Abstract
Choline is abundantly produced by eukaryotes and plays an important role as a precursor of the osmoprotectant glycine betaine. In Pseudomonas aeruginosa, glycine betaine has additional roles as a nutrient source and an inducer of the hemolytic phospholipase C, PlcH. The multiple functions for glycine betaine suggested that the cytoplasmic pool of glycine betaine is regulated in P. aeruginosa. We used 13C nuclear magnetic resonance (13C-NMR) to demonstrate that P. aeruginosa maintains both choline and glycine betaine pools under a variety of conditions, in contrast to the transient glycine betaine pool reported for most bacteria. We were able to experimentally manipulate the choline and glycine betaine pools by overexpression of the cognate catabolic genes. Depletion of either the choline or glycine betaine pool reduced phospholipase production, a result unexpected for choline depletion. Depletion of the glycine betaine pool, but not the choline pool, inhibited growth under conditions of high salt with glucose as the primary carbon source. Depletion of the choline pool inhibited growth under high-salt conditions with choline as the sole carbon source, suggesting a role for the choline pool under these conditions. Here we have described the presence of a choline pool in P. aeruginosa and other pseudomonads that, with the glycine betaine pool, regulates osmoprotection and phospholipase production and impacts growth under high-salt conditions. These findings suggest that the levels of both pools are actively maintained and that perturbation of either pool impacts P. aeruginosa physiology.
INTRODUCTION
Acquisition or synthesis of osmoprotectants is important for survival of many pathogens in the host (23, 43, 51). A number of osmoprotectants can be derived from the host, including proline, choline, carnitine, and glycine betaine (GB). As a moiety on phosphatidylcholine and sphingomyelin, choline is likely the most abundant of these compounds at infection sites. Choline must undergo metabolism to GB to afford osmoprotection (28), with a single reported exception (20). Pseudomonas aeruginosa has an efficient choline acquisition system regulated by both positive-feedback induction and choline-dependent repression (34, 46, 55). Choline is initially transported into the cell by the BetT1 and BetT3 transporters (9, 34), where it can be used to generate bacterial phosphatidylcholine and to posttranslationally modify proteins (1, 32, 35, 59). Cytoplasmic choline can also release BetI repression of betBA transcription (25), which leads to production of choline oxidase (BetA) and betaine aldehyde dehydrogenase (BetB), which sequentially oxidize choline to GB (18, 26).
P. aeruginosa can use GB as a nutrient and energy source and as an osmoprotectant, as well as to regulate transcription via GbdR (29, 30, 53, 55). GB activation of the GbdR transcription factor leads to induction of the transcripts encoding the secreted phospholipase C virulence factor PlcH (27, 31, 39, 44, 54, 57, 58), the periplasmic phosphorylcholine phosphatase, PchP (37, 55), and the CbcXWV high-capacity ABC transport system for choline and GB (9, 34). These products result in increased potential for choline acquisition from lipid precursors (55) and contribute to the formation of a positive-feedback induction loop. Choline acquisition has been proposed to promote both the survival and the virulence of P. aeruginosa due to the multiple functions of choline and GB (31). We predict that P. aeruginosa regulates intracellular pools of both choline and GB to ensure balance between the multiple intracellular roles of each metabolite.
Many bacteria rapidly accumulate cytoplasmic GB when supplied with choline or GB in a high-salt medium (7, 26, 42). This has been best studied in the Enterobacteriaceae and Bacillus subtilis, where the GB pool is actively regulated by balancing import and efflux (3, 21). Intracellular GB accumulation has physiological consequences that can be deleterious, particularly in certain mutant backgrounds, supporting the importance of proper GB regulation in the cell (16, 21). Unlike the Enterobacteriaceae and B. subtilis, many soil- and water-dwelling Proteobacteria, including P. aeruginosa, can use GB as a sole source of carbon, nitrogen, and energy through successive demethylations to glycine (5, 22, 52). As both a nutrient and an osmoprotectant, the regulation of choline's and GB's fate within P. aeruginosa cells is predicted to be more complex than in the Enterobacteriaceae. Because choline acquisition from lipid precursors, high-capacity transport under low-salt conditions, and GB metabolism are all regulated by GB activation of GbdR in P. aeruginosa (9, 34, 55, 56), we predict that misregulation of GB and choline pool sizes impact one or more of these activities.
Diab and colleagues examined the fate of exogenous GB, but not exogenous choline, in P. aeruginosa, where they demonstrated an intracellular pool of GB that was long-lived even when GB was present as the sole carbon source (12). However, the consequences of having choline as the source of GB for these metabolite pools in P. aeruginosa remained unknown. In this study, we have examined choline and GB homeostasis where supplied choline was the sole source of choline and resultant GB. Our focus on choline allowed us to uncover the existence of a novel choline pool and describe the previously unappreciated interaction between the choline and GB pools in P. aeruginosa. The presence of a choline pool in P. aeruginosa is in direct contrast to similar studies of Sinorhizobium meliloti, which maintained a GB pool only during growth on choline (15, 52, 56). To our knowledge, this is the first report of physiologic choline pools that are present in the absence of osmotic stress or betBA mutations (4, 20). We also used experimental depletion of the endogenous choline and GB pools to demonstrate that depletion of either pool alters PlcH production and growth under high-salt conditions. These findings directly link regulation of choline and GB pools with virulence and stress protection. Finally, the presence of choline and GB pools in other pseudomonads also suggests importance across this group of bacteria.
MATERIALS AND METHODS
Strains and growth conditions.
Pseudomonas aeruginosa strains PAO1 and PA14, Pseudomonas putida (ATCC 49128), Pseudomonas fluorescens (ATCC 13525), Pseudomonas syringae DC3000, Burkholderia cepacia (ATCC 25416), Sinorhizobium meliloti RM1021, and Escherichia coli strains were maintained on LB medium. When necessary, gentamicin was added to final concentrations of 10 μg/ml for E. coli, 50 μg/ml for P. aeruginosa in LB medium, and 20 μg/ml for P. aeruginosa in MOPS (morpholinepropanesulfonic acid) medium (55). Growth of all species was measured by determining optical density at 600 nm (OD600). Prior to all experiments described here, cells picked from a fresh LB plate were grown for 24 h in MOPS minimal medium with 20 mM sodium pyruvate and 5 mM glucose (15). After this overnight growth, we saw no evidence of catabolite repression of GB catabolism, presumably due to early utilization of glucose during growth. Growth experiments were performed with choline as a sole carbon source or choline as a supplemental carbon source; in these experiments the total carbon source concentration equaled 20 mM unless otherwise noted. For high-salt experiments, NaCl was added to increase the salt concentration by 700 mM unless otherwise noted, which results in a final NaCl concentration of 750 mM. l-Arabinose was added to a final concentration of 0.05% (wt/vol) to induce expression from pBAD-based expression plasmids. P. aeruginosa and B. cepacia were grown at 37°C or 30°C, as noted below, while P. putida, P. fluorescens, P. syringae, and S. meliloti were grown at 30°C.
Cell labeling, small-molecule extraction, sample preparation, and 13C-NMR.
Labeled choline was synthesized by reacting labeled [1,2-13C]dimethylethanolamine (custom synthesized by Cambridge Isotopes) with iodomethane as described previously (15). We chose to use a 13C-labeled compound because this would allow us to track the abundance and the identity of the molecule with certainty, because choline and a downstream metabolite exhibit doublets in the 13C nuclear magnetic resonance (13C-NMR) spectrum, resulting from coupling of the adjacent 13C nuclei. In P. aeruginosa, choline is sequentially demethylated to glycine (12, 56). During growth on choline, glycine is hydroxylated to serine and serine is deaminated to form pyruvate (38, 52). Once pyruvate is decarboxylated during complete metabolism of choline, the original labeled carbons are separated and thus the doublet disappears.
Cells were grown in 30 ml of MOPS with 20 mM pyruvate and 5 mM glucose, collected by centrifugation, washed with Dulbecco's phosphate-buffered saline (DPBS), and resuspended in 30 ml of MOPS with 5 mM [1,2-13C]choline and 15 mM unlabeled choline or an alternate carbon source, with or without additional NaCl. Unless otherwise noted, after 9 h of growth at the organism's preferred temperature, with shaking at 200 rpm, cells were collected by centrifugation and washed twice with DPBS. The pellet was twice extracted with 250 μl of 80% ethanol. The extracted cell pellets and ethanolic extracts were dried in a rotary evaporator. The pellets were weighed, and the dried ethanolic extract residue was resuspended in 100% deuterium oxide (D2O). [13C]methanol was added to achieve a final concentration of 20 nmol per mg (dry weight) of pellet as an internal reference standard. 13C-NMR spectra were collected on a 500-MHz Bruker AXR or a 500-MHz Varian Unity Innova high-field NMR spectrometer. The spectrometer was referenced to an external standard of 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS) in D2O. All peaks, including those of compound standards and the added methanol standard in the resuspended ethanolic extract, were consistently found 2 ppm further downfield from their published ppm values due to solvent effects. During acquisition, we used a 1- or 2-s delay to enhance the detection of 13C-labeled compounds. We used a full relaxation protocol (7-s delay) to quantify the abundance of each labeled carbon. We compared integration spectra of reference samples with a 1- or 2-s delay to those of the same samples with the full relaxation protocol, which allowed us to generate correction factors for choline and GB in relation to methanol. We used these correction factors to normalize the signals from the C-2 atoms of choline and GB to calculate the nmol per mg or the dry weight for each sample.
Construction of BetBA and GbcBA expression vectors.
The pBetBA expression vector was constructed by amplifying the betBA portion of the betIBA operon from P. aeruginosa PA14 genomic DNA using Phusion DNA polymerase with GC buffer (NEB), primer BetBA exp-F (5′-actctctactgtttctccatacccgtttttttgggctagcAAGAGAGGACTGCAACATGG-3′), and primer BetBA exp-R (5′-tcaggctgaaaatcttctctcatccgccaaaacagccaaCATGCTGGTCGAGGTGTG-3′). The lowercase letters in the primers represent regions for Saccharomyces cerevisiae homologous recombination. This product was cloned into the l-arabinose-inducible expression vector pMQ80 using yeast recombination based on the methods of Shanks and colleagues (48). After validation of the construct by sequencing, we verified that the pBetBA construct rescued the PAO1 betB::Tn5 (PAO1 library number 104 [19]) strain for growth on choline (data not shown).
The enzymes for GB demethylation, GbcA and -B, are encoded by a pair of divergently transcribed genes. In order to ensure inducible coexpression, we used a PCR strategy to construct a gbcBA operon under the control of the pBAD promoter to generate pGbcBA. The pGbcBA expression vector was constructed by amplifying gbcA with primer gbcABconstruct-Afor1 (5′-GGCATGAGGAGTTACCGATG-3′) and primer gbcABconstruct-Arev1 (5′-tgtatcaggctgaaaatcttctctcatccgccaaaacagcGCAGGTTGTCCATGACCTTT-3′) and amplifying gbcB with primer gbcABconstruct-Bfor1 (5′-ttctccatacccgtttttttgggctagcgaattcgagctcCAGGTCGCGATAAGCATGTA-3′) and primer gbcABconstruct-Brev1 (5′-ctcagggtggaagtgacgtccatcggtaactcctcatgccGCCCTCAGTAGTCGATGACC-3′) from P. aeruginosa PA14 genomic DNA using Phusion DNA polymerase in GC buffer (NEB). The lowercase letters in the primers represent regions designed for the splice overlap or yeast homologous recombination. These initial amplicons were used in a splice overlap extension PCR using primers gbcABconstruct-Bfor1 and gbcABconstruct-Arev1 to generate a product that placed gbcB in front of gbcA in a single operon. This PCR product was cloned into pMQ80 using yeast recombination based on the methods of Shanks and colleagues (48). After validation of the construct by sequencing, we verified that the pGbcBA construct rescued the ΔgbcAB mutant (56) for growth on choline (data not shown).
Phospholipase C activity assay.
Phospholipase C activity was measured by observing the hydrolysis of the synthetic substrate p-nitrophenylphosphorylcholine (NPPC) based upon the Kurioka and Matsuda method (24), modified as we have described previously (55), using a final concentration of 10 mM NPPC. P. aeruginosa was inoculated at an OD600 of 0.3 into MOPS minimal media with 20 mM pyruvate, gentamicin, and 0.05% l-arabinose, with or without 1 mM choline and with or without added NaCl. Cells were grown for 24 h at 37°C with shaking. NPPC hydrolysis was measured by monitoring absorbance at 410 nm over time using a Synergy2 spectrophotometer (BioTek). Phospholipase C activity was reported in μmol of p-nitrophenol generated per minute of reaction per optical density (OD600) per ml of culture using the extinction coefficient 17,700 M−1 cm−1 (49).
RESULTS
P. aeruginosa maintains intracellular pools of choline and GB.
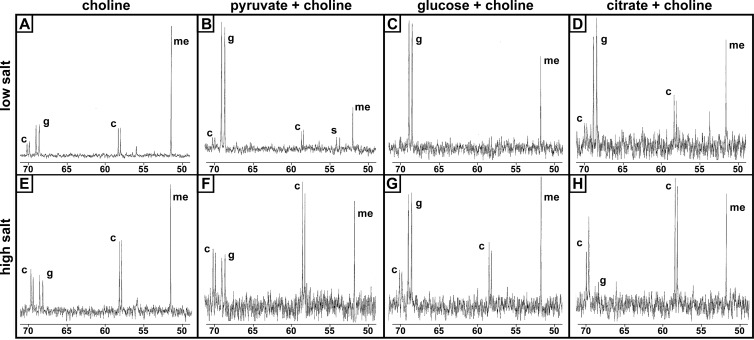
Many bacteria maintain GB pools during exposure to choline or GB, particularly under osmostress conditions (8, 21, 36). However, accumulation of a substantial choline pool has been reported in only one organism grown under high-salt conditions (20). Additional reports of long-lived choline accumulation are in the context of mutations in the choline oxidase genes (3). We previously observed that deletion or chemical inhibition of the dimethylglycine demethylase (Dgc) in P. aeruginosa led to accumulation of dimethylglycine and also a substantial accumulation of choline and GB (15). Our original hypothesis was that GB accumulated due to feedback inhibition on the GB demethylase. However, examination of subsequent data suggested the existence of physiologic pools of choline and GB during P. aeruginosa growth on choline in the absence of mutations. To address these observations and examine the proposed choline and GB pools, we grew P. aeruginosa PAO1 on 15 mM choline, pyruvate, glucose, or citrate with the addition of 5 mM [1,2-13C]choline in the presence of 50 mM NaCl. Ethanol extracts from cells harvested during exponential phase were analyzed by 13C-NMR (Fig. 1; results are quantified in Table 1). P. aeruginosa maintains nearly equivalent pools of choline and GB when growing on choline as a sole carbon source (Fig. 1A and Table 1). When choline was supplemented with other carbon sources, P. aeruginosa maintained similarly sized or larger GB pools but smaller choline pools (Fig. 1B to D and Table 1). When grown on pyruvate with labeled choline, the GB pool exceeded the choline pool nearly 5-fold (Fig. 1B). When grown with glucose or citrate, choline content was low and close to the detection limit, while GB quantities were close to those of choline-grown cells (Fig. 1C to D and Table 1). These quantities of GB are comparable to those reported for E. coli and B. subtilis in moderate salt concentrations (17, 28).
Fig 1.
13C-NMR of cell extracts showing choline and glycine betaine (GB) pools during growth on different carbon sources in the presence of high or low concentrations of salt. The carbon source for each column is noted at the top of the figure. The top row is low salt (50 mM NaCl, standard MOPS medium), and the bottom row is high salt (MOPS plus 700 mM NaCl). The x axis represents ppm. Under all conditions, cells were grown in the presence of 5 mM [1,2-13C]choline. For panels A and E, additional unlabeled choline was added for a total of 20 mM choline; therefore, the labeled compound is only one-quarter of the pool in these cells (as observed by the peak heights compared to that of the methanol standard). Abbreviations: c, choline; g, glycine betaine (GB); me, [13C]methanol; s, sarcosine. Each panel shows results representative of at least three experiments.
Table 1.
Choline and GB pool quantificationa
| NaCl concn (mM) | Carbon source(s)b | Choline pool (nmol/mg [dry wt]) (±SD) | GB pool (nmol/mg [dry wt]) (±SD) |
|---|---|---|---|
| 50 | Choline | 42.11 (29.42) | 56.23 (16.27) |
| Pyruvate + choline | 23.57 (2.64) | 112.54 (35.03) | |
| Glucose + choline | 13.12 (1.44) | 52.70 (13.34) | |
| Citrate + choline | 16.52 (5.59) | 46.05 (12.41) | |
| 750 | Cholinec | 66.61 (26.42) | 51.26 (15.24) |
| Pyruvate + choline | 36.39 (9.33) | 20.69 (1.34) | |
| Glucose + choline | 14.22 (7.70) | 166.07 (49.14) | |
| Citrate + choline | 81.15 (15.58) | 44.65 (31.14) |
Normalized to the methanol standard and quantified as described in Materials and Methods.
Carbon sources are for a total of 20 mM. In all cases, [13C]choline was added to 5 mM.
Calculations for pool sizes reflect that only 25% of the supplied choline is [13C]choline.
To determine if the pools were altered by increased salinity (total, 750 mM NaCl), we measured the choline and GB pools during growth under high-NaCl conditions in the presence of the same four carbon sources. We observed that the addition of NaCl substantially increased the choline pool in citrate and increased the GB pool in the presence of glucose. Conversely, when the organism was grown with pyruvate, the GB pool was significantly decreased during growth on high salt (Fig. 1B and F and Table 1) (P = 0.04). The other pools did not substantially change with the addition of salt. Interestingly, when cells were grown on citrate with labeled choline in hyperosmotic media, choline was nearly twice as abundant as GB (Fig. 1H and Table 1). Based on rough calculations using the various estimates from Gram-negative bacteria, only the GB pool of glucose-exposed cells in high salt were capable of providing osmoprotection as the sole osmoprotectant. It is important to note that our ability to detect compounds outside the choline catabolic pathway is limited and that there are likely de novo syntheses of other osmoprotectants under these conditions.
We began to detect choline and GB accumulation by 1 h postaddition, a time course similar to those previously reported (9, 33, 47). These pools were maintained for at least 12 h during growth on choline as a sole carbon source but were below the detection limit at 24 h (data not shown). Therefore, although the pools are present during growth, they are depleted for use as a carbon source when no additional carbon source is available.
Overexpressing the choline or GB catabolic enzymes can deplete the respective intracellular pool.
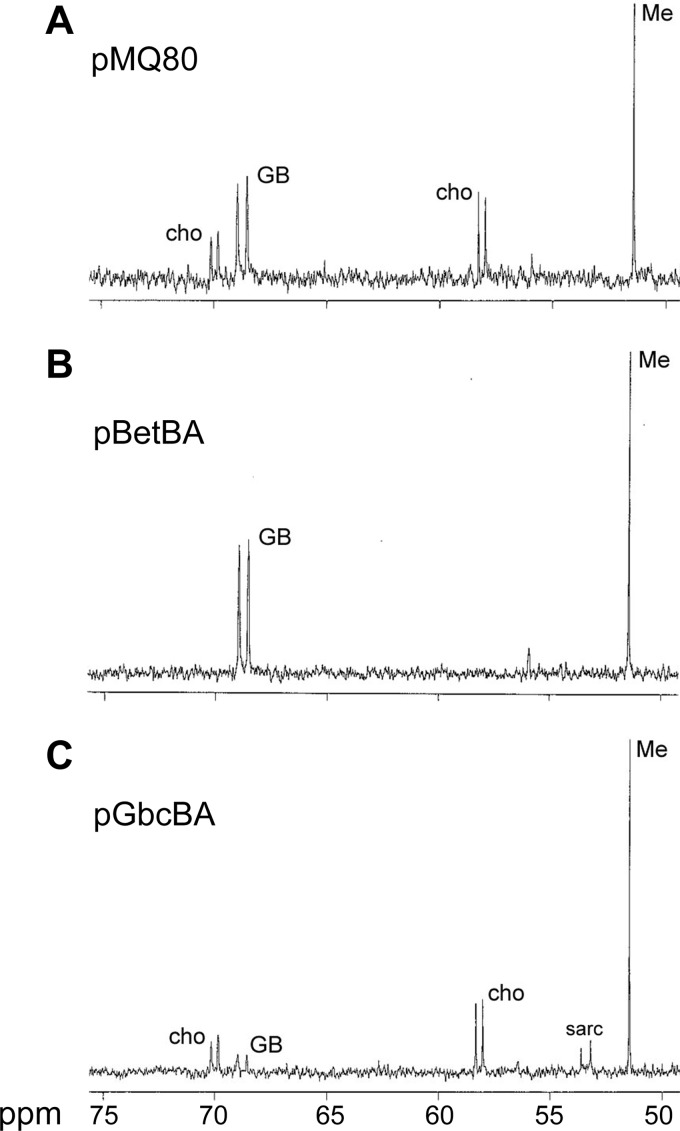
To study the functions of these intracellular metabolite pools, we tested whether the choline or GB pools could be experimentally depleted without the need to alter growth conditions. We hypothesized that overexpressing P. aeruginosa's native catabolic enzymes for choline oxidation (BetBA) or GB demethylation (GbcBA) would lead to depletion of the corresponding cytoplasmic pools. When the genes responsible for oxidizing choline to GB (betBA) were overexpressed, there was no observable choline pool (Fig. 2B) (compare to the empty vector control in Fig. 2A). It is interesting to note that when normalized to the methanol standard, the GB content increased approximately 25%, an increase of approximately 20 nmol/mg (dry weight). The original choline pool was approximately 40 nmol/mg (dry weight). Therefore, the entire choline pool did not accumulate as GB upon choline depletion, suggesting compensatory regulation of the size of the GB pool.
Fig 2.
13C-NMR of P. aeruginosa cell extracts after exposure to [1,2-13C]choline, with choline as the sole carbon source. Cells were grown as described in Materials and Methods in the presence of gentamicin, l-arabinose (0.05%), and labeled choline for 6 h. (A) Cells maintaining the pMQ80 empty vector (expresses green fluorescent protein [GFP] during arabinose induction under pBAD control); (B) cells expressing the betBA genes under pBAD control; (C) cells expressing the gbcBA genes under pBAD control. These spectra are representative of at least three biological replicates. Abbreviations: cho, choline; GB, glycine betaine; Me, [13C]methanol standard; sarc, sarcosine.
When the genes encoding the GB demethylase (gbcBA) were overexpressed, the GB pool was depleted approximately 80% compared to that with the empty vector control (Fig. 2A and C). However, the choline pool was not depleted, suggesting that P. aeruginosa does not drain the choline pool to replenish the GB pool. During depletion of the GB pool, we observed a doublet corresponding to sarcosine (Fig. 2C). We were not able to deplete the GB pool below detection, as we did for choline, but we were able to demonstrate that overexpression of the gbcBA genes leads to substantial depletion of the GB pool and creation of a detectable sarcosine pool. Addition of l-arabinose to the empty vector does not alter pools compared to those in cells without the vector (compare Fig. 1A and 2A).
Effects of overexpressing betBA or gbcBA on phospholipase C activity.
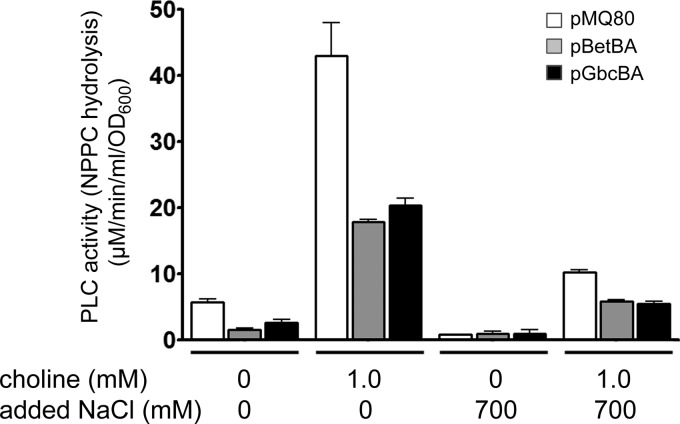
After establishing our ability to experimentally deplete each metabolite pool, we examined the functional consequences of choline and GB pool depletion. We hypothesized that modulation of choline and GB pools would impact processes related to these metabolites, including GB-induced production of the hemolytic phospholipase C, PlcH (55), and growth under hyperosmotic conditions (29). We first examined production of PlcH as measured by NPPC hydrolysis (50). Transcriptional induction of plcH by the metabolites of choline, GB and dimethylglycine, is dependent on the AraC family transcription factor GbdR (55, 56); therefore, we predicted that depletion of the GB pool by gbcBA overexpression would reduce PlcH expression due to depletion of the GbdR ligand (Fig. 2). As shown in Fig. 3, overexpression of gbcBA resulted in an approximately 50% reduction in secreted phospholipase C activity.
Fig 3.
PLC activity measured by NPPC hydrolysis activity in culture supernatants. Cells were grown in MOPS pyruvate with or without choline and with or without NaCl for 24 h as noted below the x axis. Enzyme activity was calculated as described in Materials and Methods and was normalized to that of a culture OD600. These data are representative of three independent experiments each containing three biological replicates. Error bars represent standard deviations.
We initially predicted that overexpressing the betBA genes would increase PlcH expression due to the increase in GB content in the cells and, hence, GbdR activation. However, we observed that choline depletion resulted in an approximately 50% reduction in phospholipase C activity, similar to the level of depletion of the GB pool (Fig. 3). During betBA overexpression, roughly half of the depleted choline pool is added to the GB pool (Fig. 2). We will address the potential role of the choline pool in PlcH regulation in the discussion. As previously reported, in the presence of high NaCl concentrations, P. aeruginosa produces less PLC activity overall (46). However, as with low-salt conditions, overexpression of gbcBA or betBA reduced PLC activity approximately 45% under each condition when cells were induced with choline (Fig. 3).
Overexpression of either catabolic system reduced PLC activity under the uninduced condition. We do not know whether this change is due to removal of choline and GB carried into the overnight culture from the proceeding LB plate or whether these enzymes somehow interact with the regulatory system independently of their enzymatic function.
Effects of overexpressing betBA or gbcBA on growth under high-salt conditions.
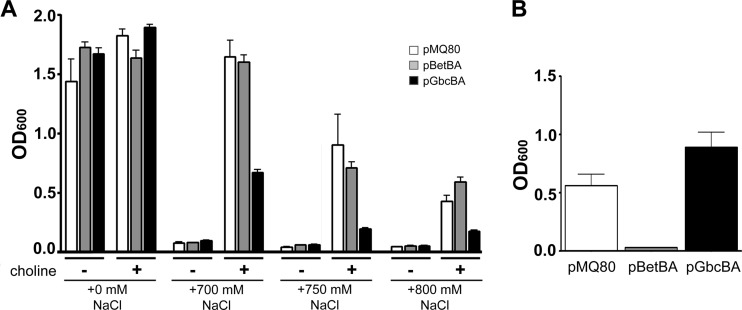
GB has been studied extensively for its ability to act as a potent osmoprotectant. Therefore, we hypothesized that depletion of the GB pool by gbcBA overexpression would result in poor growth under hyperosmotic conditions. Conversely, we predicted that intracellular GB accumulation driven by betBA overexpression would result in improved growth in hyperosmotic media. We first tested the ability of P. aeruginosa to grow under hyperosmotic conditions with glucose as the primary carbon source, with or without a low concentration of choline (250 μM). The low concentration of choline in the medium allowed us to uncouple P. aeruginosa's ability to process choline and utilize it as an osmoprotectant from its ability to gain a net benefit from choline as a carbon source, as this concentration of choline does not support substantial growth (data not shown). We assessed growth by measuring the OD600 at 24 h when cells were grown on glucose as the primary carbon source and at 40 h when cells were grown on choline as the sole carbon source. With choline as the sole carbon source in high salt, cells do not exit lag phase until ∼18 h postinoculation.
In the absence of choline, P. aeruginosa grew well on glucose in MOPS media with no added NaCl, but when NaCl was added at 700 mM or higher, there was no growth regardless of which plasmid was carried (Fig. 4). The addition of l-arabinose did not alter the osmostress response or growth under these conditions. In the presence of choline, growth under low-salt conditions was slightly increased as reported previously (12), and addition of choline allowed growth with high salt (Fig. 4A), as has been well documented for most bacteria (26). When the betBA genes were overexpressed under hyperosmotic conditions with glucose as the primary carbon source, growth was equal to, or slightly better than, that with the empty vector control at 24 h, as we predicted (Fig. 4A). Conversely, overexpressing the gbcBA genes in hyperosmotic media yielded much less growth than with the empty vector control, as predicted based on the importance of GB for osmoprotection (Fig. 4A).
Fig 4.
Effect of catabolic enzyme overexpression on P. aeruginosa growth in increasing salt concentrations. (A) P. aeruginosa carrying the specified vector grown on MOPS glucose with gentamicin and 0.05% l-arabinose for 24 h. Growth conditions were with or without 250 μM choline and with or without added NaCl, as noted below the x axis. Cells exposed to these high-salt conditions were unable to grow without added choline. (B) P. aeruginosa growth in 750 mM NaCl when cells were grown with choline as the sole carbon source for 40 h. Both panels are representative of the results of at least three experiments each containing three biological replicates. Error bars represent standard deviations.
Glucose is partially catabolite repressive to GB catabolism in P. aeruginosa (12), so we chose to examine the effect of catabolic-enzyme overexpression during high-salt growth with choline as the sole carbon source. Overexpression of either set of genes did not change growth on choline as a sole carbon source under low-salt conditions (data not shown). However, the results in Fig. 4B show that overexpression of either betBA or gbcBA significantly altered growth on choline under hyperosmotic conditions. After 40 h of growth, overexpression of gbcBA yielded much better growth than with the empty vector control, while overexpression of betBA resulted in no growth. These effects were unpredicted and will be addressed in the discussion.
Choline and GB pools in other aerobically choline-catabolizing Proteobacteria.
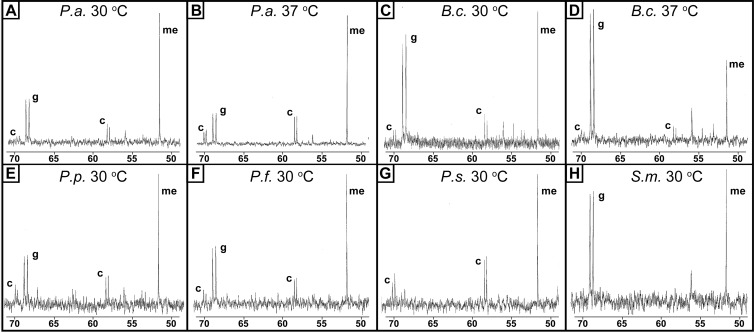
Many species of bacteria can aerobically catabolize choline and use it as a sole carbon, nitrogen, and energy source (22). Thus, we hypothesized that other soil- and water-dwelling Proteobacteria, particularly other pseudomonads, can maintain cytoplasmic pools of choline and GB during growth. We grew P. aeruginosa, P. fluorescens, P. putida, P. syringae, Burkholderia cepacia, and Sinorhizobium meliloti in 15 mM choline and 5 mM [1,2-13C]choline for 9 h and analyzed the labeled metabolites by 13C-NMR. All of the pseudomonads that we examined maintained both choline and GB pools during growth on choline (Fig. 5). At 30°C, P. aeruginosa, P. fluorescens, and P. putida maintained a larger pool of GB than choline, a pattern that was reversed in P. syringae. B. cepacia, a betaproteobacterium, maintained a substantial pool of GB and a small choline pool. As has been previously reported (52), the alphaproteobacterium and plant symbiont S. meliloti maintained a large GB pool and no detectable choline pool. To standardize conditions for these comparisons, we grew all the species at 30°C. When we compared the P. aeruginosa results to those of our 37°C experiments, we noted that the choline pool was ∼30% smaller at 30°C than at 37°C, with a GB-to-choline ratio of ∼3 at 30°C, compared to ∼1.5 at 37°C (Fig. 5A and B). The other bacterium in our comparison capable of growth at both temperatures was B. cepacia. In contrast to P. aeruginosa, B. cepacia did not substantially alter its choline pool, but the GB pool was smaller at the lower temperature (Fig. 5C and D). These changes suggest some form of temperature-dependent regulation of the choline and GB pools in both P. aeruginosa and B. cepacia.
Fig 5.
Accumulation of choline and GB in a set of Proteobacteria capable of aerobic choline catabolism as measured by 13C-NMR when the organisms were grown on choline as a sole carbon source. P. aeruginosa (P.a.) and Burkholderia cepacia (B.c.) were grown at both 30°C and 37°C. P. putida (P.p.), P. fluorescens (P.f.), P. syringae (P.s.), and Sinorhizobium meliloti (S.m.) were grown at 30°C. Panel B is identical to Fig. 1A and is our representative image for this condition in P. aeruginosa. Data are representative of at least three independent experiments. Abbreviations: c, choline; g, glycine betaine; me, [13C]methanol. Results in each panel are representative of at least three experiments.
DISCUSSION
We are interested in eukaryote-derived molecules that bacteria sense to alter their physiology and regulatory processes to impact cross-kingdom associations. Bacterial detection of eukaryotes is a key component of a variety of interactions, including host-pathogen, commensal-host, and symbiont-host relationships. One molecule that appears to participate in large number of such associations is choline (1, 5, 11, 13, 14, 32, 36, 40). Choline is a small molecule produced abundantly by eukaryotes, and some prokaryotes, both in its free form and as a moiety on phosphatidylcholine, sphingomyelin, and choline-O-sulfate. We and others have hypothesized that choline is a signal that denotes the presence of a eukaryote for P. aeruginosa (31, 50, 56). Based on this model, choline-specific phospholipase C functions as a secreted mediator to detect eukaryotes due to release of phosphorylcholine from eukaryotic membranes. Likewise, catabolic genes that deplete this signal function to turn off the detection system or modulate its response. In the context of this model, we are interested in how the catabolism of choline and its metabolites alters bacterial physiology and virulence.
In the Enterobacteriaceae, choline import and subsequent GB accumulation occur only under osmostress conditions (26). Thus, GB appears to serve a solely osmoprotective role in these bacteria. In contrast, many soil- and water-dwelling bacteria can use choline and GB as a sole source of carbon, nitrogen, and energy (22). For these organisms, transport and regulation of GB fate do not depend solely on osmostress (9, 34). For example, it has been demonstrated that the uptake of choline occurs regardless of salinity in S. meliloti and that pools of GB are present under both high- and low-salt conditions (52). In S. meliloti, glycine betaine methyltransferase (GBMT) was shown to be osmotically regulated, thereby controlling flux through the GB catabolic pathway and allowing greater GB accumulation under high-salt conditions (52). This observation established the model that regulation of the catabolic enzymes in this pathway governs the GB pool size. Here we have shown that unlike S. meliloti, P. aeruginosa and other pseudomonads maintain pools of both choline and GB during growth, which we could deplete by overexpression of the cognate catabolic enzymes. Thus, it appears that, like S. meliloti, one way that P. aeruginosa can control osmoprotectant flux is via regulation of catabolism. However, one difference between the catabolic pathways in these two organisms is that P. aeruginosa uses an N-demethylating oxygenase to demethylate GB instead of the GBMT family of enzymes (56). We do not currently know the level at which GB demethylation is regulated in P. aeruginosa, but our previous data suggest that regulation is likely posttranscriptional (56).
The presence of a stable choline pool has been reported in only a few cases (4, 20); however, our report of the P. aeruginosa choline pool represents a new observation for bacteria capable of choline catabolism as a carbon source. Choline is not a useful osmoprotectant, with one known exception, in bacteria (20). Therefore, the regulated accumulation of choline, even during osmostress (Fig. 1), was surprising given the presence of a functional choline catabolic locus. Importantly, while depletion of the GB pool led to the predicted decline in PlcH production and growth in high salt, depletion of the choline pool led to two unexpected findings. First, we predicted that choline pool depletion would lead to improved growth during osmostress. This hypothesis was supported by data from growth in high salt with glucose as the primary carbon source (Fig. 4A). However, when choline was the sole carbon source, choline pool depletion resulted in little or no growth, while GB pool depletion led to improved growth (Fig. 4B). Our second prediction was that the increasing flux to GB via choline pool depletion would increase the production of the virulence factor PlcH through enhanced GbdR activation. However, here we show that depletion of the choline pool reduces the production of PlcH (Fig. 3), suggesting that the choline pool has an important role in virulence regulation. From our high-salt and phospholipase induction results, we hypothesize that the choline pool may work to regulate transport and perhaps other genes via the BetI transcriptional repressor (25, 34, 45). Thus, competency for BetI regulation may be important for controlling the size and fate of the choline pool, thereby regulating PlcH production and the proper expression of catabolic and transport proteins.
One important component of osmoprotectant pool regulation is an efflux mechanism that can sense GB (or stress-related changes) and control the maximum cytoplasmic accumulation of GB, which has been best described in Salmonella enterica (16, 21). The stable maintenance of pool levels—even during blockage in GB catabolism (15)—strongly suggests that P. aeruginosa also possesses a regulated efflux mechanism or, alternatively, a tightly regulated import system. Preferential efflux of GB from supplied choline has been demonstrated in a mucoid P. aeruginosa strain isolated from cystic fibrosis sputum (2), but the molecular identity of the GB efflux system in P. aeruginosa is currently unknown.
Our data, combined with our previous studies, lead to a model where P. aeruginosa carefully regulates flux through two sequential catabolic steps to modulate cytoplasmic pools of choline and GB. To understand the role of these two pools in P. aeruginosa biology, it is important to note that each of these steps is regulated by its respective substrate via the cognate transcriptional regulators BetI and GbdR. We have previously pointed out that GB can function as an osmoprotectant, a nutrient source, and an inducer of gene transcription via GbdR. We know less about the direct roles of choline in P. aeruginosa biology and virulence. We know that choline positively induces choline import via release of BetI repression (25, 45) at the betT1 and betT3 promoters (34), that choline can be covalently attached to elongation factor Tu (1), and that it functions as a substrate for the synthesis of bacterial phosphatidylcholine (59). Maintenance of a choline pool may regulate one or more of these activities. Importantly, we do not know the full range of BetI targets in P. aeruginosa, so choline may contribute to as-yet-unknown biology. Recently, Nau-Wagner and colleagues reported the regulation of choline to GB flux by the Bacillus subtilis GbsR transcription factor (41). They demonstrated that GbsR senses choline directly and proposed that GbsR indirectly senses GB to tune the levels of choline oxidase production (41). It is probable that the functionally analogous transcription factor in P. aeruginosa, BetI, works in a similar manner as part of the mechanism to regulate the choline and GB pools.
We also noted the temperature-dependent regulation of choline and GB pools in P. aeruginosa. Such an association of choline and GB with temperature is not surprising given the ability of GB to function as both a thermo- and a cryoprotectant (6, 10). We do not currently know the molecular basis of this regulation, but the differences between the P. aeruginosa regulatory systems and the B. cepacia regulatory systems may provide insights into potential mechanisms for temperature-dependent regulation of the choline and GB pools.
In conclusion, P. aeruginosa and other Gram-negative bacteria maintain stable GB pools. Unlike S. meliloti and B. cepacia, however, the pseudomonads establish and maintain abundant choline pools during growth on choline. P. aeruginosa changes the abundance and relative proportions of its choline and GB pools depending on the main carbon source, medium salinity, and temperature, and both metabolite pools impact P. aeruginosa physiology via osmoprotection and PlcH production. We propose that these two pools may regulate aspects of P. aeruginosa biology and virulence beyond those described here, potentially involving other BetI targets or additional transcriptional regulators.
ACKNOWLEDGMENTS
We thank P. Bruce Deker for advice and discussion regarding NMR methodology and Mary Tierney for providing Pseudomonas syringae DC3000. We also thank Bonnie Cross, Adam Nock, Jamie Meadows, and Annette LaBauve for careful reading of the manuscript and discussions.
This project was supported by grants from the National Center for Research Resources (5P20RR021905 to -07) and the National Institute of General Medical Sciences of the National Institutes of Health (8 P20 GM103496 to -07) supporting the Vermont Center for Immunology and Infectious Diseases and by pilot funding from the National Center for Research Resources and the National Institute of General Medical Sciences of the National Institutes of Health (grant P30 GM103532) supporting the Vermont Lung Center.
Footnotes
Published ahead of print 29 June 2012
REFERENCES
- 1. Barbier M, et al. 2008. Novel phosphorylcholine-containing protein of Pseudomonas aeruginosa chronic infection isolates interacts with airway epithelial cells. J. Infect. Dis. 197:465–473 [DOI] [PubMed] [Google Scholar]
- 2. Behrends V, Ryall B, Wang X, Bundy JG, Williams HD. 2010. Metabolic profiling of Pseudomonas aeruginosa demonstrates that the anti-sigma factor MucA modulates osmotic stress tolerance. Mol. Biosyst. 6:562–569 [DOI] [PubMed] [Google Scholar]
- 3. Boch J, Kempf B, Bremer E. 1994. Osmoregulation in Bacillus subtilis: synthesis of the osmoprotectant glycine betaine from exogenously provided choline. J. Bacteriol. 176:5364–5371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boch J, Kempf B, Schmid R, Bremer E. 1996. Synthesis of the osmoprotectant glycine betaine in Bacillus subtilis: characterization of the gbsAB genes. J. Bacteriol. 178:5121–5129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boncompagni E, Osteras M, Poggi MC, le Rudulier D. 1999. Occurrence of choline and glycine betaine uptake and metabolism in the family Rhizobiaceae and their roles in osmoprotection. Appl. Environ. Microbiol. 65:2072–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Caldas T, Demont-Caulet N, Ghazi A, Richarme G. 1999. Thermoprotection by glycine betaine and choline. Microbiology 145(Part 9):2543–2548 [DOI] [PubMed] [Google Scholar]
- 7. Canovas D, Vargas C, Csonka LN, Ventosa A, Nieto JJ. 1998. Synthesis of glycine betaine from exogenous choline in the moderately halophilic bacterium Halomonas elongata. Appl. Environ. Microbiol. 64:4095–4097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Canovas M, Bernal V, Torroglosa T, Ramirez JL, Iborra JL. 2003. Link between primary and secondary metabolism in the biotransformation of trimethylammonium compounds by Escherichia coli. Biotechnol. Bioeng. 84:686–699 [DOI] [PubMed] [Google Scholar]
- 9. Chen C, Malek AA, Wargo MJ, Hogan DA, Beattie GA. 2010. The ATP-binding cassette transporter Cbc (choline/betaine/carnitine) recruits multiple substrate-binding proteins with strong specificity for distinct quaternary ammonium compounds. Mol. Microbiol. 75:29–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cleland D, Krader P, McCree C, Tang J, Emerson D. 2004. Glycine betaine as a cryoprotectant for prokaryotes. J. Microbiol. Methods 58:31–38 [DOI] [PubMed] [Google Scholar]
- 11. de Rudder KE, Sohlenkamp C, Geiger O. 1999. Plant-exuded choline is used for rhizobial membrane lipid biosynthesis by phosphatidylcholine synthase. J. Biol. Chem. 274:20011–20016 [DOI] [PubMed] [Google Scholar]
- 12. Diab F, et al. 2006. Succinate-mediated catabolite repression control on the production of glycine betaine catabolic enzymes in Pseudomonas aeruginosa PAO1 under low and elevated salinities. Microbiology 152:1395–1406 [DOI] [PubMed] [Google Scholar]
- 13. Domenech CE, Garrido MN, Lisa TA. 1991. Pseudomonas aeruginosa cholinesterase and phosphorylcholine phosphatase: two enzymes contributing to corneal infection. FEMS Microbiol. Lett. 66:131–135 [DOI] [PubMed] [Google Scholar]
- 14. Fan X, Pericone CD, Lysenko E, Goldfine H, Weiser JN. 2003. Multiple mechanisms for choline transport and utilization in Haemophilus influenzae. Mol. Microbiol. 50:537–548 [DOI] [PubMed] [Google Scholar]
- 15. Fitzsimmons LF, et al. 2011. Small-molecule inhibition of choline catabolism in Pseudomonas aeruginosa and other aerobic choline-catabolizing bacteria. Appl. Environ. Microbiol. 77:4383–4389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gutierrez JA, Csonka LN. 1995. Isolation and characterization of adenylate kinase (adk) mutations in Salmonella typhimurium which block the ability of glycine betaine to function as an osmoprotectant. J. Bacteriol. 177:390–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hoffmann T, Bremer E. 2011. Protection of Bacillus subtilis against cold stress via compatible-solute acquisition. J. Bacteriol. 193:1552–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ikuta S, Matuura K, Imamura S, Misaki H, Horiuti Y. 1977. Oxidative pathway of choline to betaine in the soluble fraction prepared from Arthrobacter globiformis. J. Biochem. 82:157–163 [DOI] [PubMed] [Google Scholar]
- 19. Jacobs MA, et al. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 100:14339–14344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kets EPW, Groot MN, Galinski EA, DeBont JAM. 1997. Choline and acetylcholine: novel cationic osmolytes in Lactobacillus plantarum. Appl. Microbiol. Biotechnol. 48:94–98 [Google Scholar]
- 21. Koo SP, Higgins CF, Booth IR. 1991. Regulation of compatible solute accumulation in Salmonella typhimurium: evidence for a glycine betaine efflux system. J. Gen. Microbiol. 137:2617–2625 [DOI] [PubMed] [Google Scholar]
- 22. Kortstee GJ. 1970. The aerobic decomposition of choline by microorganisms. I. The ability of aerobic organisms, particularly coryneform bacteria, to utilize choline as the sole carbon and nitrogen source. Arch. Mikrobiol. 71:235–244 [PubMed] [Google Scholar]
- 23. Kunin CM, Hua TH, Van Arsdale White L, Villarejo M. 1992. Growth of Escherichia coli in human urine: role of salt tolerance and accumulation of glycine betaine. J. Infect. Dis. 166:1311–1315 [DOI] [PubMed] [Google Scholar]
- 24. Kurioka S, Matsuda M. 1976. Phospholipase C assay using p-nitrophenylphosphoryl-choline together with sorbitol and its application to studying the metal and detergent requirement of the enzyme. Anal. Biochem. 75:281–289 [DOI] [PubMed] [Google Scholar]
- 25. Lamark T, Rokenes TP, McDougall J, Strøm AR. 1996. The complex bet promoters of Escherichia coli: regulation by oxygen (ArcA), choline (BetI), and osmotic stress. J. Bacteriol. 178:1655–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Landfald B, Strøm AR. 1986. Choline-glycine betaine pathway confers a high level of osmotic tolerance in Escherichia coli. J. Bacteriol. 165:849–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lanotte P, Mereghetti L, Lejeune B, Massicot P, Quentin R. 2003. Pseudomonas aeruginosa and cystic fibrosis: correlation between exoenzyme production and patient's clinical state. Pediatr. Pulmonol. 36:405–412 [DOI] [PubMed] [Google Scholar]
- 28. Le Rudulier D, Strøm AR, Dandekar AM, Smith LT, Valentine RC. 1984. Molecular biology of osmoregulation. Science 224:1064–1068 [DOI] [PubMed] [Google Scholar]
- 29. Lisa TA, Garrido MN, Domenech CE. 1983. Induction of acid phosphatase and cholinesterase activities in Ps. aeruginosa and their in-vitro control by choline, acetylcholine and betaine. Mol. Cell. Biochem. 50:149–155 [DOI] [PubMed] [Google Scholar]
- 30. Lisa TA, Garrido MN, Domenech CE. 1984. Pseudomonas aeruginosa acid phosphatase and cholinesterase induced by choline and its metabolic derivatives may contain a similar anionic peripheral site. Mol. Cell. Biochem. 63:113–118 [DOI] [PubMed] [Google Scholar]
- 31. Lisa TA, Lucchesi GI, Domenech CE. 1994. Pathogenicity of Pseudomonas aeruginosa and its relationship to the choline metabolism through the action of cholinesterase, acip phosphatase, and phospholipase C. Curr. Microbiol. 29:193–199 [DOI] [PubMed] [Google Scholar]
- 32. Lopez-Lara IM, Geiger O. 2001. Novel pathway for phosphatidylcholine biosynthesis in bacteria associated with eukaryotes. J. Biotechnol. 91:211–221 [DOI] [PubMed] [Google Scholar]
- 33. Lucchesi GI, Pallotti C, Lisa AT, Domenech CE. 1998. Constitutive choline transport in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 162:123–126 [DOI] [PubMed] [Google Scholar]
- 34. Malek AA, Chen C, Wargo MJ, Beattie GA, Hogan DA. 2011. Roles of three transporters, CbcXWV, BetT1, and BetT3, in Pseudomonas aeruginosa choline uptake for catabolism. J. Bacteriol. 193:3033–3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Malek AA, Wargo MJ, Hogan DA. 2012. Absence of membrane phosphatidylcholine does not affect virulence and stress tolerance phenotypes in the opportunistic pathogen Pseudomonas aeruginosa. PLoS One 7(2):e30829 doi:10.1371/journal.pone.0030829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mandon K, et al. 2003. The Sinorhizobium meliloti glycine betaine biosynthetic genes (betlCBA) are induced by choline and highly expressed in bacteroids. Mol. Plant Microbe Interact. 16:709–719 [DOI] [PubMed] [Google Scholar]
- 37. Massimelli MJ, et al. 2005. Identification, cloning, and expression of Pseudomonas aeruginosa phosphorylcholine phosphatase gene. Curr. Microbiol. 50:251–256 [DOI] [PubMed] [Google Scholar]
- 38. Meskys R, Harris RJ, Casaite V, Basran J, Scrutton NS. 2001. Organization of the genes involved in dimethylglycine and sarcosine degradation in Arthrobacter spp.: implications for glycine betaine catabolism. Eur. J. Biochem. 268:3390–3398 [DOI] [PubMed] [Google Scholar]
- 39. Meyers DJ, et al. 1992. In vivo and in vitro toxicity of phospholipase C from Pseudomonas aeruginosa. Toxicon 30:161–169 [DOI] [PubMed] [Google Scholar]
- 40. Moscoso M, Garcia E, Lopez R. 2006. Biofilm formation by Streptococcus pneumoniae: role of choline, extracellular DNA, and capsular polysaccharide in microbial accretion. J. Bacteriol. 188:7785–7795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nau-Wagner G, et al. 2012. Genetic control of osmoadaptive glycine betaine synthesis in Bacillus subtilis through the choline-sensing and glycine betaine-responsive GbsR repressor. J. Bacteriol. 194:2703–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Perroud B, Le Rudulier D. 1985. Glycine betaine transport in Escherichia coli: osmotic modulation. J. Bacteriol. 161:393–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Price CT, Bukka A, Cynamon M, Graham JE. 2008. Glycine betaine uptake by the ProXVWZ ABC transporter contributes to the ability of Mycobacterium tuberculosis to initiate growth in human macrophages. J. Bacteriol. 190:3955–3961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rahme LG, et al. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899–1902 [DOI] [PubMed] [Google Scholar]
- 45. Rokenes TP, Lamark T, Strøm AR. 1996. DNA-binding properties of the BetI repressor protein of Escherichia coli: the inducer choline stimulates BetI-DNA complex formation. J. Bacteriol. 178:1663–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sage AE, Vasil AI, Vasil ML. 1997. Molecular characterization of mutants affected in the osmoprotectant-dependent induction of phospholipase C in Pseudomonas aeruginosa PAO1. Mol. Microbiol. 23:43–56 [DOI] [PubMed] [Google Scholar]
- 47. Salvano MA, Lisa TA, Domenech CE. 1989. Choline transport in Pseudomonas aeruginosa. Mol. Cell. Biochem. 85:81–89 [DOI] [PubMed] [Google Scholar]
- 48. Shanks RM, Caiazza NC, Hinsa SM, Toutain CM, O'Toole GA. 2006. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl. Environ. Microbiol. 72:5027–5036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shikita M, Fahey JW, Golden TR, Holtzclaw WD, Talalay P. 1999. An unusual case of ‘uncompetitive activation’ by ascorbic acid: purification and kinetic properties of a myrosinase from Raphanus sativus seedlings. Biochem. J. 341(Part 3):725–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shortridge VD, Lazdunski A, Vasil ML. 1992. Osmoprotectants and phosphate regulate expression of phospholipase C in Pseudomonas aeruginosa. Mol. Microbiol. 6:863–871 [DOI] [PubMed] [Google Scholar]
- 51. Sleator RD, Francis GA, O'Beirne D, Gahan CGM, Hill C. 2003. Betaine and carnitine uptake systems in Listeria monocytogenes affect growth and survival in foods and during infection. J. Appl. Microbiol. 95:839–846 [DOI] [PubMed] [Google Scholar]
- 52. Smith LT, Pocard JA, Bernard T, Le Rudulier D. 1988. Osmotic control of glycine betaine biosynthesis and degradation in Rhizobium meliloti. J. Bacteriol. 170:3142–3149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vasil ML, Vasil AI, Shortridge VD. 1994. Phosphate and osmoprotectants in the pathogenesis of Pseudomonas aeruginosa, p 126–132 In Torriani-Gorrini E, Yagil E, Silver S. (ed), Phosphate in microorganisms: cellular and molecular biology. ASM Press, Washington, DC [Google Scholar]
- 54. Wargo MJ, et al. 2011. Hemolytic phospholipase C inhibition protects lung function during Pseudomonas aeruginosa infection. Am. J. Respir. Crit. Care Med. 184:345–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wargo MJ, Ho TC, Gross MJ, Whittaker LA, Hogan DA. 2009. GbdR regulates Pseudomonas aeruginosa plcH and pchP transcription in response to choline catabolites. Infect. Immun. 77:1103–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wargo MJ, Szwergold BS, Hogan DA. 2008. Identification of two gene clusters and a transcriptional regulator required for Pseudomonas aeruginosa glycine betaine catabolism. J. Bacteriol. 190:2690–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wieland CW, et al. 2002. Pulmonary inflammation induced by Pseudomonas aeruginosa lipopolysaccharide, phospholipase C, and exotoxin A: role of interferon regulatory factor 1. Infect. Immun. 70:1352–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wiener-Kronish JP, et al. 1993. Alveolar epithelial injury and pleural empyema in acute P. aeruginosa pneumonia in anesthetized rabbits. J. Appl. Physiol. 75:1661–1669 [DOI] [PubMed] [Google Scholar]
- 59. Wilderman PJ, Vasil AI, Martin WE, Murphy RC, Vasil ML. 2002. Pseudomonas aeruginosa synthesizes phosphatidylcholine by use of the phosphatidylcholine synthase pathway. J. Bacteriol. 184:4792–4799 [DOI] [PMC free article] [PubMed] [Google Scholar]