Abstract
Many members of the Omp85 family of proteins form essential β-barrel outer membrane protein (OMP) biogenesis machinery in Gram-negative bacteria, chloroplasts, and mitochondria. In Escherichia coli, BamA, a member of the Omp85 family, folds into an outer membrane-embedded β-barrel domain and a soluble periplasmic polypeptide-transport-associated (POTRA) domain. Although the high-resolution structures of only the BamA POTRA domain of E. coli are available, the crystal structure of FhaC, an Omp85 family member and a component of the two-partner secretion system in Bordetella pertussis, suggests that the BamA β-barrel likely folds into a 16-stranded β-barrel. The FhaC β-barrel is occluded by an N-terminal α-helix and a large β-barrel loop, L6, which carries residues that are highly conserved among the Omp85 family members. Deletion of L6 in FhaC did not affect its biogenesis but abolished its secretion function. In this study, we tested the hypothesis that the conserved residues of the putative L6 loop, which presumably folds back into the lumen of the BamA β-barrel like the FhaC counterpart, play an important role in OMP and/or BamA biogenesis. The conserved 641RGF643 residues of L6 were either deleted or replaced with alanine in various permutations. Phenotypic and biochemical characterization of various BamA L6 mutants revealed that the conserved RGF residues are critical for OMP biogenesis. Moreover, three BamA L6 alterations, ΔRGF, AAA, and AGA, produced a conditional lethal phenotype, concomitant with severely reduced BamA levels and folding defects. Thus, the conserved 641RGF643 residues of the BamA L6 loop are important for BamA folding and biogenesis.
INTRODUCTION
Omp85/BamA and Sam50/Tob55 are the central components of the hetero-oligomeric β-barrel outer membrane protein (OMP) assembly machinery in Gram-negative bacteria and mitochondria, respectively (12, 22, 32). Chloroplasts contain two essential Omp85/BamA homologs, Toc75-III and OEP80 (Toc75-V) (15), of which the latter has been suggested to be involved in β-barrel OMP assembly (14). Interestingly, it appears that these highly conserved core components are the only ones common in the β-barrel assembly machinery (BAM) of Gram-negative bacteria and eukaryotic organelles (for recent reviews, see references 1, 2, 11, 13, and 14).
The N terminus of the Omp85/BamA superfamily of proteins forms a soluble polypeptide-transported-associated (POTRA) domain (25), while the C terminus is embedded in the outer membrane and is predicted to fold into a 16-stranded β-barrel (19). High-resolution structures of the POTRA domain have been solved (9, 10, 16, 38), showing a β1α1α2β2β3 fold for each POTRA domain (16). The numbers of POTRA domains vary greatly: typically five in Omp85/BamA, three in OEP80, and only one in Sam50/Tob55 (15). The POTRA domain is thought to interact with substrate OMPs via β-augmentation and initiate their folding into β-barrels (16, 17, 23). In Escherichia coli, the POTRA domain is also the site of interactions with other lipoprotein BAM members (37) and the periplasmic chaperone SurA (3, 36). Deletion of POTRA domains 1 and 2 of E. coli BamA is possible; cells expressing a BamA variant with POTRA domain 2 deleted grow normally, while those with a deletion of POTRA domain 1 grow poorly and have reduced OMP levels (3, 16). Cells with a deletion of POTRA domains 3 and 4 do not survive the depletion of the wild-type copy of BamA, indicating that these two POTRA domains are essential (16). Deletion of POTRA domain 5 causes toxicity even in the presence of wild-type BamA (16). Interestingly, a deletion of the only POTRA domain from Sam50 of Saccharomyces cerevisiae affects neither cell growth nor β-barrel OMP biogenesis (18). Thus, while POTRA domains are highly conserved, their roles may vary in bacteria, mitochondria, and chloroplasts.
The roles of the β-barrel domain of Omp85/BamA, Sam50/Tob55, and OEP80/Toc75-V are even less well understood than those of the POTRA domain. In 2007, the crystal structure of FhaC was solved (6). It contains a 16-stranded β-barrel and two POTRA domains. FhaC is a member of the Omp85 superfamily and belongs to the two-partner secretion (Tps) system that mediates the secretion of filamentous hemagglutinin (FHA) in Bordetella pertussis (30). The β-barrel of FhaC is occluded by an N-terminal α-helix (H1) and a long loop (L6), which connects β-strands 11 and 12 of the C-terminal β-barrel domain (6). Deletion of the α-helix H1 did not affect FHA secretion, but deletion of the entire L6 loop abolished FHA secretion (6). Notably, the L6 loop deletion did not affect the FhaC level or its localization to the outer membrane, indicating that L6 bears importance in FhaC function only (6). Subsequent mutational analysis revealed the functional importance of the highly conserved VRGY(F) motif (20) located at the tip of the L6 loop (7).
Recently, our laboratory described the isolation and characterization of BamA mutants in which single amino acid substitutions in the β-barrel domain overcame the growth and OMP biogenesis defects in a genetic background simultaneously lacking BamB and BamE (29), the two nonessential lipoproteins of the E. coli BAM complex (5, 27, 33, 37). Four of the six substitutions mapped in the presumed L6 loop of the E. coli BamA β-barrel but did not alter the conserved VRGY(F) motif (29). Since the L6 loop is the site of the functionally important motif of FhaC, and changes in this region modulate BamA's function to improve OMP biogenesis without BamB and BamE, we set out to test the importance of the conserved VRGY(F) motif in BamA's function.
We took a two-step mutagenesis approach. In the first step, three of the four conserved L6 motif residues, 641RGF643, were either deleted or simultaneously replaced with alanine residues to see if these changes interfere with BamA's function. (Residue numbers are relative to the mature BamA protein sequence.) The data showed that the AAA-substituted BamA protein was unable to support bacterial growth on rich medium and was severely defective in promoting OMP assembly. Subsequently, we systematically restored the AAA residues to the wild-type residues to assess the relative importance of the three residues in OMP assembly. The data showed that the RGF residues of L6 are critical for OMP assembly, including that of BamA itself, and that R is most important of the three residues, followed by F and G.
MATERIALS AND METHODS
Bacterial strains and media.
Bacterial strains used in this study are listed in Table 1. Luria broth (LB), Luria broth agar (LBA), and M63 salt-based minimal media were prepared as previously described (26). Minimal media were supplemented with glycerol and Casamino Acids (0.4% and 0.1%, respectively). When appropriate, growth media were also supplemented with l-arabinose (0.2%, wt/vol), chloramphenicol (12.5 μg/ml), and kanamycin (25 μg/ml).
Table 1.
Bacterial strains used in this study
| Strain | Characteristics | Reference or source |
|---|---|---|
| MC4100 | F− araD139 Δ(argF-lac)U139 rspL150 relA1 flbB5301 ptsF25 deoC1 thi-1 rbsR | 4 |
| RAM1292 | MC4100 Δara714 | 35 |
| RAM1431 | MC4100 Δara714 ΔbamA::scar pBAD33-bamA (BamA-WT) | 3 |
| RAM1967 | RAM1431 recA::scar | This study |
| RAM1969 | RAM1967 pZS21-bamA6His (BamA-WT) | This study |
| RAM1971 | RAM1967 pZS21-bamA6His (BamAEGF) | This study |
| RAM1973 | RAM1967 pZS21-bamA6His (BamAΔRGF) | This study |
| RAM1974 | RAM1967 pZS21-bamA6His (BamAAAA) | This study |
| RAM1975 | RAM1967 pZS21-bamA6His (BamAΔR44) | This study |
| RAM1982 | RAM1967 pZS21-bamA6His (BamAAGF) | This study |
| RAM1984 | RAM1969 cured of pBAD33-bamA (BamA-WT) | This study |
| RAM1985 | RAM1971 cured of pBAD33-bamA (BamA-WT) | This study |
| RAM1987 | RAM1982 cured of pBAD33-bamA (BamA-WT) | This study |
| RAM1988 | RAM1973 cured of pBAD33-bamA (BamA-WT) | This study |
| RAM1989 | RAM1974 cured of pBAD33-bamA (BamA-WT) | This study |
| RAM1991 | RAM1975 cured of pBAD33-bamA (BamA-WT) | This study |
| RAM2052 | RAM1967 pZS21-bamA6His (BamARAA) | This study |
| RAM2053 | RAM1967 pZS21-bamA6His (BamAAGA) | This study |
| RAM2054 | RAM1967 pZS21-bamA6His (BamAAAF) | This study |
| RAM2055 | RAM1967 pZS21-bamA6His (BamARGA) | This study |
| RAM2057 | RAM2052 cured of pBAD33-bamA (BamA-WT) | This study |
| RAM2058 | RAM2053 cured of pBAD33-bamA (BamA-WT) | This study |
| RAM2059 | RAM2054 cured of pBAD33-bamA (BamA-WT) | This study |
| RAM2060 | RAM2055 cured of pBAD33-bamA (BamA-WT) | This study |
Antibiotic sensitivity assays.
Sensitivities to antibiotics were analyzed by placing either presoaked rifampin disks (5 μg/ml; Becton, Dickinson) or blank paper disks soaked with vancomycin (75 μg/ml) on M63 minimal plates overlaid with 4 ml of soft agar containing 100 μl of overnight-grown bacterial cultures. Plates were incubated for 24 h at 30°C, after which the diameters of inhibition zones were measured. Two independent cultures were used, and each culture was tested with two duplicate antibiotic disks. The zones were measured with a professional decimal circle template (Staedtler-Mars) with cutout measurements in thousandths of a millimeter.
DNA methods.
Mutant bamA alleles used in this study were created using the QuikChange Lightning site-directed mutagenesis kit (Agilent Technologies) per the manufacturer's instructions. Mutations were created on the pZS21-bamA6His plasmid template and were confirmed by DNA sequencing of the entire bamA gene. Primers used for mutagenesis and sequencing are available upon request.
Plasmid curing.
Strains containing pBAD33-bamA (BamA-WT; Cmr) and pZS21-bamA6His (Kmr) were cured of the former by streaking colonies on M63 minimal medium containing kanamycin only. After 24 h of incubation at 30°C, Kmr colonies were tested for sensitivity to chloramphenicol. This procedure was repeated until a Kmr Cms colony lacking the pBAD33-bamA (BamA-WT) plasmid was obtained. In most cases, strains were cured of pBAD33-bamA (BamA-WT) during the first round of incubation.
Protein methods.
OMPs were analyzed from purified envelopes by Coomassie blue staining after sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE). For envelope isolation, bacterial cell lysis was achieved by the French press method (3). Whole-cell envelopes were isolated by centrifuging cell-free lysates for 1 h at 105,000 × g. Envelope pellets were resuspended in SDS sample buffer. To better resolve OmpC and OmpF bands, 4 M urea was added in the SDS-polyacrylamide running gel. The heat modifiability test to assess BamA's folding status was carried out using purified whole-cell envelopes. Envelope samples containing 5 μg of proteins were solubilized in SDS sample buffer. Prior to SDS-PAGE analysis, SDS buffer-solubilized envelope samples were either heated in a boiling water bath for 5 min or left unheated at room temperature. Protein concentrations were determined using the Coomassie (Bradford) protein assay kit from Thermo Scientific.
For Western blot analysis, proteins were transferred onto polyvinylidene difluoride (PVDF) membranes (Immobilon-Millipore). The membrane blots were incubated with appropriate primary antibodies for 1.5 h, followed by incubation with goat anti-rabbit alkaline phosphatase-conjugated IgG secondary antibodies for 1 h. Finally, membrane blots were incubated with luminol substrate for 5 min and protein bands were visualized using a chemiluminescence imager (Bio-Rad). Primary rabbit antibodies and dilutions used were α-AcrA at 1:16,000 and α-BamA at 1:5,000.
RESULTS AND DISCUSSION
Mutagenesis of the conserved L6 tip residues of BamA: rationale and strategy.
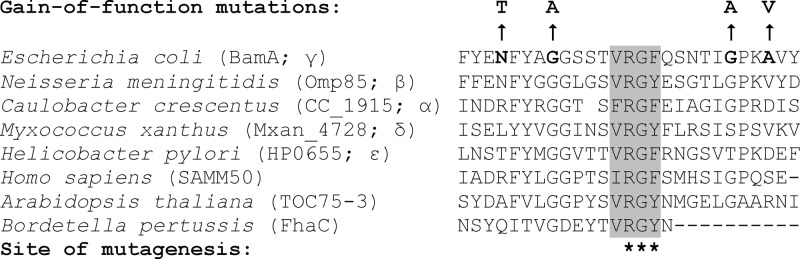
Two observations drew our attention to the L6 loop of BamA. First, the mutagenesis studies on FhaC showed that certain changes in the conserved L6 tip residues—VRGY—or the deletion of the entire L6 loop significantly diminished FhaC's ability to secrete FHA (6). This pointed to a crucial role for the L6 loop in FhaC-mediated secretion of FHA. Second, our own recent work found compensatory alterations in the putative L6 loop of BamA that overcame the conditional lethal phenotype of an E. coli strain simultaneously lacking the BamB and BamE lipoproteins (29). Interestingly, none of the compensatory alterations mapping in the L6 loop altered the highly conserved VRGF motif of BamA, indicating that other regions of the L6 loop may also be involved in BamA's activity (Fig. 1).
Fig 1.
Amino acid sequence alignment of the putative loop 6 region of the BamA β-barrel. The conserved VRGF motif is shaded. Greek letters represent different classes of the phylum Proteobacteria. The gain-of-function alterations in BamA that reverse the growth and OMP assembly defects of a strain simultaneously lacking BamB and BamE are indicated by arrows (29). Residues altered by site-directed mutagenesis in this study are marked by asterisks.
In this study, we focused on the 641RGF643 residues of the conserved VRGF motif of BamA (Fig. 1). As a general strategy, we carried out site-directed mutagenesis of bamA present in a low-copy-number plasmid, pZS21, and then transformed the mutagenized pZS21-bamA plasmid into a ΔbamA ΔrecA strain containing pBAD33-bamA (BamA-WT). Without arabinose, the survival of the host is entirely dependent on the pZS21-bamA plasmid. In some instances, survival without arabinose required that the strains be streaked on minimal glycerol medium and grown at 30°C. We presume that these growth conditions sufficiently slowed bacterial growth and thus allowed for even more defective BamA expressed from pZS21 to support bacterial growth. Subsequently, pBAD33-bamA (BamA-WT) plasmid was cured by streaking the cells on minimal glycerol medium not supplemented with chloramphenicol and arabinose and incubating them at 30°C. The cured ΔbamA ΔrecA strains expressed BamA exclusively from the pZS21 replicon.
Phenotypic characterization of the BamA L6 mutants.
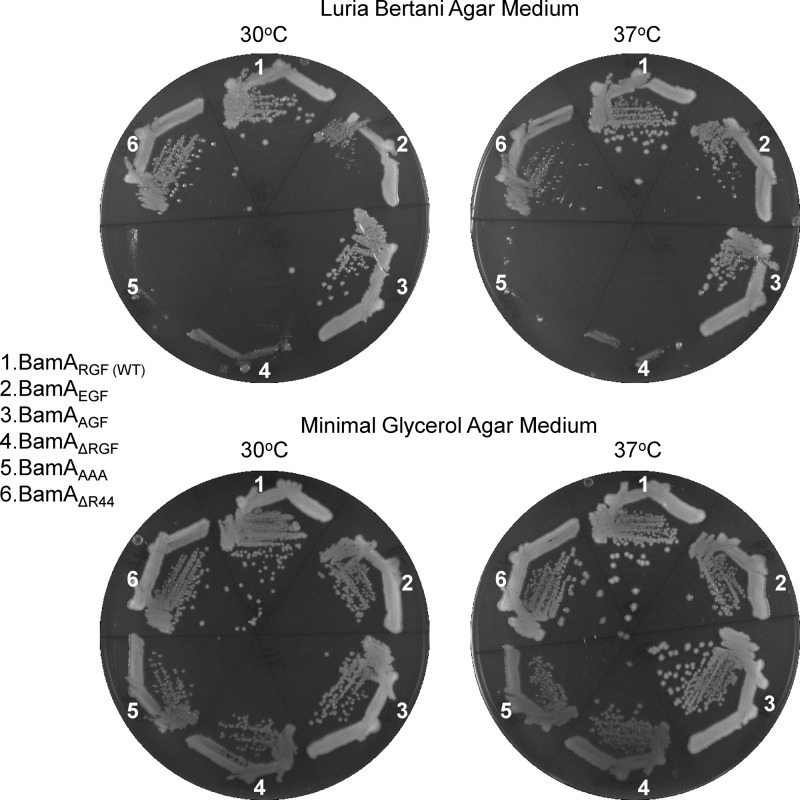
Initially, we constructed four BamA L6 mutants: in two mutants, 641RGF643 was either deleted or replaced with alanine residues (ΔRGF and AAA), and in the other two mutants, R641 was replaced with an E or A (EGF and AGF). Because the curing of pBAD33-bamA (BamA-WT) from strains expressing BamAAAA and BamAΔRGF from pZS21was possible only on minimal glycerol medium, we anticipated that the cured strains would show growth defects on the rich medium. Indeed, Fig. 2 shows that both BamA variants failed to form colonies on LBA at 30°C and 37°C. Although these strains formed single colonies on the minimal glycerol medium, their sizes were smaller than that of the isogenic strain expressing wild-type BamA. The strain expressing BamAAGF showed no apparent growth defect, while that expressing BamAEGF grew more poorly at 30°C than at 37°C. This cold-sensitive growth phenotype of BamAEGF was not apparent on the minimal glycerol medium. A previously characterized POTRA domain 1 mutant, BamAΔR44 (3), confers a temperature-sensitive growth phenotype on rich medium (Fig. 2), in contrast to the cold-sensitive phenotype of BamAEGF.
Fig 2.
Growth phenotypes of strains expressing wild-type BamA (WT; sector 1) or various mutant BamA proteins (sectors 2 to 6) on LBA or glycerol minimal agar plates. Plates were incubated for 24 h (LBA) or 36 h (M63 minimal medium) at 30°C and 37°C. BamA was expressed from the pZS21 plasmid replicon.
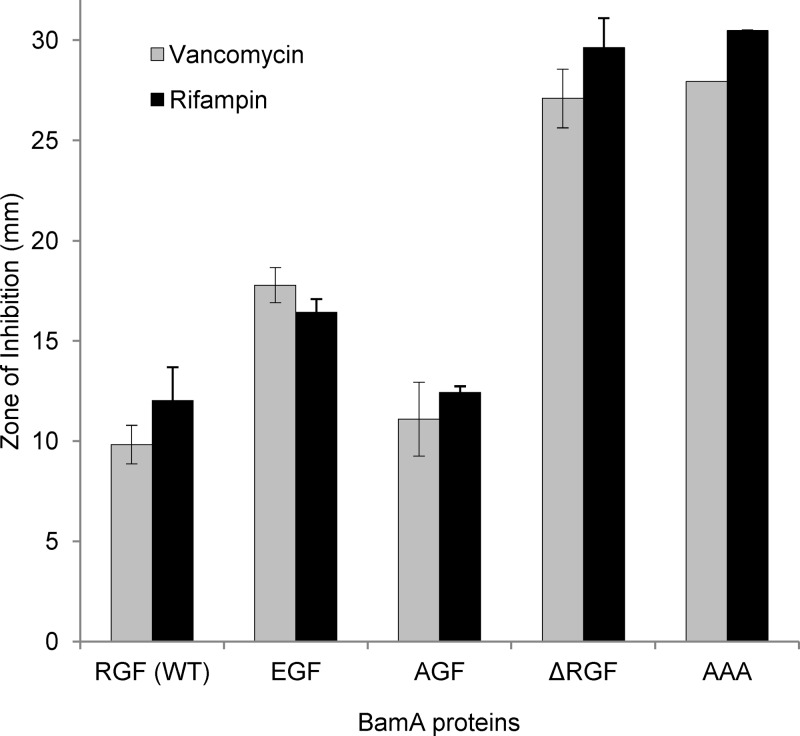
The antibiotic sensitivity test is a convenient means of assessing the status of the outer membrane permeability barrier that normally precludes the entry of noxious compounds, such as vancomycin and rifampin, into the cell (21, 31). If a mutant BamA protein fails to correctly assemble itself or other β-barrel OMPs, including the lipopolysaccharide (LPS) transporter LptD, then cells expressing the mutant BamA protein may have a compromised outer membrane permeability barrier resulting in vancomycin and rifampin sensitivity. Consistent with this view, we have recently reported a BamA POTRA domain mutant with increased sensitivity toward vancomycin and rifampin (36). Because cells expressing BamAAAA or BamAΔRGF grow best on glycerol minimal medium at 30°C (Fig. 2), antibiotic sensitivity tests were carried out under these conditions. Cells expressing BamAAAA or BamAΔRGF were hypersensitive to both vancomycin and rifampin, while those expressing BamAEGF and BamAAGF showed intermediate and wild-type-level sensitivity, respectively, to these antibiotics (Fig. 3). These data corroborated well with the growth data (Fig. 2) and further indicated the severity of defects for some BamA mutants while less so for others.
Fig 3.
Antibiotic sensitivities of strains expressing wild-type (WT) or mutant BamA proteins. Antibiotic sensitivities were measured from overnight cultures grown in glycerol minimal medium at 30°C. Zones of inhibition from two independent cultures were measured after 24 h of incubation at 30°C. Paper disks (6.5 mm in diameter) were either presoaked with rifampin (5 μg/ml) or soaked with 10 μl of vancomycin (75 μg/ml). Error bars show standard deviations.
Biochemical characterization of the BamA L6 mutants.
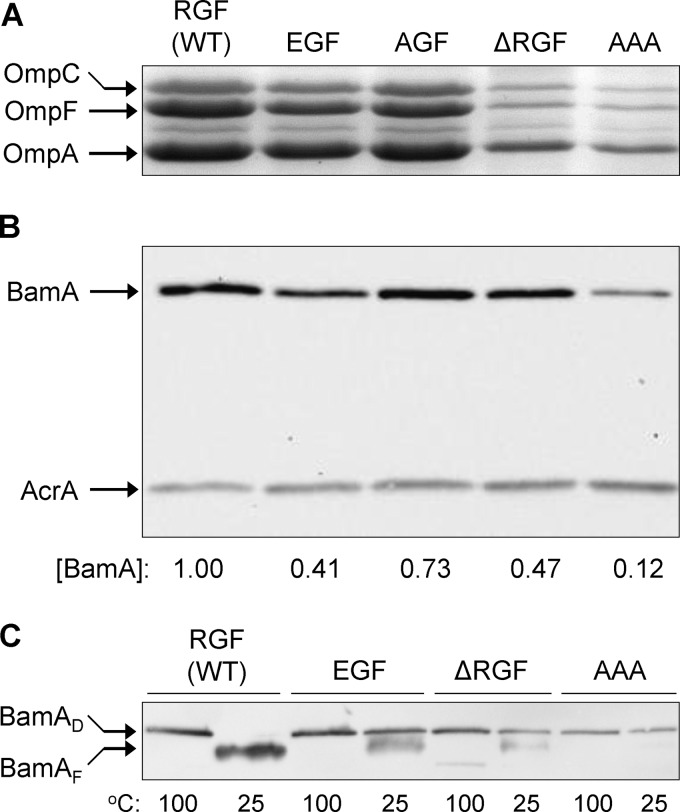
We proceeded to analyze envelopes from strains expressing wild-type and mutant BamA proteins to assess the levels of major OMPs—OmpA, OmpC, and OmpF—which assemble in a BamA-dependent manner (3). Envelopes extracted from cells expressing BamAAAA or BamAΔRGF had dramatically reduced levels of the three major OMPs (Fig. 4). Their levels were also reduced in BamAEGF envelopes, but the mutant expressing BamAAGF had the same levels of the major OMPs as the wild-type had (Fig. 4). Thus, the OMP data corroborate well with the phenotypic data.
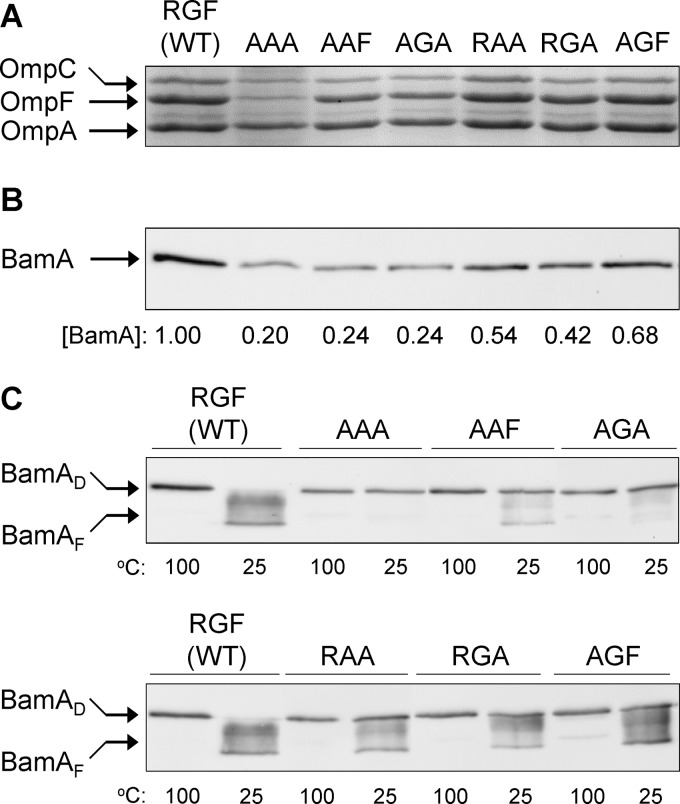
Fig 4.
Assessment of OMP and BamA levels and BamA's folding status. (A) OMPs were visualized from purified envelopes by Coomassie blue staining of an SDS (urea)-polyacrylamide gel. Envelopes were purified from overnight-grown cultures in glycerol minimal liquid medium at 30°C. Each lane contained 5 μg of proteins. (B) BamA levels were determined by Western blots from purified envelopes. Membrane blots were probed with antibodies specific to BamA and AcrA, with the latter serving as a gel loading control. BamA levels were determined relative to AcrA and then normalized to the wild-type (WT) value of 1. (C) BamA's folding status was determined by assessing its heat modifiability. Purified envelopes were analyzed as for panel B except that prior to SDS-PAGE analysis, SDS-solubilized samples were either left at room temperature (25°C) or heated (100°C). BamA was detected by Western blot analysis. BamAD and BamAF are denatured and folded forms of BamA.
Since the mutations created in this study directly alter BamA, its biogenesis is expected to be affected by them. The levels and folding status of BamA were examined, and the data are shown in Fig. 4B and C. The levels of BamA bearing EGF, ΔRGF, and AAA alterations were significantly reduced compared to the wild-type BamA levels. In contrast, a relatively little reduction in the level of BamAAGF was observed.
The folding status of BamA was assessed by conducting the heat modifiability test, in which the correctly folded form of BamA is distinguished from the unfolded (denatured) form based on the faster mobility of the former on an SDS-polyacrylamide gel (16, 28, 29, 36). As expected, wild-type BamA was correctly folded, as no slowly migrating (denatured) species was present in the unheated envelope sample (Fig. 4C). In contrast, the three BamA mutants whose levels were significantly reduced (Fig. 4B) showed folding defects, with BamAAAA displaying the most pronounced defect (Fig. 4C). On the other hand, BamAAGF, whose level was only modestly reduced, also showed only a modest folding defect (see Fig. 7C). Thus, among the four BamA mutants, there was a positive correlation between the OMP biogenesis defect and the BamA level and folding defect.
Fig 7.
Assessment of OMP (A) and BamA (B) levels and BamA's folding status (C). These assays were carried out as described in the Fig. 4 legend. BamAD and BamAF are denatured and folded forms of BamA. Please note that the migration of folded forms of BamA in multiple bands is caused by subtle changes in the pH of the running SDS-PAGE buffer.
Together, the phenotypic and biochemical data revealed the importance of the three conserved L6 loop residues in the structure and function of BamA. Besides shortening the length of L6, the deletion of 641RGF643 abolishes both the backbone- and side chain-mediated interactions of the L6 loop tip region. On the other hand, the replacement of 641RGF643 by AAA would principally affect the side chain-mediated interactions. With glycine having no side chain, the effect of its replacement by an alanine would likely be structural. Thus, a severe phenotype of the BamAAAA mutant is the result of the combined effects of structural perturbation (G642 to A) and the loss of side chain-mediated interactions (R641 to A and F643 to A). Interestingly, while the R641-to-A substitution produced no phenotype, the growth phenotype of the R641-to-E substitution was apparent on LBA at both 30°C and 37°C. A lack of phenotype when R641 was replaced with a small neutral residue and the appearance of a phenotype when replaced with a residue of an opposite charge suggest that R641 possibly forms a salt bridge. However, since the R641-mediated salt bridge may be dispensable, the R641 residue can be replaced by an alanine without producing an obvious phenotype. Notably, as shown below, the importance of the R641-mediated salt bridge becomes apparent when the neighboring residues are simultaneously altered.
Finer analysis of the 641RGF643 residues of BamA.
Having established that 641RGF643 residues of the putative L6 loop of BamA are important for its folding and OMP biogenesis, we carried out a finer mutational analysis of these residues to rank the relative importance of the side chain of the individual residues. For this, we began with the BamAAAA mutant, which showed the most drastic defects (Fig. 2 to 4), and systematically restored the individual alanine residues to the corresponding wild-type residues. In all, four additional mutants were created: AAF, AGA, RAA, and RGA mutants. We also included the 641AGF643 mutant used in experiments described above as a control. As before, mutations were created in the pZS21-bamA plasmid replicon, which was then transformed into a ΔbamA recA::scar (pBAD33-bamA [BamA-WT]) strain. The pBAD33-bamA (BamA-WT) plasmid was subsequently cured by growth on minimal glycerol medium at 30°C without chloramphenicol and arabinose. The resulting cured strains expressed BamA exclusively from the pZS21 replicon.
Phenotypic analyses of the BamA (position 641 to 643) alanine mutants.
The BamA AAF, AGA, RAA, RGA, and AGF alanine mutants were tested for growth and antibiotic sensitivity phenotypes (Fig. 5 and 6). On rich medium, mutants expressing BamAAAF and BamAAGA showed drastic growth defects, with the BamAAGA mutant being almost as defective as the starting BamAAAA mutant (Fig. 5). In contrast, mutants expressing BamARAA, BamARGA, and BamAAGF grew just as well as the strain expressing wild-type BamA (Fig. 5). With the exception of the BamAAAA mutant and, to a lesser degree, the mutant expressing BamAAGA, no other BamA mutants showed growth defects on the minimal medium (Fig. 5).
Fig 5.
Growth phenotypes of strains expressing wild-type (WT) BamA (sectors 1 and 5) or various mutant BamA proteins (sectors 2 to 4 and 6 to 8) on LBA or glycerol minimal agar plates. Plates were incubated for 24 h (LBA) or 36 h (M63 minimal medium) at 30°C and 37°C. BamA was expressed from the pZS21 plasmid replicon.
Fig 6.
Antibiotic sensitivities of strains expressing wild-type (WT) or mutant BamA proteins. The assays were carried out as described in the Fig. 3 legend.
In terms of antibiotic sensitivity, restoration of any one of the three wild-type residues of the 641RGF643 motif did not significantly lower antibiotic sensitivity, although the inhibition zones were slightly smaller than when all three wild-type residues were replaced with an alanine (Fig. 6). On the other hand, restoration of the first two or the last two residues to the wild type brought the antibiotic sensitivity down to the wild-type level. From these results, combined with the growth data (Fig. 5), it can be deduced that R641 is the most important of the three residues, followed by F643 and G642.
Biochemical analyses of the BamA (position 641 to 643) alanine mutants.
The levels of major OMPs as well as BamA were analyzed from envelopes purified from the BamA AAF, AGA, RAA, RGA, and AGF mutants and strains expressing the parental BamAAAA and wild-type BamA proteins. The OMP levels were determined by direct Coomassie blue staining of the SDS (urea)-polyacrylamide gel (Fig. 7A), while BamA levels were determined by Western blot analysis using HisProbe-horseradish peroxidase (HisProbe-HRP) (Fig. 7B). The levels of OmpA, OmpC, and OmpF were significantly down in the BamAAAF and BamAAGA mutants, but not as much in the starting BamAAAA mutant (Fig. 7A). On the other hand, the OMP levels in BamARAA and the remaining two single alanine mutants were similar to the wild-type level (Fig. 7A). The level of BamA in the mutants followed a pattern similar to that observed for the major OMPs, except that all mutants have somewhat lower BamA levels than the strain expressing wild-type BamA (Fig. 7B).
Figure 7C shows the folding status of the BamA mutants. In general, BamAAAF and BamAAGA mostly migrated at the denatured position, with only small amounts migrating at the folded position. On the other hand, a significant amount of folded BamA species was detectable from envelopes prepared from BamARAA, BamARGA, and BamAAGF mutants. Thus, the folding status of BamA correlated well with the BamA levels (Fig. 7B) and the levels of the major OMPs (Fig. 7A).
Since the BamAAGA mutant shows an overall defect similar to that of the BamAAAA mutant, the conserved G642 residue by itself does not appear to be very critical for BamA structure and function. However, its significance becomes apparent when R641 or F643 is simultaneously replaced with an alanine. Of the three possible double alanine combinations affecting the conserved 641RGF643 motif, the BamAAGA mutation produces the most defects, followed by BamAAAF and BamARAA. Again, this shows that R641 plays the most critical role, followed by F643 and G642. A similar conclusion was reached for the FhaC protein (7).
All of the four assays carried out were extremely useful in dissecting the structure-function defects of the BamA mutants, with the folding assay being the most sensitive of all. In general, the strong phenotypes of BamA mutants reflected correspondingly strong biogenesis defects of BamA and, subsequently, of OMPs. There were cases where an apparent BamA folding defect did not always result in a functional or phenotypic defect. For example, BamARGA and BamAAGF showed modest folding defects and had somewhat reduced BamA levels, but the mutants expressing these proteins showed no appreciable growth or outer membrane permeability defects. Thus, to a certain degree, BamA folding and level defects can be tolerated without causing an apparent loss in BamA function.
Role of the BamA L6 loop in BamA and OMP biogenesis.
From the mutagenesis work, it is abundantly clear that the highly conserved 641RGF643 residues of the putative L6 loop of BamA are important for BamA and OMP biogenesis. It is interesting to note, however, that while the alterations of the conserved L6 loop residues affect BamA biogenesis, similar changes and the deletion of the entire L6 loop do not affect FhaC stability and localization (6, 7). Based on these findings, we speculate that a defect in BamA's biogenesis may not be due to the destabilization of the mutant BamA β-barrel structure per se. Instead, we consider the possibility that BamA is involved in its own assembly, besides assembling other β-barrel OMPs, and that the L6 loop plays a critical role in this process. The inability of some BamA L6 mutants to assemble the nascent BamA molecules would lead to misfolding and degradation of the majority of the incoming mutant BamA molecules and to the formation of dysfunctional BAM complexes. The idea that BamA is involved in its own assembly was first proposed by us based on the analysis of a POTRA domain 1 deletion mutant (3).
We have recently reported the isolation and characterization of a novel set of BamA mutants that overcome the simultaneous deficiency of BamB and BamE (29). A ΔbamB ΔbamE strain grows poorly and shows a severe BamA and OMP biogenesis defect, but the second-site alterations in BamA reverse the growth and BamA/OMP biogenesis defects. Four of the six different compensatory alterations affected the T631, G635, G649, and A652 residues of the BamA L6 loop (Fig. 1). The fact that changes in these sites overcome the requirement for BamB and BamE suggests that interactions of the Bam lipoproteins with BamA may allosterically influence the BamA β-barrel region and, more specifically, the L6 loop to facilitate OMP assembly. Accordingly, without BamB and BamE, the BamA β-barrel is unable to adapt the necessary “active” conformation, which, in part, can be achieved by compensatory changes in the β-barrel domain of BamA. The fact that the L6 loop can be the site of both advantageous and deleterious alterations indicates that this region of the protein plays an intricate role in BamA's function.
Note that envelope analyses and antibiotic sensitivity assays were carried out with bacterial cultures grown under permissive conditions, i.e., at 30°C in minimal medium. This was done because strains expressing BamAAAA, BamAΔRGF, or BamAAGA do not grow in rich medium (Fig. 2 and 5). However, for other mutants that can grow, e.g., BamAEGF and BamAAAF, more drastic OMP and antibiotic sensitivity phenotypes would be expected in rich medium, in which the mutant BamA proteins may not be able to keep up with a greater demand for envelope biogenesis under faster growth conditions.
Concluding remarks.
The deletion of the FhaC L6 loop but not the α-helix H1 abolished FHA secretion, thus showing a critical role for L6 in FhaC's secretion function (6). In addition, the L6 deletion mutant showed increased sensitivity to large antibiotics, supporting the idea that in the resting state, FhaC's channel is plugged by the L6 loop (6). Therefore, a reasonable model for FHA secretion envisions the complete displacement of the L6 loop from the β-barrel during the secretion process (6). Several BamA L6 loop mutants described here also confer increased sensitivity to large antibiotics that normally cannot penetrate the intact E. coli outer membrane. This increased antibiotic sensitivity could be due to the entry of antibiotics through the mutant BamA β-barrel, although there is no evidence for or against this notion. Our OMP data do suggest that the increase in antibiotic sensitivity stems in part from the altered outer membrane composition. Since OMP levels are significantly reduced in these BamA mutant backgrounds, including presumably that of the LPS transporter, LptD, it is possible that this macromolecular void is filled by increased levels of phospholipids in the outer leaflet of the outer membrane, thus creating patches of phospholipid bilayers. The creation of such phospholipid-rich patches in the outer membrane is thought to be the hallmark of the “leaky” outer membrane of the deep-rough LPS mutants (21). The increased vancomycin sensitivity phenotype of the BamA mutants studied here resembles more that of the LPS synthesis-defective lpxA2 mutant (8, 34) than that of the heptose-deficient deep-rough LPS mutant (34). Therefore, it is the lower LPS levels rather than the presence of an altered species of LPS in the outer membrane that causes hypersensitivity to vancomycin. The pathway by which the hydrophilic glycopeptide antibiotic vancomycin or the hydrophilic polypeptide antibiotic bacitracin crosses the outer membrane of LPS biogenesis-defective E. coli mutants such as the lpxA2 (34) and imp (24) mutants is not known. Our data suggest that increased antibiotic sensitivity of BamA mutants may be the result of both altered BamA channel properties and reduced LPS levels.
In conclusion, our analysis highlighted the functional importance of the conserved 641RGF643 residues of the putative L6 loop of BamA in β-barrel OMP biogenesis. In some instances, changes at these conserved residues also affected BamA's folding and levels, thus coupling the biogenesis of BamA and substrate OMPs to BamA's L6 loop. The dynamics of the L6 loop of BamA are not known and will be the subject of future studies.
ACKNOWLEDGMENTS
We are indebted to Phu Voung for critically reading the manuscript. We thank Dante Ricci and Tom Silhavy for insightful discussion of their BamA barrel mutants.
This work was supported by a grant (GM048167) from the National Institutes of Health.
Footnotes
Published ahead of print 29 June 2012
REFERENCES
- 1. Andrès C, Agne B, Kessler F. 2010. The TOC complex: preprotein gateway to the chloroplast. Biochim. Biophys. Acta 1803:715–723 [DOI] [PubMed] [Google Scholar]
- 2. Becker T, Böttinger L, Pfanner N. 2012. Mitochondrial protein import: from transport pathways to an integrated network. Trends Biochem. Sci. 37:85–91 [DOI] [PubMed] [Google Scholar]
- 3. Bennion D, Charlson ES, Coon E, Misra R. 2010. Dissection of β-barrel outer membrane protein assembly pathways through characterizing BamA POTRA 1 mutants of Escherichia coli. Mol. Microbiol. 77:1153–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Casadaban MJ. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104:541–555 [DOI] [PubMed] [Google Scholar]
- 5. Charlson ES, Werner JN, Misra R. 2006. Differential effects of yfgL mutation on Escherichia coli outer membrane proteins and lipopolysaccharide. J. Bacteriol. 188:7186–7194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Clantin B, et al. 2007. Structure of the membrane protein FhaC: a member of the Omp85-TpsB transporter superfamily. Science 317:957–961 [DOI] [PubMed] [Google Scholar]
- 7. Delattre A-S, et al. 2010. Functional importance of a conserved sequence motif in FhaC, a prototypic member of the TpsB/Omp85 superfamily. FEBS J. 277:4756–4765 [DOI] [PubMed] [Google Scholar]
- 8. Galloway SM, Raetz CRH. 1990. A mutant of Escherichia coli defective in the first step of endotoxin biosynthesis. J. Biol. Chem. 265:6394–6402 [PubMed] [Google Scholar]
- 9. Gatzeva-Topalova PZ, Walton TA, Sousa MC. 2008. Crystal structure of YaeT: conformational flexibility and substrate recognition. Structure 16:1873–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gatzeva-Topalova PZ, Warner LR, Pardi A, Sousa MC. 2010. Structure and flexibility of the complete periplasmic domain of BamA: the protein insertion machine of the outer membrane. Structure 18:1492–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gebert N, Ryan MT, Pfanner N, Wiedemann N, Stojanovski D. 2011. Mitochondrial protein import machineries and lipids: a functional connection. Biochim. Biophys. Acta 1808:1002–1011 [DOI] [PubMed] [Google Scholar]
- 12. Gentle I, Gabriel K, Beech P, Waller R, Lithgow T. 2004. The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J. Cell Biol. 164:19–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hagan CL, Silhavy TJ, Kahne D. 2011. β-Barrel membrane protein assembly by the Bam complex. Annu. Rev. Biochem. 80:189–210 [DOI] [PubMed] [Google Scholar]
- 14. Hsu S-C, Inoue K. 2009. Two evolutionarily conserved essential β-barrel proteins in the chloroplast outer envelope membrane. Biosci. Trends 3:168–178 [PubMed] [Google Scholar]
- 15. Huang W, Ling Q, Bédard J, Lilley K, Jarvis P. 2011. In vivo analyses of the roles of essential Omp85-related proteins in the chloroplast outer envelope membrane. Plant Physiol. 157:147–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim S, et al. 2007. Structure and function of an essential component of the outer membrane protein assembly machine. Science 317:961–964 [DOI] [PubMed] [Google Scholar]
- 17. Knowles TJ, et al. 2008. Fold and function of polypeptide transport-associated domains responsible for delivering unfolded proteins to membranes. Mol. Microbiol. 68:1216–1227 [DOI] [PubMed] [Google Scholar]
- 18. Kutik S, et al. 2008. Dissecting membrane insertion of mitochondrial β-barrel proteins. Cell 132:1011–1024 [DOI] [PubMed] [Google Scholar]
- 19. Misra R. 2007. First glimpse of the crystal structure of YaeT's POTRA domains. ACS Chem. Biol. 2:649–651 [DOI] [PubMed] [Google Scholar]
- 20. Moslavac S, et al. 2005. Conserved pore-forming regions in polypeptide transporting proteins. FEBS J. 272:1367–1378 [DOI] [PubMed] [Google Scholar]
- 21. Nikaido H, Vaara M. 1985. Molecular basis of bacterial outer membrane permeability. Microbiol. Rev. 49:1–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Paschen SA, et al. 2003. Evolutionary conservation of biogenesis of β-barrel membrane proteins. Nature 426:862–866 [DOI] [PubMed] [Google Scholar]
- 23. Remaut H, Waksman G. 2006. Protein-protein interaction through β-strand addition. Trends Biochem. Sci. 31:436–444 [DOI] [PubMed] [Google Scholar]
- 24. Sampson BA, Misra R, Benson SA. 1989. Identification and characterization of a new gene of Escherichia coli K-12 involved in outer membrane permeability. Genetics 122:491–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sanchez-Pulido L, Devos D, Genevrois S, Vicente M, Valencia A. 2003. POTRA: a conserved domain in the FtsQ family and a class of β-barrel outer membrane proteins. Trends Biochem. Sci. 28:523–526 [DOI] [PubMed] [Google Scholar]
- 26. Silhavy TJ, Berman ML, Enquist LW. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 27. Sklar JG, et al. 2007. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 104:6400–6405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stegmeier JF, Andersen C. 2006. Characterization of pores formed by YaeT (Omp85) from Escherichia coli. J. Biochem. 140:275–283 [DOI] [PubMed] [Google Scholar]
- 29. Tellez R, Jr, Misra R. 2012. Substitutions in the BamA β-barrel domain overcome the conditional lethal phenotype of a ΔbamB ΔbamE strain of Escherichia coli. J. Bacteriol. 194:317–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tuomanen E, Weiss A. 1985. Characterization of two adhesins of Bordetella pertussis for human ciliated respiratory-epithelial cells. J. Infect. Dis. 152:118–125 [DOI] [PubMed] [Google Scholar]
- 31. Vaara M. 1993. Antibiotic-supersusceptible mutants of Escherichia coli and Salmonella typhimurium. Antimicrob. Agents Chemother. 37:2255–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J. 2003. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299:262–265 [DOI] [PubMed] [Google Scholar]
- 33. Vuong P, Bennion D, Mantei J, Frost D, Misra R. 2008. Analysis of YfgL and YaeT interactions through bioinformatics, mutagenesis, and biochemistry. J. Bacteriol. 190:1507–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vuorio R, Vaara M. 1992. The lipid A biosynthesis mutation lpxA2 of Escherichia coli results in drastic antibiotic supersusceptibility. Antimicrob. Agents Chemother. 36:826–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Werner J, Misra R. 2005. YaeT (Omp85) affects the assembly of lipid-dependent and lipid-independent outer membrane proteins of Escherichia coli. Mol. Microbiol. 57:1450–1459 [DOI] [PubMed] [Google Scholar]
- 36. Workman P, et al. 2012. Genetic, biochemical, and molecular characterization of the BamA POTRA domain of Escherichia coli. J. Bacteriol. 194:3512–3521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wu T, et al. 2005. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121:235–245 [DOI] [PubMed] [Google Scholar]
- 38. Zhang H, et al. 2011. High-resolution structure of a new crystal form of BamA POTRA4-5 from Escherichia coli. Acta Crystallogr. 67:734–738 [DOI] [PMC free article] [PubMed] [Google Scholar]