derepression by the TetR family transcriptional repressors, ligand-binding induces the repositioning of the DNA-binding domain relative to the ligand-binding domain of each monomer to increase the distance between the DNA recognition helices, α3 and α3′ (26, 35, 41). The cognate ligands predominantly interact with one subunit of the dimer in the cases of TetR and ActR (26, 41). In contrast, in another derepression mechanism, ligand binding does not induce such dispositions of the domains in each monomer; instead, it stabilizes the relative positions of the two monomers in the dimer so they are incompatible with DNA binding, as reported for SimR (18). In this case, the cognate ligand makes substantial contacts with both monomers in the dimer, and the two extra α-helices, which are not present in most typical TetR family proteins (42), play a role in the conformational stabilization (18). The tunnel-like pocket formed at the center of the PfmR molecule is possibly a ligand-binding site because the position and the residues comprising the pocket are similar to those of the characterized TetR family regulators (42). Since the pocket is predominantly composed of the residues derived from one monomer and since PfmR does not contain extra α-helices, unlike the case of SimR, the derepression mechanism by PfmR might be similar to the first mode, involving ligand-mediated repositioning. PfmR did not bind a medium-to-long-chain fatty acyl-CoA, unlike FadR, and it also did not recognize malonyl-CoA, a ligand for B. subtilis FapR, which is a transcriptional regulator of the FA biosynthesis pathway (22). Moreover, PA-CoA, a ligand of T. thermophilus PaaR, had no effect on the activity of PfmR. Some unidentified compound related to the PAA degradation, FA degradation, and FA biosynthesis pathways might be the ligand of PfmR. Further research on PfmR and the PfmR-regulated gene products will be necessary to elucidate the cellular roles of this transcriptional regulator, its mechanisms of transcriptional repression and derepression, and the metabolism of organic compounds in T. thermophilus HB8.
Supplementary Material
ACKNOWLEDGMENTS
We thank Aimi Osaki for construction of the plasmid pUC-ΔpfmR, Kayoko Matsumoto and Toshi Arima for protein purification, and Toshi Arima for crystallization. We also thank Hitoshi Iino and Kenji Fukui for the data collection at SPring-8.
This work was supported by a Grant-in-Aid for Scientific Research (C), 22510208, from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Footnotes
Published ahead of print 29 June 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Agari Y, Agari K, Sakamoto K, Kuramitsu S, Shinkai A. 2011. TetR family transcriptional repressor Thermus thermophilus FadR controls fatty acid degradation. Microbiology 157:1589–1601 [DOI] [PubMed] [Google Scholar]
- 2. Agari Y, Kashihara A, Yokoyama S, Kuramitsu S, Shinkai A. 2008. Global gene expression mediated by Thermus thermophilus SdrP, a CRP/FNR family transcriptional regulator. Mol. Microbiol. 70:60–75 [DOI] [PubMed] [Google Scholar]
- 3. Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U. S. A. 98:10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brünger AT, et al. 1998. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905–921 [DOI] [PubMed] [Google Scholar]
- 5. Collaborative Computational Project Number 4 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763 [DOI] [PubMed] [Google Scholar]
- 6. Cowtan K. 2006. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 62:1002–1011 [DOI] [PubMed] [Google Scholar]
- 7. Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 8. Gouet P, Robert X, Courcelle E. 2003. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 31:3320–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Grkovic S, Brown MH, Roberts NJ, Paulsen IT, Skurray RA. 1998. QacR is a repressor protein that regulates expression of the Staphylococcus aureus multidrug efflux pump QacA. J. Biol. Chem. 273:18665–18673 [DOI] [PubMed] [Google Scholar]
- 10. Grkovic S, Brown MH, Schumacher MA, Brennan RG, Skurray RA. 2001. The staphylococcal QacR multidrug regulator binds a correctly spaced operator as a pair of dimers. J. Bacteriol. 183:7102–7109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hashimoto Y, Yano T, Kuramitsu S, Kagamiyama H. 2001. Disruption of Thermus thermophilus genes by homologous recombination using a thermostable kanamycin-resistant marker. FEBS Lett. 506:231–234 [DOI] [PubMed] [Google Scholar]
- 12. Hinrichs W, et al. 1994. Structure of the Tet repressor-tetracycline complex and regulation of antibiotic resistance. Science 264:418–420 [DOI] [PubMed] [Google Scholar]
- 13. Kabsch W, Sander C. 1983. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22:2577–2637 [DOI] [PubMed] [Google Scholar]
- 14. Kisker C, Hinrichs W, Tovar K, Hillen W, Saenger W. 1995. The complex formed between Tet repressor and tetracycline-Mg2+ reveals mechanism of antibiotic resistance. J. Mol. Biol. 247:260–280 [DOI] [PubMed] [Google Scholar]
- 15. Kuramitsu S, Hiromi K, Hayashi H, Morino Y, Kagamiyama H. 1990. Pre-steady-state kinetics of Escherichia coli aspartate aminotransferase catalyzed reactions and thermodynamic aspects of its substrate specificity. Biochemistry 29:5469–5476 [DOI] [PubMed] [Google Scholar]
- 16. Larkin MA, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 17. Le TB, Schumacher MA, Lawson DM, Brennan RG, Buttner MJ. 2011. The crystal structure of the TetR family transcriptional repressor SimR bound to DNA and the role of a flexible N-terminal extension in minor groove binding. Nucleic Acids Res. 39:9433–9447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Le TB, et al. 2011. Structures of the TetR-like simocyclinone efflux pump repressor, SimR, and the mechanism of ligand-mediated derepression. J. Mol. Biol. 408:40–56 [DOI] [PubMed] [Google Scholar]
- 19. LeMaster DM, Richards FM. 1985. 1H-15N heteronuclear NMR studies of Escherichia coli thioredoxin in samples isotopically labeled by residue type. Biochemistry 24:7263–7268 [DOI] [PubMed] [Google Scholar]
- 20. Luengo JM, Garcia JL, Olivera ER. 2001. The phenylacetyl-CoA catabolon: a complex catabolic unit with broad biotechnological applications. Mol. Microbiol. 39:1434–1442 [DOI] [PubMed] [Google Scholar]
- 21. Marchler-Bauer A, et al. 2002. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 30:281–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Martinez MA, et al. 2010. A novel role of malonyl-ACP in lipid homeostasis. Biochemistry 49:3161–3167 [DOI] [PubMed] [Google Scholar]
- 23. Minezaki Y, Homma K, Nishikawa K. 2005. Genome-wide survey of transcription factors in prokaryotes reveals many bacteria-specific families not found in archaea. DNA Res. 12:269–280 [DOI] [PubMed] [Google Scholar]
- 24. Ohtani N, Tomita M, Itaya M. 2012. The third plasmid pVV8 from Thermus thermophilus HB8: isolation, characterization, and sequence determination. Extremophiles 16:237–244 [DOI] [PubMed] [Google Scholar]
- 25. Orth P, et al. 1998. Conformational changes of the Tet repressor induced by tetracycline trapping. J. Mol. Biol. 279:439–447 [DOI] [PubMed] [Google Scholar]
- 26. Orth P, Schnappinger D, Hillen W, Saenger W, Hinrichs W. 2000. Structural basis of gene regulation by the tetracycline inducible Tet repressor-operator system. Nat. Struct. Biol. 7:215–219 [DOI] [PubMed] [Google Scholar]
- 27. Oshima T, Imahori K. 1974. Description of Thermus thermophilus (Yoshida and Oshima) com. nov., a non-sporulating thermophilic bacterium from a Japanese thermal spa. Int. J. Syst. Bacteriol. 24:102–112 [Google Scholar]
- 28. Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326 [DOI] [PubMed] [Google Scholar]
- 29. Pepper SD, Saunders EK, Edwards LE, Wilson CL, Miller CJ. 2007. The utility of MAS5 expression summary and detection call algorithms. BMC Bioinformatics 8:273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ramos JL, et al. 2005. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 69:326–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reichheld SE, Yu Z, Davidson AR. 2009. The induction of folding cooperativity by ligand binding drives the allosteric response of tetracycline repressor. Proc. Natl. Acad. Sci. U. S. A. 106:22263–22268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rodionov DA. 2007. Comparative genomic reconstruction of transcriptional regulatory networks in bacteria. Chem. Rev. 107:3467–3497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sakamoto K, Agari Y, Kuramitsu S, Shinkai A. 2011. Phenylacetyl coenzyme A is an effector molecule of the TetR family transcriptional repressor PaaR from Thermus thermophilus HB8. J. Bacteriol. 193:4388–4395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schumacher MA, et al. 2002. Structural basis for cooperative DNA binding by two dimers of the multidrug-binding protein QacR. EMBO J. 21:1210–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schumacher MA, et al. 2001. Structural mechanisms of QacR induction and multidrug recognition. Science 294:2158–2163 [DOI] [PubMed] [Google Scholar]
- 36. Shinkai A, et al. 2007. Transcription activation mediated by a cyclic AMP receptor protein from Thermus thermophilus HB8. J. Bacteriol. 189:3891–3901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Storey JD. 2002. A direct approach to false discovery rates. J. R. Stat. Soc. Series B Stat. Methodol. 64:479–498 [Google Scholar]
- 38. Terwilliger TC, Berendzen J. 1999. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55:849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Teufel R, et al. 2010. Bacterial phenylalanine and phenylacetate catabolic pathway revealed. Proc. Natl. Acad. Sci. U. S. A. 107:14390–14395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ueno G, et al. 2006. RIKEN structural genomics beamlines at the SPring-8; high throughput protein crystallography with automated beamline operation. J. Struct. Funct. Genomics 7:15–22 [DOI] [PubMed] [Google Scholar]
- 41. Willems AR, et al. 2008. Crystal structures of the Streptomyces coelicolor TetR-like protein ActR alone and in complex with actinorhodin or the actinorhodin biosynthetic precursor (S)-DNPA. J. Mol. Biol. 376:1377–1387 [DOI] [PubMed] [Google Scholar]
- 42. Yu Z, Reichheld SE, Savchenko A, Parkinson J, Davidson AR. 2010. A comprehensive analysis of structural and sequence conservation in the TetR family transcriptional regulators. J. Mol. Biol. 400:847–864 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.