Abstract
Daptomycin is a lipopeptide antibiotic used clinically for the treatment of certain types of Gram-positive infections, including those caused by methicillin-resistant Staphylococcus aureus (MRSA). Details of the mechanism of action of daptomycin continue to be elucidated, particularly the question of whether daptomycin acts on the cell membrane, the cell wall, or both. Here, we use fluorescence microscopy to directly visualize the interaction of daptomycin with the model Gram-positive bacterium Bacillus subtilis. We show that the first observable cellular effects are the formation of membrane distortions (patches of membrane) that precede cell death by more than 30 min. Membrane patches are able to recruit the essential cell division protein DivIVA. Recruitment of DivIVA correlates with membrane defects and changes in cell morphology, suggesting a localized alteration in the activity of enzymes involved in cell wall synthesis that could account for previously described effects of daptomycin on cell wall morphology and septation. Membrane defects colocalize with fluorescently labeled daptomycin, DivIVA, and fluorescent reporters of peptidoglycan biogenesis (Bocillin FL and BODIPY FL-vancomycin), suggesting that daptomycin plays a direct role in these events. Our results support a mechanism for daptomycin with a primary effect on cell membranes that in turn redirects the localization of proteins involved in cell division and cell wall synthesis, causing dramatic cell wall and membrane defects, which may ultimately lead to a breach in the cell membrane and cell death. These results help resolve the longstanding questions regarding the mechanism of action of this important class of antibiotics.
INTRODUCTION
Daptomycin is a cyclic lipopeptide natural product produced by Streptomyces roseosporus and approved for the treatment of skin and skin structure infections by Gram-positive bacteria and of bacteremia and right-sided endocarditis caused by Staphylococcus aureus (10). In vitro, daptomycin displays excellent potency and rapid bactericidal activity against a variety of key Gram-positive pathogens, including methicillin-resistant S. aureus (MRSA) and vancomycin-resistant Enterococcus (VRE). The antibacterial activity of daptomycin is strictly dependent on the presence of physiological levels of free calcium ions (11). The peptide moiety of daptomycin is anionic (net charge, −3), but in the presence of calcium, it shares many properties with the larger class of cationic antimicrobial peptides (53).
Despite nearly 25 years of study and 8 years of clinical use, aspects of the mechanism of action of daptomycin remain poorly understood. Daptomycin disrupts Gram-positive cytoplasmic membrane function, causing leakage of potassium (and potentially other) ions, ultimately leading to loss of membrane potential and cell death (50). Membrane function is compromised in the absence of cell lysis or the creation of large pores (32). Studies using artificial membranes have shown that daptomycin can act directly on the lipid bilayer in the absence of any bacterial protein or other cell surface component (26, 27). More detailed studies have demonstrated the ability of daptomycin, in the presence of calcium ions, to destabilize phosphatidylglycerol-containing membrane bilayers in vitro, inducing both lipid “flip-flop” within membranes and vesicle fusion between membranes (25). Biophysical studies also suggest that daptomycin has an intrinsic ability to alter membrane curvature (24). These biophysical studies provide a plausible mechanism by which daptomycin might disrupt the function of bacterial membranes, though flip-flop and curvature have not been demonstrated in vivo to date.
The role of the membrane as a central target is further supported by studies of daptomycin-resistant mutants in multiple species (2, 15, 18, 38). Many of the mutations that alter susceptibility to daptomycin have been shown to directly affect membrane lipid composition. MprF, for example, is a membrane protein responsible for synthesizing a positively charged phospholipid, lysylphosphatidylglycerol. In S. aureus, mutations that eliminate the expression of the mprF gene sensitize cells to daptomycin, and mutations that confer resistance are believed to be “gain-of-function” alleles that increase the amount of lysylphosphatidylglycerol (15, 47, 57). Interestingly, the specific membrane modifications that confer resistance vary by species: in enterococci, changes in cardiolipin synthesis are associated with resistance (2, 38), while in Bacillus subtilis, resistance may be linked to the overall phosphatidylglycerol content due to mutations in pgsA (18).
Despite significant evidence pointing to action against the cell membrane in susceptible bacteria, the cell wall has also been suspected to be an important target of daptomycin. Transcriptional-profiling studies show that daptomycin induces the cell wall stress response in S. aureus and B. subtilis, which is typically induced by cell wall-active antibiotics, like vancomycin, oxacillin, and fruilimicin (17, 37). This suggests that daptomycin causes significant cell wall damage. Electron microscopy (EM) studies show that daptomycin treatment induces abnormal septation events in many different species, including Staphyloccocus and Enterococcus faecalis (7; J. Silverman, unpublished observations). Moreover, alterations in cell wall physiology have been suggested to alter daptomycin susceptibility in S. aureus strains displaying reduced susceptibility to vancomycin, though this remains controversial (8, 56). In addition, mutations conferring daptomycin resistance in S. aureus have been reported in the yycG gene product, a histidine kinase believed to coordinate peptidoglycan synthesis and cell division (15, 16). Daptomycin is structurally related to fruilimicin and MX-2401, each of which has been shown to inhibit peptidoglycan synthesis by binding to the key intermediate bactoprenol phosphate (46, 49). However, daptomycin does not inhibit any of the enzymatic steps in cell wall biosynthesis that have been tested (49). Therefore, it has remained unclear why daptomycin induces cell wall stress response pathways and what role effects on the cell wall might play in its mechanism.
Here, we directly examine the interaction of daptomycin with B. subtilis. We show that one of the earliest observable effects on the cell is distortion of the cell membrane, which precedes death by more than 30 min, and that the distortions reflect local alterations in membrane curvature. Daptomycin localizes to the sites of membrane distortion, as does DivIVA, an essential and highly conserved cell division protein in Gram-positive bacteria, and other components of the cell wall synthesis machinery. Our results led us to propose a refined model that may resolve the longstanding confusion about daptomycin's mechanism of action and may pave the way for understanding cationic peptides whose mechanisms of action are still poorly understood.
MATERIALS AND METHODS
Bacterial strains.
Bacillus subtilis strain PY79 and its derivatives were used for all experiments (58). Strain KR541 expresses DivIVA-green fluorescent protein (GFP) from the IPTG (isopropyl-β-d-thiogalactopyranoside)-dependent Pspac promoter at the amyE locus (44). Strain KR318 expresses SpoVM-GFP from the IPTG-dependent Pspac promoter at the amyE locus (43). Strain KR515 expresses DivIVA-GFP at native levels (13, 44). Strains KR515, KR541, and KR318 were kindly provided by Kumaran Ramamurthi.
Media and growth conditions.
Cells were grown in LB medium at either 30°C or 37°C, as indicated. Growth curves were conducted on cells growing in LB medium in a 250-ml baffled-bottom flask with shaking at 37°C. Samples were removed every 15 min, and the optical density at 600 nm (OD600) was measured. MICs for daptomycin were determined using the serial-dilution method. Sporulation was induced using the Sterlini and Mandelstam method of resuspension (51). Samples (0.4 ml) of sporulating culture were removed after 1.5 h and 2 h of sporulation, stained with 1 μg/ml FM 4-64 (40), concentrated 10-fold by centrifugation, and placed on a coverslip treated with polylysine. To visualize DivIVA-GFP or SpoVM-GFP produced by strain KR541 or KR318, respectively, cells were grown with 500 μM IPTG for 2 h at 30°C. To visualize DivIVA-GFP produced at native levels, strain KR515 was grown at 30°C.
Microscopy.
For time lapse microscopy experiments, cells were grown in a heated chamber at 30°C on a 1% agarose pad containing 0.2% glucose and 10% LB (40). The vital stain FM 4-64 was added to the media at 1 μg/ml, a concentration that has no deleterious effect on growth or sporulation (40). The vital stain sytox green (Invitrogen) was added to agarose pads at 0.4 μg/ml (41). Images were collected using a Delta Vision Spectris Deconvolution microscope equipped with a Cool Snap camera using standard filter sets for DAPI (4′,6-diamidino-2-phenylindole) (360- to 400-nm excitation and 457- to 507-nm emission), rhodamine (for FM 4-64 visualization, 555- to 583-nm excitation and 617- to 690-nm emission), and fluorescein isothiocyanate (FITC) (461- to 489-nm excitation and 501- to 549-nm emission), as previously described (40). Optical sections were collected spaced 0.15 μm apart. Control experiments demonstrated that there was no significant crossover between red and green channels for the given amount of fluorescent reporters and exposure times used in these experiments.
For labeling with fluorescently labeled daptomycin, daptomycin-BODIPY was added to a 1-ml culture of cells at a final concentration of 16 μg/ml and allowed to incubate at 37°C for 10 min (41). Daptomycin-BODIPY is 2 to 4 times less active than the unmodified molecule. Cells were similarly labeled for 10 min with Bocillin FL (Life Technologies) at a final concentration of 1 μg/ml and with BODIPY FL-vancomycin (Van-BDP) (Life Technologies) at a final concentration of 0.5 μg/ml. Excess unincorporated label was removed by washing the cells three times in LB medium. The cells were concentrated 10-fold at the last washing step, and a small aliquot (3 μl) was placed on a polylysine-treated coverslip.
RESULTS
Time course of exposure to daptomycin.
To gain insight into the bactericidal activity of daptomycin, we used fluorescence microscopy to image B. subtilis growing on an agarose pad containing a lethal dose (10 μg/ml) of daptomycin (under these conditions, MIC = 5 μg/ml) (Fig. 1A). Vital stains were included in the pad to allow visualization of how daptomycin affected the cell membrane structure (FM4-64) and to determine when cells lost membrane integrity (sytox green). The membrane stain FM 4-64 accumulates in the membrane and uniformly stains wild-type cells and is a sensitive indicator of membrane rearrangements (40). Sytox green is a nucleic acid stain that poorly penetrates cells containing an intact membrane but intensely stains the nucleoids once the membrane barrier has collapsed (45). Exponentially growing B. subtilis cells were placed on an agarose pad containing daptomycin, and different fields of cells were imaged over time (Fig. 1C to H). At the first two time points (7 and 10 min), the cells appeared identical to the control sample: FM 4-64 membrane staining was completely uniform, and sytox green was largely excluded from the cell, showing a low level of green fluorescence (Fig. 1C and D). However, as the cells continued to grow in the presence of daptomycin, they accumulated membrane defects at apparently random positions throughout the cell and then later lost viability. The membrane defects were clearly visible at 15 min and appeared as bright foci or protrusions (Fig. 1E, 15 min, arrow) and became more pronounced by 20 and 30 min. As shown Fig. 1E, F, and G, cells with clear membrane distortions displayed normal sytox green staining, indicating that the membranes were still intact. Quantitation of the loss of membrane integrity over time (Fig. 1B) showed that only 11% of the cells (n = 314) examined between the 20- and 30-min time points had bright sytox staining, even though most of them (>90%) displayed membrane defects. Between the 45- and 60-min time points, the majority (94%; n = 446) of nucleoids stained brightly with sytox green (Fig. 1H, 45 min), indicating a loss of membrane barrier function and cell death. These results suggest that membrane damage occurs rapidly (within 15 min) and is an early precursor of cell death. We note that at later time points (150 min), we observed a drop in the OD600, likely due to cell lysis (Fig. 1A). Similar lysis was not observed in S. aureus (7).
Fig 1.
Microscopic evaluation of the cell biological effects of daptomycin exposure. (A) Growth curve of cells in different concentrations of daptomycin demonstrating that cells grown in 1 μg/ml, 2 μg/ml, or 3 μg/ml daptomycin have growth rates identical to those of cells grown without antibiotic. In contrast, cells grown with 10 μg/ml daptomycin fail to grow and lyse after approximately 2.5 h. Note that B. subtilis is prone to lysis due to the activation of autolysins, whereas S. aureus does not exhibit lysis in the presence of daptomycin. (B) The percentage of cells containing bright sytox green staining increases over time when the cells grow in a lethal concentration of daptomycin (10 μg/ml). The cells displaying bright sytox green staining were counted and binned into groups, and the percentages of cells displaying bright sytox green staining were plotted. Zero to 10 min, 0% sytox green-positive cells, n = 146; 11 to 20 min, 7% sytox green-positive cells, n = 300; 21 to 30 min, 11% sytox green-positive cells, n = 314; 31 to 45 min, 74% sytox green-positive cells, n = 57; 46 to 60 min, 94% sytox green-positive cells, n = 446. (C to H) Time course of cells grown on an agarose pad containing FM 4-64, sytox green, and a lethal dose of daptomycin (10 μg/ml) demonstrating that cells suffer membrane deformations and ultimately lose their membrane integrity and stain brightly with sytox green. The cells in panels C and D have normal membrane and sytox green staining. The cells in panels E to G have membrane defects but normal (low) sytox green staining. The cells in panel H have high sytox green staining. (I to L) Time lapse microscopy demonstrating that membrane deformations precede cell lysis. (I) After 25 min of growth in the presence of daptomycin, a cell contains a large membrane protrusion (I) but displays low sytox green staining (K). (J and L) Five minutes later, the membrane has become permeable and the cell stains brightly with sytox green. The top images in panels I and J show membranes only. The bottom images show membranes and sytox green. (K) Quantification of sytox fluorescence intensity for the cell in panel I. (L) Quantification of sytox fluorescence intensity for the cell in panel J.
Time lapse microscopy of B. subtilis during exposure to daptomycin.
Time lapse microscopy was performed to directly follow the fate of individual cells as they grew in the presence of daptomycin. One example is shown in Fig. 1I to L, in which a cell containing a membrane bulge after 25 min of daptomycin exposure was still capable of excluding sytox green (Fig. 1I and K; see Movie S1 in the supplemental material). However, 5 min later, the cell stained brightly with sytox green, suggesting a loss of membrane integrity (Fig. 1J and L). In contrast, untreated cells grew and divided normally under these conditions and exhibited completely uniform membrane staining (see Movie S2 in the supplemental material). Taken together with the time course experiment, these results suggest that membrane deformations precede the collapse of membrane integrity and cell death. We conclude that membrane defects are an early and therefore likely direct effect of daptomycin binding to cells.
Daptomycin-BODIPY preferentially binds to the leading edges of invaginating membranes.
A fluorescent derivative of daptomycin that retains antibacterial activity was used to gain insight into its cellular targets and mechanism of action (17). An exponentially growing culture of B. subtilis was labeled for 10 min with daptomycin-BODIPY, stained with the membrane stain FM 4-64, and imaged with a fluorescence microscope. Labeling with daptomycin-BODIPY produced two distinct patterns of localization: faint foci that decorated the cylindrical portion of the cell and intense bands of staining at the midpoints of cells corresponding to the sites of septation (Fig. 2A to G). Serial optical sectioning demonstrated that sidewall foci are randomly distributed around the cell so that different foci are observable at random positions in different focal planes (Fig, 2D and E; see Movie S3 in the supplemental material). Three-dimensional fluorescence intensity plots of daptomycin-BODIPY fluorescence show that septal staining is significantly (∼3- to 4-fold) more intense than that of the sidewall (Fig. 2F), demonstrating a strong preference for interacting with the septum. Quantitation of 23 septa and 57 sidewall foci revealed that septa contained on average a 4-fold increase in fluorescence intensity per unit area. Three-dimensional reconstructions of daptomycin-BODIPY staining at midcell demonstrated that the apparent bands of fluorescence are actually rings that circumnavigate the cell along with the invaginating membrane (Fig. 2H and I; see Movie S4 in the supplemental material). These results are consistent with an earlier report by Hachmann et al. examining daptomycin-BODIPY labeling, where a preference for septal staining was also observed (17).
Fig 2.
Daptomycin-BODIPY staining of B. subtilis during growth. Cells were labeled for 10 min with 16 μg/ml daptomycin-BODIPY (green) and stained with FM 4-64 (red) during vegetative growth. All panels are at the same scale (bar, 1 μm). Daptomycin labels the sidewalls as discrete foci and intensely stains active division sites. (A to C) A single cell was labeled with FM 4-64 (A), daptomycin-BODIPY (C), or both (B). (D and E) A single cell showing that medial (D) and top (E) focal planes reveal different foci surrounding the cell. (F) Three-dimensional (3D) intensity plot corresponding to the region boxed in panel E showing that septal staining (large peaks) is more intense than sidewall staining. (G) Middle focal plane of a chain of cells labeled with daptomycin-BODIPY (green). (H and I) A single nascent septum labeled with daptomycin-BODIPY (green). The boxed region was reconstructed in three dimensions and then rotated through 90° (panel I, from left to right), demonstrating that the daptomycin-BODIPY fluorescence at division sites circumnavigates the cell.
Because of the potential significance of daptomcyin binding to the septum, we investigated the intensity and distribution of daptomycin-BODIPY staining relative to the degree of septal progression. We compared the accumulation of daptomycin-BODIPY in complete versus incomplete septa (Fig. 3). Daptomycin-BODIPY preferentially stains nascent septa (100%; n = 37) (Fig. 3D to F) and is generally absent from the majority (76%; n = 55) of completed septa (Fig. 3G to I), suggesting that daptomycin has an affinity for active division sites.
Fig 3.

Demonstration that daptomycin-BODIPY (DAP-Bodipy) intensely stains nascent septa but is generally absent from complete septa. Cells were labeled for 10 min with 16 μg/ml daptomycin-BODIPY as described in Materials and Methods. (A to C) Images showing cell membranes stained with FM 4-64 (A), daptomycin-BODIPY (B), or both (C). Scale bar, 1 μm. (D to F) Fluorescence intensity plots of an incomplete septum (1 in panel A, enlarged in panel F) showing that the peaks of membrane fluorescence corresponding to septal invagination (D) coincide with peaks of daptomycin-BODIPY fluorescence (E). Scale bar, 1 μm. (G to I) Fluorescence intensity plots for a complete septum (2 in panel A, enlarged in panel I) showing that the peaks of daptomycin-BODIPY fluorescence (H) do not coincide with the peaks of membrane fluorescence (G).
We investigated the nature of daptomycin septal staining by comparing the width of daptomcyin-BODIPY rings to the width of nascent septa (Fig. 4). Intensity plots of daptomycin-BODIPY staining for three septa at different stages of constriction are shown in Fig. 4F. When septa are just beginning to form, daptomcyin-BODIPY forms a bright ring of fluorescence that is equal in width to the cell (Fig. 4, septum A, 0.85 μm, and graph F1). As septa constrict, daptomycin-BODIPY staining appears as continuously smaller rings of fluorescence (Fig. 4, septum B, 0.72 μm, and septum C, 0.60 μm). Once septa have constricted to a minimal size, rings of daptomycin-BODIPY staining can no longer be resolved, and the staining appears as a broad band of fluorescence (Fig. 4, septum D, 0.40 μm, and graph F2). In septa that are nearly complete, daptomycin-BODIPY staining appears as a tight focus (Fig. 4, septum E, 0.20 μm, and graph F3). Quantitation showed that only 24% (n = 55) of septa judged to be complete based on FM 4-64 staining contained foci or small bands of fluorescence, as in Fig. 4D and E. These results demonstrate a strong correlation between the distribution of daptomycin-BODIPY staining at the septum and the degree to which septation is completed, suggesting that daptomycin stains the leading edges of the invaginating membranes.
Fig 4.
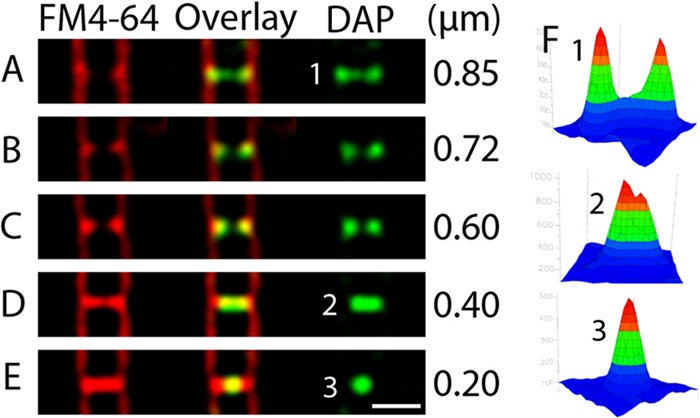
Five cells containing septa at various stages of constriction. Daptomycin-BODIPY stains nascent septa first as a ring, then as a smaller ring, then as a small band of fluorescence, and finally, when the septum appears to be complete, as a bright focus. The width of the daptomycin-BODIPY (DAP) labeling is provided at the right of each panel. Note that most septa (76%; n = 55) that appear to be complete, as in panel E, have no daptomycin-BODIPY staining. Scale bar, 1 μm. All panels are at the same scale.
The pattern of daptomycin-BODIPY staining changes during sporulation.
Our results suggested that daptomycin binds preferentially to the leading edges of the invaginating membranes, where the membranes are highly curved and active peptidoglycan synthesis occurs. To confirm the observation that daptomycin prefers to localize to membranes undergoing rearrangement, we examined the localization of daptomycin-BODIPY during sporulation. At the onset of sporulation, B. subtilis assembles two sites at which cell division can occur, one near each pole of the cell (12, 29, 48). These polar cell division complexes initiate septal biogenesis sequentially, and only one successfully completes division (40). The first septum to complete polar division activates a transcription factor in the newly created forespore cell, which then generates a signal to inactivate the second potential division site in the larger mother cell (30, 40). Cell-specific gene expression in each of these cells produces proteins required for the phagocytosis-like process of engulfment, in which the mother cell membranes migrate around and engulf the forespore (1, 5, 12, 30). Engulfment was recently shown to require active peptidoglycan biogenesis at the leading edge of the engulfing membrane (34). Peptidoglycan-biosynthetic enzymes and fluorescent peptidoglycan analogs localize to both the polar septa and the leading edges of the engulfing membranes during sporulation (34).
If daptomycin also prefers to bind to regions of active membrane rearrangement and peptidoglycan biogenesis, as suggested above, then the position of daptomycin-BODIPY staining would be expected to switch from the division septa to the leading edges of the engulfing membranes during sporulation. To test this idea, we examined daptomycin-BODIPY binding in cells that had been sporulating for 1.5 h. We found that cells at various stages of sporulation showed a daptomycin-BODIPY staining pattern (Fig. 5A to C) dramatically different from that of vegetatively growing cells, just as predicted. The cell shown in Fig. 5A has just entered the sporulation pathway and has a partially constricted polar septum near one end. The cell has two rings of daptomycin-BODIPY staining (blue arrows), with the brightest ring occurring at the nascent septum (right end of the cell). Cells that had completed polar septation and had begun engulfment (Fig. 5B and C) displayed bright daptomycin-BODIPY septal staining, in contrast to completed septa in vegetative cells, which incorporated little stain. As the cells progressed through engulfment, daptomycin-BODIPY stained the leading edges of the membranes, where peptidoglycan biogenesis is the most active and membranes are most sharply curved (Fig. 5B and C, white arrows). In addition to staining sporulation septa and engulfing membranes, daptomycin-BODIPY also formed foci at random positions along the sidewalls, similar to vegetative cells. We found that 99% (n = 117) of cells that had entered sporulation displayed these dramatic changes in daptomycin-BODIPY localization. These results demonstrate that the pattern of daptomcyin-BODIPY staining changes during sporulation in a manner that mirrors the relocalization of the peptidoglycan biogenesis apparatus and the reorganization of the membranes.
Fig 5.

Daptomycin-BODIPY localization during sporulation. The arrows indicate the positions of intense labeling. At 1.5 h (A and B) and 2 h (C) after the onset of sporulation, cells were labeled with 16 μg/ml daptomycin-BODIPY (green) and stained with FM 4-64 (red) as described in Materials and Methods. (A) Cells that have not yet synthesized a polar sporulation septum contain two polar daptomycin-BODIPY rings (blue arrows). (B and C) Cells that have completed polar septation and initiated engulfment show intense staining at the leading edge of the engulfing mother cell membrane (white arrows). Scale bar, 1 μm. All panels are at the same scale.
Sublethal doses of daptomycin induced dramatic changes in cell shape.
Daptomycin is bactericidal, raising the possibility that some of the cellular effects that we observed upon treatment were a secondary consequence of cell death rather than a direct action of the drug. We reasoned that we may able to gain additional insight into the cell biological effects of daptomcyin by observing the effects of sublethal concentrations (below the MIC) on cell growth. We therefore grew cells in LB containing 1 μg/ml daptomycin and imaged them after 2 h. Surprisingly, we found that cells grown in the presence of sublethal concentrations of daptomycin had a striking change in morphology (Fig. 6A to F). The cells were severely bent and occasionally swollen at one end and exhibited patches of extra membranes. Nearly every cell in the population was either slightly too long, too wide, or bent in a dramatic way, despite the fact that the growth rate was completely unaffected, as measured by optical density (Fig. 1A). Since the cell wall determines the shape of the cell, these results indicate that sublethal doses of daptomycin interfere (directly or indirectly) with both the membranes and proper formation of the cell wall. If daptomycin were directly responsible for causing these changes, we would expect daptomycin to localize to the bent positions. To test this idea, we grew cells in a sublethal concentration of daptomycin-BODIPY (10 μg/ml; MIC = 16 μg/ml) for 2 h at 37°C and then imaged the cells in the fluorescence microscope. Approximately 75% (n = 90) of the cells were misshapen and contained membrane patches similar to those of cells that were treated with sublethal concentrations of unlabeled daptomycin. Four different cells from this experiment with various degrees of membrane and cell shape defects are shown in Fig. 6G to J. Strikingly, fluorescence from daptomycin-BODIPY always colocalized with the membrane patches at bent regions (Fig. 6G to J). These results suggest that daptomycin is directly responsible for the cell shape and membrane alterations observed at sublethal concentrations. The remaining 25% of the cells in the population had lysed, indicating that they had received a lethal dose of drug.
Fig 6.

Cells grown in the presence of sublethal concentrations of daptomycin have a striking change in morphology. (A to F) Images of B. subtilis (PY79) after 2 h of growth in the presence of a sublethal concentration of daptomycin (1 μg/ml). The cell membranes are stained red with FM 4-64. (G to J) Daptomycin (Dap)-BODIPY localizes to membrane patches and to the membrane at the interiors of curved regions at sublethal concentrations. The panels show membrane staining (top), daptomycin-BODIPY labeling (bottom), or both (middle). PY79 was grown in LB with the sublethal dose of 10 μg/ml daptomycin-BODIPY for 2 h at 37°C. All panels are at the same scale. Scale bars, 1 μm.
DivIVA-GFP targets membrane patches formed during treatment with sublethal doses of daptomycin.
Daptomycin induced the formation of aberrant membrane patches at both lethal (Fig. 1) and sublethal (Fig. 6) doses. Membrane patches preceded cell death at lethal doses and had no effect on the rate of growth at sublethal doses, suggesting that they contained intact membranes with areas of altered curvature. Since membrane curvature can be a key determinant for protein localization, we reasoned that daptomycin-induced membrane patches might cause the aberrant recruitment of essential cell envelope proteins. This would disrupt cell wall biogenesis and contribute directly to the mechanism of lethality. To test this model, we examined two proteins (DivIVA and SpoVM) whose localization is highly sensitive to membrane curvature (22, 42). DivIVA is an essential cell division protein that regulates cell division assembly in B. subtilis (4). DivIVA-GFP targets negatively curved membranes and localizes to the midcell (28, 44), forming a pair of closely spaced rings by associating with the sharply curved membranes that occur at the site of septation (13). In contrast, SpoVM targets regions of positive membrane curvature (43). These two proteins might provide sensitive markers for gaining insight into the structure of the membrane patches induced by daptomycin. Since DivIVA is an essential component of the cell division apparatus, its localization may also provide insight into the mechanism of daptomycin-mediated alterations in cell wall biogenesis described above (Fig. 1 and 6).
We examined DivIVA-GFP in cells treated with sublethal concentrations of daptomycin (ranging from 1.5 μg/ml to 2.5 μg/ml). In untreated cells, DivIVA-GFP localized to the site of cell division, as observed previously (Fig. 7A). Growth in the presence of a sublethal dose of daptomycin resulted in cells with severe cell shape defects and membrane patches (Fig. 7B to D). Strikingly, DivIVA-GFP also localized to these membrane patches and to the bent regions (Fig. 7B to D). Three-dimensional fluorescence intensity plots show that DivIVA-GFP specifically colocalized with the daptomycin-induced membrane patches (Fig. 7B). We observed a 99% (n = 80) correspondence between the presence of abnormal membrane patches and the presence of DivIVA-GFP. These results suggest a strong relationship between cell damage caused by daptomycin and mislocalization of DivIVA-GFP. We obtained identical results when DivIVA-GFP was expressed from its native promoter on the chromosome, indicating that these results are not an artifact of DivIVA-GFP overexpression (not shown).
Fig 7.
DivIVA-GFP targets positions of membrane rearrangement and bent regions of sub-MIC daptomycin-treated cells, but SpoVM-GFP is specifically excluded. (A) In wild-type cells, DivIVA-GFP localizes to membrane invaginations during and after cell division. The fluorescence intensity graphs for membranes (FM 4-64) and DivIVA-GFP correspond to the boxed region. (B) DivIVA-GFP localizes to membrane patches formed in cells grown in a sublethal concentration of daptomycin (2 μg/ml). The three-dimensional graphs of fluorescence intensity corresponding to the boxed region show that regions containing concentrated membranes recruit DivIVA-GFP. (C and D) Two additional examples of cells grown in sublethal concentrations of daptomycin (2 μg/ml) showing that DivIVA-GFP is localized to the bent regions of the cells (arrows), where membrane patches occur. (E) SpoVM-GFP uniformly decorates the membrane in wild-type cells. Three-dimensional fluorescence intensity graphs of SpoVM-GFP and FM 4-64 are shown for the boxed region. (F and G) SpoVM-GFP is excluded from daptomycin-induced membrane patches. The three-dimensional fluorescence intensity graphs show that SpoVM-GFP is uniformly distributed around the cell and does not accumulate within the membrane patches. All panels are at the same scale. Scale bars, 1 μm.
We tested if SpoVM-GFP is also targeted to these same membrane patches. In untreated control cells, SpoVM-GFP uniformly localized to all membranes (Fig. 7E). Strikingly, SpoVM-GFP did not target the membrane patches formed by sublethal doses of daptomycin (Fig. 7F and G). Instead, quantitation of the images demonstrated that SpoVM-GFP is evenly distributed around the periphery, i.e., it is not intensified in areas of membrane accumulation, indicating that it is excluded from the daptomycin-induced membrane patches. Taken together, these results suggest that daptomycin induces changes in the membrane structure so that it resembles structures formed at the site of cell division that recruit DivIVA. Since daptomycin also localizes to these patches (Fig. 6G to J), daptomycin is likely to be directly responsible for the cell shape and membrane alterations and the subsequent mislocalization of essential cell division proteins, such as DivIVA.
We used fluorescent reporters of peptidoglycan biogenesis to explore the possible functional consequences of DivIVA mislocalization. Cells were grown either with or without a sublethal dose of daptomycin and then exposed to Van-BDP or Bocillin FL. Van-BDP has been used extensively to detect the location of peptidoglycan synthesis in Gram-positive bacteria (9, 14, 17, 31, 39, 54). Bocillin FL is a fluorescent penicillin V derivative that binds tightly to penicillin binding proteins (PBPs) (35, 59). In control cultures (without daptomycin), Van-BDP intensely stained both complete and incomplete septa and only faintly stained the sidewalls (Fig. 8A), as expected (9, 17, 54). In cells grown in a sublethal dose of daptomycin, Van-BDP also intensely stained membrane patches and bent regions (Fig. 8B). We obtained similar results for Bocillin FL (Fig. 9), which intensely stained the actively dividing septa of control cells (Fig. 9A to D) and strikingly stained the membrane patches and bent regions of cells grown in sublethal doses of daptomycin (Fig. 9E to H). Quantitation showed that the daptomycin-induced membrane patches and bent regions contained intense Bocillin FL staining in 96% (n = 72) of cases and intense Van-BDP staining in 99% (n = 101) of cases. Taken together, these results suggest a correlation between the membrane patches that are induced by the presence of daptomycin and the mislocalization of DivIVA and the peptidoglycan synthesis machinery.
Fig 8.

Fluorescent vancomycin targets positions of active peptidoglycan biogenesis, accumulating within septa in control cells and also in membrane patches and bent regions of sub-MIC daptomycin-treated cells. Cells were stained with the vital membrane stains FM 4-64 (1 μg/ml), DAPI (2 μg/ml), and a fluorescent derivative of vancomycin, as described in Materials and Methods. (A) In cells not treated with daptomycin, Van-BDP (0.5 μg/ml; ∼1× MIC) intensely stains septa and only very faintly stains the sidewalls, as previously shown (9, 17, 54). The septum in the middle of the cell is just beginning to divide and accumulates Van-BDP as a ring of fluorescence, which appears as two dots in the medial focal plan. Van-BDP forms an intense band at completed septa, as previously shown (9, 17, 54). Scale bar, 1 μm. (B) Two examples showing that Van-BDP localizes to the membrane patches and bent regions that are formed in cells grown in a sublethal concentration of daptomycin (2 μg/ml). Scale bar, 1 μm.
Fig 9.
The fluorescent penicillin V derivative Bocillin FL targets positions of active peptidoglycan biogenesis, accumulating within septa in control cells and in bent regions of sub-MIC daptomycin-treated cells. Cells were stained with the vital membrane stains FM 4-64 (1 μg/ml), DAPI (2 μg/ml), and Bocillin FL as described in Materials and Methods. (A) In cells not treated with daptomycin, Bocillin FL more intensely stains regions actively undergoing septation, as expected. The cell on the left is just beginning to divide and accumulates fluorescent Bocillin as a ring of fluorescence. The cell on the right has nearly completed cell division, and Bocillin FL accumulates as a small focus in the center of the septum. Scale bar, 1 μm. (B) Bocillin FL intensely stains the membrane patches and bent regions formed in cells grown in a sublethal concentration of daptomycin (2 μg/ml). Two examples are shown. Scale bar, 1 μm.
DISCUSSION
Daptomycin is a clinically important lipopeptide antibiotic whose ability to interact with the membrane has been well studied in vitro and is presumed to be essential to its mode of action (3). Previous models have suggested that daptomycin alters membrane structure (and possibly curvature) to allow leakage of essential ions from the cell in the absence of large pores (50). Here, we have used a variety of cell biological assays to provide new insight into the mechanism of action. We show that the first effect of daptomycin on the cell is the generation of randomly positioned membrane patches at both lethal and sublethal doses. The membrane patches colocalize with a fluorescent daptomycin derivative, suggesting that they are a direct result of daptomycin insertion into the membrane. We show that daptomycin-induced membrane patches specifically recruit proteins thought to be capable of recognizing negative membrane curvature, such as the conserved cell division protein DivIVA. Finally, we provide evidence that daptomycin has a dramatic local effect on the cell wall by showing that growth in sublethal doses of antibiotic leads to cells with severely altered cell shapes and altered localization of fluorescent reporters of peptidoglycan biogenesis. Taken together, our results demonstrate a direct correlation between daptomycin binding, membrane curvature, localization of the cell division protein DivIVA, and alterations in the cell wall. Importantly, these results bridge two previously disparate lines of data regarding the mechanism of daptomycin and may explain how a membrane-active antibiotic produces changes in cell wall physiology.
Based on these findings, we propose a unified model for the action of daptomycin at sub- and supra-MIC doses that links demonstrated effects on cell membranes to effects on cell wall architecture (Fig. 10). Insertion of daptomycin into bacterial membranes requires the presence of phosphatidylglycerol (25). Following insertion, daptomycin aggregates in the membrane (36). Viewed as a lipid, daptomycin has a large headgroup volume and a short, small-volume tail; aggregation of such “conical” lipids will produce local alterations in membrane curvature (24, 33, 53). (Note that for the purposes of this model, daptomycin is assumed to remain in the outer leaflet of the bilayer; this has not been definitively demonstrated, though it is suggested by studies with electron spin resonance probes [W. E. Trommer, personal communication].) At sub-MIC doses, the local alteration of outer-leaflet curvature is met with a local alteration in inner-leaflet curvature (potentially by the localization of cardiolipin). The localized region of membrane distortion and altered curvature is recognized by DivIVA (28, 44), leading the cell to incorrectly identify the location as a site of potential cell division. Recruitment of DivIVA leads to local changes in peptidoglycan biogenesis, including localized inhibition of lateral cell wall biosynthesis. Because no corresponding change is made around the cell wall, lateral extension continues on the opposite side of the cell, leading to asymmetric extension and, in the case of a rod-shaped cell, bending. We note that in S. aureus, the effect of sub-MIC daptomycin is induction of aberrant and asymmetric division septa (7), suggesting that differences in cell wall biosynthesis regulation in these species produce a different morphological outcome from a common insult.
Fig 10.
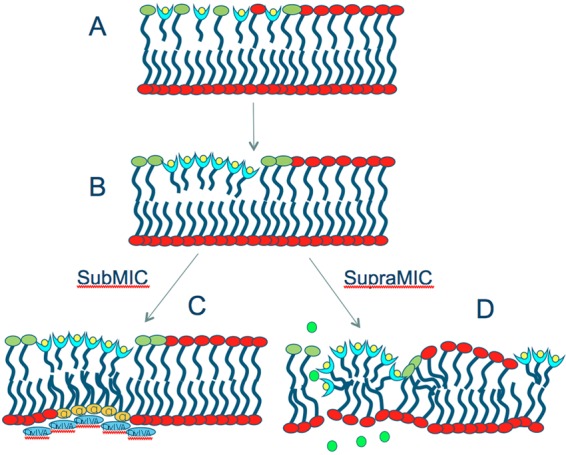
A refined model for the mechanism of action of daptomycin (light-blue “cups” with yellow circles). See the text for details. Shown are phosphatidylglycerol (green lipids), cardiolipin (yellow lipids), potassium ions (green circles), and DivIVA (blue ovals).
At supra-MIC doses, multiple sites of local membrane curvature are induced, presumably overwhelming the ability of the cell to respond through compensating membrane changes. Discontinuities in the membrane at the site of daptomycin insertion lead to slow leakage of ions and loss of membrane potential. These changes may also result in local dysregulation of the cell division or cell wall-biosynthetic machinery, including activation of autolysins, via local reduction in the proton motive force (PMF) (23, 52). We also speculate that there may be other integral membrane proteins whose functions are dependent on either proper localization or normal bilayer architecture, resulting in a highly pleiotropic effect of daptomycin on cell physiology.
Daptomycin inserts preferentially in the leading edges of the septal and forespore membranes. Interestingly, the topology of curvature of these two sites is inverted, i.e., daptomycin inserts into a highly positively curved membrane in one case and a highly negatively curved one in the other. In either setting, higher degrees of membrane strain may facilitate the insertion of exogenous lipid-like molecules. Counterintuitively, we propose that the effect of high daptomycin concentrations at this site is relatively benign and may even accelerate the cell division process because the local effect is to induce further curvature and recruitment of cell division machinery. Consistent with this hypothesis, treatment with a sub-MIC dose of daptomycin leads to the formation of additional septa in S. aureus (7), and in B. subtilis, we have not observed a strong inhibition of septation at either sub-MIC or supra-MIC doses (see Fig. S1 in the supplemental material).
The localization of daptomycin to the leading edges of the invaginating septa and the engulfing membranes might also reflect an affinity for a localized component of cell wall biosynthesis. While this is an attractive hypothesis, there is little direct evidence to support such a model, as daptomycin does not inhibit any of the enzymatic steps in cell wall biosynthesis that have been tested so far (19, 46, 49). Studies of daptomycin-resistant mutants isolated in a number of different species also generally support the idea that the membrane is the major target for daptomycin antibacterial activity. However, it is worth noting that one gene commonly mutated in daptomycin-resistant strains is yycG, encoding an essential histidine kinase that coordinates cell division and peptidoglycan biogenesis (16). Both daptomycin and YycG localize to the septum, which would be consistent with the suggestion that YycG interacts with daptomycin (16). We favor the idea that mutations in yycG arise in daptomycin-resistant mutants due to its ability to regulate the expression of genes involved in cell wall biogenesis rather than indicating direct inhibition, but we cannot explicitly rule out the latter.
The punctate sidewall staining of fluorescent daptomycin (Fig. 2) likely reflects its ability to insert into and oligomerize within the membrane (36). Oligomerization leads to the formation of curved membrane patches (as predicted by in vitro studies [24]) that recruit the cell division protein DivIVA, but not SpoVM, suggesting that they likely mimic the structure of the membranes at the division site. The formation of membrane patches leads to the misdirection and possibly localized alteration of the cell wall-biosynthetic machinery at these positions, generating a small but lethal rupture in both the cell membrane and the cell wall, consistent with the extruding membrane blebs that we observed in time lapse experiments (Fig. 1) and with previous EM studies (6, 55; J. Silverman, unpublished data).
Membrane geometry has recently been recognized as an important determinant of both lipid (21) and protein (22) localization. In Gram-positive bacteria, the essential cell division protein DivIVA relies upon changes in membrane geometry to regulate its localization and activity (13, 28, 44). If this mechanism of protein localization is both conserved and essential in bacteria, it is possible that some antibiotics may have evolved to exploit it as a novel Achilles' heel. Here, we show that an antibiotic previously proposed to interact with and alter the conformation of membranes is responsible for causing the mislocalization of DivIVA. These results raise the intriguing possibility that daptomycin might exploit the bacterial cells' reliance on membrane geometry as a novel type of molecular target. In Streptomyces, the position of DivIVA assembly determines the site of hyphal-branch development (20). DivIVA is therefore thought to play a direct role in controlling the position of peptidoglycan synthesis. Since daptomycin is produced by S. roseosporus, it is tempting to speculate that daptomycin normally serves as a diffusible morphogen, an ecological role that could provide insight into how the antibiotic evolved. The effect of daptomycin on Streptomyces species present in the soil may be to stimulate the formation of DivIVA patches and initiate hyphal branching, while its effect on other, nonbranching Gram-positive soil bacteria, such as Bacillus species, would be cell killing.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by NIH grant GM073898 to J.P. and by a grant from Cubist Pharmaceuticals to J.P.
We are grateful to W. E. Trommer for communicating results prior to publication and to Yong He and Andre Pearson for the synthesis of BODIPY-daptomycin.
Footnotes
Published ahead of print 1 June 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Abanes-De Mello A, Sun YL, Aung S, Pogliano K. 2002. A cytoskeleton-like role for the bacterial cell wall during engulfment of the Bacillus subtilis forespore. Genes Dev. 16:3253–3264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arias CA, et al. 2011. Genetic basis for in vivo daptomycin resistance in enterococci. N. Engl. J. Med. 365:892–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baltz RH. 2009. Daptomycin: mechanisms of action and resistance, and biosynthetic engineering. Curr. Opin. Chem. Biol. 13:144–151 [DOI] [PubMed] [Google Scholar]
- 4. Bramkamp M, van Baarle S. 2009. Division site selection in rod-shaped bacteria. Curr. Opin. Microbiol. 12:683–688 [DOI] [PubMed] [Google Scholar]
- 5. Broder DH, Pogliano K. 2006. Forespore engulfment mediated by a ratchet-like mechanism. Cell 126:917–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Conrad RS, Howard MJ, Garrison RC, Winters S, Henderson DA. 1998. The effects of daptomycin on chemical composition and morphology of Staphylococcus aureus. Proc. Okla. Acad. Sci. 78:15–22 [Google Scholar]
- 7. Cotroneo N, Harris R, Perlmutter N, Beveridge T, Silverman JA. 2008. Daptomycin exerts bactericidal activity without lysis of Staphylococcus aureus. Antimicrob. Agents Chemother. 52:2223–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cui L, Tominaga E, Neoh HM, Hiramatsu K. 2006. Correlation between reduced daptomycin susceptibility and vancomycin resistance in vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 50:1079–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Daniel RA, Errington J. 2003. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell 113:767–776 [DOI] [PubMed] [Google Scholar]
- 10. Eisenstein BI, Oleson FB, Jr, Baltz RH. 2010. Daptomycin: from the mountain to the clinic, with essential help from Francis Tally, MD. Clin. Infect. Dis. 50(Suppl. 1):S10–S15 [DOI] [PubMed] [Google Scholar]
- 11. Eliopoulos GM, Thauvin C, Gerson B, Moellering RC., Jr 1985. In vitro activity and mechanism of action of A21978C1, a novel cyclic lipopeptide antibiotic. Antimicrob. Agents Chemother. 27:357–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Errington J. 2003. Regulation of endospore formation in Bacillus subtilis. Nat. Rev. Microbiol. 1:117–126 [DOI] [PubMed] [Google Scholar]
- 13. Eswaramoorthy P, et al. 2011. Cellular architecture mediates DivIVA ultrastructure and regulates Min activity in Bacillus subtilis. MBio 2:e00257–11 doi:10.1128/mBio.00257-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Flardh K. 2003. Essential role of DivIVA in polar growth and morphogenesis in Streptomyces coelicolor A3(2). Mol. Microbiol. 49:1523–1536 [DOI] [PubMed] [Google Scholar]
- 15. Friedman L, Alder JD, Silverman JA. 2006. Genetic changes that correlate with reduced susceptibility to daptomycin in Staphylococcus aureus. Antimicrob. Agents Chemother. 50:2137–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fukushima T, Szurmant H, Kim EJ, Perego M, Hoch JA. 2008. A sensor histidine kinase co-ordinates cell wall architecture with cell division in Bacillus subtilis. Mol. Microbiol. 69:621–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hachmann AB, Angert ER, Helmann JD. 2009. Genetic analysis of factors affecting susceptibility of Bacillus subtilis to daptomycin. Antimicrob. Agents Chemother. 53:1598–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hachmann AB, et al. 2011. Reduction in membrane phosphatidylglycerol content leads to daptomycin resistance in Bacillus subtilis. Antimicrob. Agents Chemother. 55:4326–4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hashizume H, et al. 2011. Tripropeptin C blocks the lipid cycle of cell wall biosynthesis by complex formation with undecaprenyl pyrophosphate. Antimicrob. Agents Chemother. 55:3821–3828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hempel AM, Wang SB, Letek M, Gil JA, Flardh K. 2008. Assemblies of DivIVA mark sites for hyphal branching and can establish new zones of cell wall growth in Streptomyces coelicolor. J. Bacteriol. 190:7579–7583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huang KC, Mukhopadhyay R, Wingreen NS. 2006. A curvature-mediated mechanism for localization of lipids to bacterial poles. PLoS Comput. Biol. 2:e151 doi:10.1371/journal.pcbi.0020151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang KC, Ramamurthi KS. 2010. Macromolecules that prefer their membranes curvy. Mol. Microbiol. 76:822–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jolliffe LK, Doyle RJ, Streips UN. 1981. The energized membrane and cellular autolysis in Bacillus subtilis. Cell 25:753–763 [DOI] [PubMed] [Google Scholar]
- 24. Jung D, Powers JP, Straus SK, Hancock RE. 2008. Lipid-specific binding of the calcium-dependent antibiotic daptomycin leads to changes in lipid polymorphism of model membranes. Chem. Phys. Lipids 154:120–128 [DOI] [PubMed] [Google Scholar]
- 25. Jung D, Rozek A, Okon M, Hancock RE. 2004. Structural transitions as determinants of the action of the calcium-dependent antibiotic daptomycin. Chem. Biol. 11:949–957 [DOI] [PubMed] [Google Scholar]
- 26. Lakey JH, Lea EJ. 1986. The role of acyl chain character and other determinants on the bilayer activity of A21978C an acidic lipopeptide antibiotic. Biochim. Biophys. Acta 859:219–226 [DOI] [PubMed] [Google Scholar]
- 27. Lakey JH, Ptak M. 1988. Fluorescence indicates a calcium-dependent interaction between the lipopeptide antibiotic LY146032 and phospholipid membranes. Biochemistry 27:4639–4645 [DOI] [PubMed] [Google Scholar]
- 28. Lenarcic R, et al. 2009. Localisation of DivIVA by targeting to negatively curved membranes. EMBO J. 28:2272–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Levin PA, Losick R. 1996. Transcription factor SpoOA switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Genes Dev. 10:478–488 [DOI] [PubMed] [Google Scholar]
- 30. Losick R, Stragier P. 1992. Crisscross regulation of cell-type-specific gene expression during development in Bacillus subtilis. Nature 355:601–604 [DOI] [PubMed] [Google Scholar]
- 31. Lunde CS, Rexer CH, Hartouni SR, Axt S, Benton BM. 2010. Fluorescence microscopy demonstrates enhanced targeting of telavancin to the division septum of Staphylococcus aureus. Antimicrob. Agents Chemother. 54:2198–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mascio CT, Alder JD, Silverman JA. 2007. Bactericidal action of daptomycin against stationary-phase and nondividing Staphylococcus aureus cells. Antimicrob. Agents Chemother. 51:4255–4260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McMahon HT, Gallop JL. 2005. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature 438:590–596 [DOI] [PubMed] [Google Scholar]
- 34. Meyer P, Gutierrez J, Pogliano K, Dworkin J. 2010. Cell wall synthesis is necessary for membrane dynamics during sporulation of Bacillus subtilis. Mol. Microbiol. 76:956–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Moisan H, Pruneau M, Malouin F. 2010. Binding of ceftaroline to penicillin-binding proteins of Staphylococcus aureus and Streptococcus pneumoniae. J. Antimicrob. Chemother. 65:713–716 [DOI] [PubMed] [Google Scholar]
- 36. Muraih JK, Pearson A, Silverman J, Palmer M. 2011. Oligomerization of daptomycin on membranes. Biochim. Biophys. Acta 1808:1154–1160 [DOI] [PubMed] [Google Scholar]
- 37. Muthaiyan A, Silverman JA, Jayaswal RK, Wilkinson BJ. 2008. Transcriptional profiling reveals that daptomycin induces the Staphylococcus aureus cell wall stress stimulon and genes responsive to membrane depolarization. Antimicrob. Agents Chemother. 52:980–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Palmer KL, Daniel A, Hardy C, Silverman J, Gilmore MS. 2011. Genetic basis for daptomycin resistance in enterococci. Antimicrob. Agents Chemother. 55:3345–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pereira PM, Filipe SR, Tomasz A, Pinho MG. 2007. Fluorescence ratio imaging microscopy shows decreased access of vancomycin to cell wall synthetic sites in vancomycin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 51:3627–3633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pogliano J, et al. 1999. A vital stain for studying membrane dynamics in bacteria: a novel mechanism controlling septation during Bacillus subtilis sporulation. Mol. Microbiol. 31:1149–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ptacin JL, et al. 2008. Sequence-directed DNA export guides chromosome translocation during sporulation in Bacillus subtilis. Nat. Struct. Mol. Biol. 15:485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ramamurthi KS. 2010. Protein localization by recognition of membrane curvature. Curr. Opin. Microbiol. 13:753–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ramamurthi KS, Lecuyer S, Stone HA, Losick R. 2009. Geometric cue for protein localization in a bacterium. Science 323:1354–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ramamurthi KS, Losick R. 2009. Negative membrane curvature as a cue for subcellular localization of a bacterial protein. Proc. Natl. Acad. Sci. U. S. A. 106:13541–13545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Roth BL, Poot M, Yue ST, Millard PJ. 1997. Bacterial viability and antibiotic susceptibility testing with SYTOX green nucleic acid stain. Appl. Environ. Microbiol. 63:2421–2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rubinchik E, et al. 2011. Mechanism of action and limited cross-resistance of new lipopeptide MX-2401. Antimicrob. Agents Chemother. 55:2743–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rubio A, et al. 2011. Regulation of mprF by antisense RNA restores daptomycin susceptibility to daptomycin-resistant isolates of Staphylococcus aureus. Antimicrob. Agents Chemother. 55:364–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ryter A. 1965. Etude morphologie de la sporulation de Bacillus subtilis. Ann. Inst. Pasteur (Paris) 108:40–60 [PubMed] [Google Scholar]
- 49. Schneider T, et al. 2009. The lipopeptide antibiotic Friulimicin B inhibits cell wall biosynthesis through complex formation with bactoprenol phosphate. Antimicrob. Agents Chemother. 53:1610–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Silverman JA, Perlmutter NG, Shapiro HM. 2003. Correlation of daptomycin bactericidal activity and membrane depolarization in Staphylococcus aureus. Antimicrob. Agents Chemother. 47:2538–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sterlini JM, Mandelstam J. 1969. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem. J. 113:29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Strahl H, Hamoen LW. 2010. Membrane potential is important for bacterial cell division. Proc. Natl. Acad. Sci. U. S. A. 107:12281–12286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Straus SK, Hancock RE. 2006. Mode of action of the new antibiotic for Gram-positive pathogens daptomycin: comparison with cationic antimicrobial peptides and lipopeptides. Biochim. Biophys. Acta 1758:1215–1223 [DOI] [PubMed] [Google Scholar]
- 54. Tiyanont K, et al. 2006. Imaging peptidoglycan biosynthesis in Bacillus subtilis with fluorescent antibiotics. Proc. Natl. Acad. Sci. U. S. A. 103:11033–11038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wale LJ, Shelton AP, Greenwood D. 1989. Scanning electronmicroscopy of Staphylococcus aureus and Enterococcus faecalis exposed to daptomycin. J. Med. Microbiol. 30:45–49 [DOI] [PubMed] [Google Scholar]
- 56. Yang SJ, et al. 2010. Cell wall thickening is not a universal accompaniment of the daptomycin nonsusceptibility phenotype in Staphylococcus aureus: evidence for multiple resistance mechanisms. Antimicrob. Agents Chemother. 54:3079–3085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yang SJ, et al. 2009. Regulation of mprF in daptomycin-nonsusceptible Staphylococcus aureus strains. Antimicrob. Agents Chemother. 53:2636–2637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Youngman P, Perkins JB, Sandman K. (ed). 1984. New genetic methods, molecular cloning strategies, and gene fusion techniques for Bacillus subtilis which take advantage of Tn917 insertional mutagenesis. Academic Press, New York, NY [Google Scholar]
- 59. Zhao G, Meier TI, Kahl SD, Gee KR, Blaszczak LC. 1999. BOCILLIN FL, a sensitive and commercially available reagent for detection of penicillin-binding proteins. Antimicrob. Agents Chemother. 43:1124–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.






