Abstract
Through exome resequencing, we identified two unique mutations in recombination signal binding protein for immunoglobulin kappa J (RBPJ) in two independent families affected by Adams-Oliver syndrome (AOS), a rare multiple-malformation disorder consisting primarily of aplasia cutis congenita of the vertex scalp and transverse terminal limb defects. These identified mutations link RBPJ, the primary transcriptional regulator for the Notch pathway, with AOS, a human genetic disorder. Functional assays confirmed impaired DNA binding of mutated RBPJ, placing it among other notch-pathway proteins altered in human genetic syndromes.
Main Text
Signaling through the Notch pathway regulates cell proliferation, death, differentiation, and acquisition of specific fates in a context-dependent manner.1 Aberrant gain or loss of function of notch-signaling components has been implicated in human disease. Mutations in notch1 ligand JAG1 (MIM 601920) and notch2 receptor NOTCH2 (MIM 600275) cause Alagille syndrome types 1 (ALGS1 [MIM 118450]) and 2 (ALGS2 [MIM 610205]), respectively.2 Mutations in JAG1 can also lead to tetralogy of Fallot3 (TOF [MIM 187500]), whereas mutations in NOTCH3 (MIM 600276) can result in cerebral autosomal-dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL [MIM 125310]).1
Notch signaling induces cleavage of the Notch intracellular domain (NICD), which translocates to the nucleus and combines with RBPJ to form a transcriptional complex. RBPJ, the principal DNA-binding partner of the NICD, is an evolutionarily conserved protein that coordinates transcriptional activation of Notch-target genes through the assembly of protein complexes containing coactivators. To date, there are no germline RPBJ (MIM 147183) mutations reported to cause a genetic disorder in humans.
Adams-Oliver syndrome (AOS [MIM 100300]) was first described as the combination of vertex scalp defects and terminal limb defects.4 The clinical features are highly variable in both anatomic site and severity of expression. The most common defects are terminal limb malformations (in 84% of cases), including osseous syndactyly, rudimentary bones, or completely absent digits. Congenital cutis aplasia, the second-most-common defect (in 75% of cases), usually occurs over the posterior parietal region. Underlying bone defects can be present, and tortuous veins can occur on the posterior scalp.5,6 Congenital heart defects, microcephaly, esotropia, microphthalmia, cleft lip with or without cleft palate, Poland sequence, accessory nipples, an imperforate vaginal hymen, developmental delay, and various digital anomalies are frequent.4,5 AOS usually shows autosomal-dominant inheritance;4,5,7–9 however, sporadic cases and families appearing autosomal recessive are known.10–12
The AOS-affected individuals in our study belong to one of two families; family 1 was diagnosed by us, and family 2 was diagnosed by our review of medical records and photographs (Figure 1A). In family 1, the proband (individual III-1) was identified at birth to have cutis aplasia. Her extremities showed syndactyly of her second and third toes and were otherwise normal. She has microcephaly and short palpebral fissures. She was mildly delayed in gross motor milestones. Her father (II-2) has short distal phalanges of his fingers and has absent toes and short metatarsals bilaterally. He has microcephaly, intellectual deficits, and is at the borderline of mild mental retardation. Neither individual has heart defects, immune defects, cutis marmorata, or other associated abnormalities.
Figure 1.
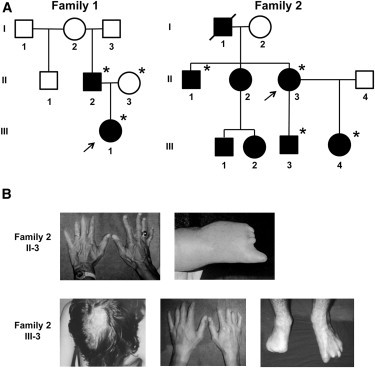
Pedigree Structure and Clinical Features of Families
(A) Pedigrees exhibiting autosomal-dominant inheritance of AOS and deleterious mutation of RBPJ. Individuals who were tested via exome sequencing are designated with an asterisk. Arrows point to the index cases in each family.
(B) Photographs of features diagnostic for AOS in family 2. The top row shows the hands and foot of subject II-3. The second row shows the cutis aplasia scar, hands, and feet of subject III-3.
In family 2, individual II-1 has normal intelligence and mild limb reductions of his hands (Figure 1B). Individual II-3 has shortened distal phalanges of her left hand, bilateral reduction of her toes, and normal intelligence. Individual III-3 has a large scarred area that is the result of cutis aplasia at birth, asymmetric shortening of the hands bilaterally, asymmetric reductions of the feet, and intellectual deficits. Individual III-4 is mildly affected with fifth-finger nail hypoplasia, fifth-toe shortening, and normal development. There is no report of congenital heart defects, other associated anomalies, or immune defects in the family.
To identify AOS-causing mutations, we performed exome resequencing by using a variant-filtering strategy. After informed consent was obtained (protocol 09567 from the institutional review board at the University of Oklahoma Health Sciences Center), blood samples were drawn from all participants and DNA was isolated from six affected individuals and one unaffected first-degree family member in each of the two unrelated families. Exome enrichment was performed with the TruSeq Exome Enrichment Kit (Illumina, San Diego, CA) as per the manufacturer's protocol. Sequencing was performed on an Illumina HiSeq 2000 instrument with paired-end 100 bp reads. Sequence reads were mapped to the human reference genome (hg18) with the Burrows-Wheeler Aligner (BWA).13 Local realignment around problematic areas and empirical base-quality-score recalibration were done with the Genome Analysis Tool Kit (GATK).14 SNPs were identified with the GATK Unified Genotyper. Variants were excluded from further analyses if they failed to meet any of the following parameters: an alignment quality higher than 30, a read depth of at least 8, a quality-by-depth score greater than 2.5, presence within a homopolymer run of 5 bases or fewer, a strand-bias score less than −10.0, and a map-quality score greater than 25.
We used Annovar to functionally annotate variants prior to filtering.15 We screened variants to first remove synonymous variants and then to remove variants within segmental duplications and variants outside highly conserved regions. Variants previously seen in the 1000 Genomes Project and the dbSNP130 variant database were removed. Remaining variants were then mapped back to their respective genes. Lists of these genes were assembled for each sample on the basis of a dominant or recessive model; the dominant model required only one variant per gene, and the recessive model required two or more variants per gene. Because AOS is a rare condition and was expressed in multiple generations in both families, the dominant model was considered most plausible. Lists of genes by individual within each family were compared to those of other members of their respective family for the identification of family-wide, nonsynonymous variants that appeared to segregate with AOS affectation status (Tables S1 and S2, available online). We identified 44 genes with mutations unique to AOS-affected individuals in family 1 and 26 genes in family 2. The gene lists from each family were compared, which identified only one gene in common, RBPJ (RefSeq accession number NM_005349) (Table 1). The raw alignment of reads covering the respective variants for each individual was then inspected with the Integrated Genome Viewer and determined to be sufficient for the variant call.16 We then confirmed the presence of the mutations in each individual by using Sanger sequencing (Figure S1).
Table 1.
Gene-Filtering Strategy Used for Identifying RBPJ in AOS Pedigrees
|
Family 1 |
Family 2 |
||||||
|---|---|---|---|---|---|---|---|
| II-2 | II-3 | III-1 | II-1 | II-3 | III-3 | III-4 | |
| Number of genes with ≥1 nsSNP | 6,072 | 5,903 | 6,069 | 5,817 | 6,007 | 5,895 | 5,273 |
| From row 1, number of genes with ≥1 nsSNP evolutionarily conserveda | 2,613 | 2,497 | 2,616 | 2,496 | 2,568 | 2,531 | 2,213 |
| From row 2, number of genes not in 1000 Genomes Project | 329 | 316 | 335 | 309 | 314 | 303 | 279 |
| From row 3, number of genes not in dbSNP130 | 187 | 177 | 183 | 180 | 187 | 167 | 173 |
| From row 4, number of genes shared in AOS within pedigree | 44 | − | − | 26 | − | − | − |
| From row 5, number of genes shared in AOS between pedigrees | 1 (RBPJ) | − | − | − | − | − | − |
The following abbreviations are used: nsSNP, nonsynonymous SNP; and AOS, Adams-Oliver syndrome.
Evolutionary conservation was determined by comparison to PhastCon 44 vertebrate data.
In family 1, the RBPJ mutation, an A to G transition (c.188A>G) (RefSeq NM_005349.2), resulted in a heterozygous glutamic acid to glycine amino acid change (p.Glu63Gly) (RefSeq NM_005349). The mutation in family 2 was an A to G transition (c.505A>G) (RefSeq NM_005349.2) as well and resulted in a heterozygous lysine to glutamic acid amino acid change (p.Lys169Glu) (RefSeq NM_005349) (Figure S2). Comparison of the human RBPJ amino acid sequence with the highly conserved homologous CSL protein from C. elegans located both mutations in the DNA-binding domain (Figure S3).17 The amino acid positions are either within two positions of (p.Glu63Gly) or adjacent to (p.Lys169Glu) amino acids that interact directly with bound DNA (Figure S2), and both were predicted to be damaging by SIFT18 and Polyphen2.19
Given the location of the mutations in each family, we hypothesized that the mutated forms of RBPJ would demonstrate defects in DNA binding. To test this hypothesis, we made FLAG-tagged expression constructs containing wild-type RBPJ or the two mutated forms (Table S3 and Figure S4) and performed an electrophoretic mobility shift assay (EMSA) with an oligonucleotide corresponding to a canonical RBPJ binding site in the promoter of human homolog of hairy and enhancer of split 1 (Drosophila), HES1 (MIM 139605) (Table S4). Wild-type RBPJ formed a specific complex with the probe in an amount proportional to that of nuclear extract used (Figure 2A). In contrast, compared with the negative control, both mutant forms (p.Glu63Gly and p.Lys169Glu) did not exhibit any specific binding complex. Densitometric measurement of band intensity from multiple independent experiments showed statistically significant differences between wild-type RBPJ and mutants of RBPJ (Figure 2B). Using a supershift assay with anti-FLAG tag antibody, we verified that recombinant RBPJ bound to the HES1 probe was present in the shifted protein complex (Figure 2A). Binding specificity of RBPJ to the HES1 sequence was confirmed by cold competition (Figure S5).
Figure 2.
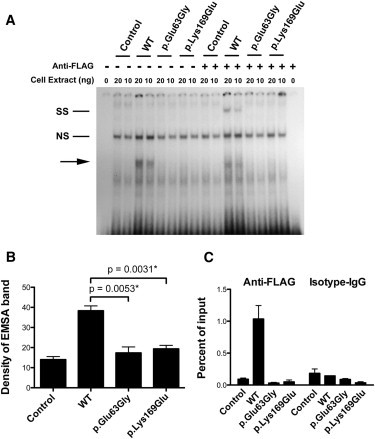
Functional Characterization of RBPJ Alterations Identified in AOS-Affected Families
(A) A representative EMSA from three independent experiments. The first lane shows a free probe derived from the HES1 promoter; subsequent lanes show nuclear protein extract (20 ng and 10 ng) that was prepared either from HEK 293T cells transfected with FLAG-tagged wild-type or mutant RBPJ or from untransfected HEK 293T cells as indicated. The shifted band corresponding to recombinant RBPJ constructs is labeled with an arrow. Super shift was performed by the addition of an antibody to the FLAG epitope expressed on the recombinant RBPJ proteins and is labeled in the figure as “SS.” A nonspecific band is labeled “NS.”
(B) We performed densitometric quantification of the EMSA bands from four independent experiments. The mean densities from untransfected HEK 293T cells and HEK 293T cells transfected with various RBPJ constructs are shown in the columns. Error bars represent the mean ± standard error of the mean (SEM). Statistical differences between the means were calculated with the unpaired t test.
(C) ChIP was performed with anti-FLAG and anti-isotype control IgG on chromatin prepared from HEK 293T cells transfected with RBPJ constructs. Quantification of enriched DNA fragments was performed with quantitative RT-PCR with primers flanking a known RBPJ binding site in the HES1 promoter. Shown is a representative ChIP-qPCR from two independent experiments. Each experiment was composed of three technical replicates. Mean enrichment as a percentage of input chromatin is displayed. Error bars correspond to the mean ± SEM.
To determine whether these mutations affect the interaction between RBPJ and the endogenous HES1 promoter in live cells, we examined RBPJ binding by chromatin immunoprecipitation (ChIP) quantitative PCR (qPCR) in cells expressing similar levels of recombinant wild-type RBPJ and mutants (Figure S6 and Table S5). Compared with wild-type RPBJ, RBPJ mutants showed decreased binding to the HES1 promoter (Figure 2C). The qPCR product was verified by Sanger sequencing to be the target sequence from the HES1 promoter (data not shown). The reduced binding affinity of RBPJ mutants also resulted in decreased expression of HES1 (Figure S7).
To evaluate whether mutations in RBPJ affect protein interaction with the NICD, we cotransfected human embryonic kidney (HEK) 293T cell lines with NICD and RBPJ constructs. Coimmunoprecipitation (Co-IP) confirmed that neither of the RBPJ mutations altered protein-protein interaction with NICD given that similar amounts of RBPJ and NICD were detected in each sample (Figure S8).
In this study, we have identified two mutations in RPBJ, the key transcriptional regulator for Notch, in two independent kindreds affected by AOS. These mutations are located in highly conserved protein regions that are predicted to make contact with DNA. Functional studies confirmed that both mutations reduce the affinity of RBPJ binding to a canonical sequence in the HES1 promoter. A previous site-directed mutagenesis screen of RBPJ identified two lysine residues that are directly adjacent to p.Lys169Glu and that are important for DNA binding, supporting our findings.20
A link between Notch and AOS had been previously hypothesized on the basis of the association between cardiovascular malformations and aplasia cutis congenital.21 RBPJ-mediated NOTCH signaling is important for mesenchymal cell proliferation and skeletal formation,22 epidermis and hair-follicle development,23 and vascular-structure formation.24 Furthermore, RBPJ-deficient mice have defective cranial-bone formation,25 and RBPJ-conditional-knockout mice have arteriovenous malformations.26 These findings in model systems overlap the phenotypic spectrum of AOS.
Mutations in ARHGAP31 (MIM 610911) and DOCK6 (MIM 614194) have been reported in AOS-affected kindreds with both autosomal-dominant and autosomal-recessive inheritance.9,10 The mechanism by which these genes influence AOS has been reported to be through inactivation of Rac1 and Cdc42; inactivation of these proteins leads to impaired cytoskeletal homeostasis.9,10 Our discovery of mutations in RBPJ adds to the genetic heterogeneity of AOS and underscores the hypothesis that AOS is a multigene, multipathway disorder. It is tempting to postulate that these genes converge on a final common pathway that results in the manifestations of AOS; however, existing data are insufficient for drawing this conclusion at this time. No deleterious mutations in ARHGAP31 or DOCK6 were found in our AOS-affected families.
In summary, our data provide genetic evidence that alterations in the Notch transcription factor RBPJ predispose to malformations in humans. Given the phenotypic heterogeneity of AOS, our results support RBPJ mutational screening for individuals presenting with congenital malformations consistent with AOS and related disorders.
Acknowledgments
We thank the individuals who participated in this research through contribution of personal health information and samples without the expectation of personal gain, as well as fellow medical professionals who referred them. We thank Hong Chen and Courtney Griffin for providing advice on the experimental design. We thank Xiao-Hong Sun for critically reading and providing advice on the content of the manuscript. This work was funded through an Oklahoma Medical Research Foundation Research Grant (9138-12 to P.M.G.).
Contributor Information
Susan J. Hassed, Email: susan-hassed@OUHSC.edu.
Patrick M. Gaffney, Email: gaffneyp@omrf.org.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
1000 Genomes Project, http://www.1000genomes.org/
Online Mendelian Inheritance in Man (OMIM), http://omim.org/
References
- 1.Hofmann J.J., Iruela-Arispe M.L. Notch signaling in blood vessels: Who is talking to whom about what? Circ. Res. 2007;100:1556–1568. doi: 10.1161/01.RES.0000266408.42939.e4. [DOI] [PubMed] [Google Scholar]
- 2.Gridley T. Notch signaling and inherited disease syndromes. Hum. Mol. Genet. 2003;12(Spec No 1):R9–R13. doi: 10.1093/hmg/ddg052. [DOI] [PubMed] [Google Scholar]
- 3.Eldadah Z.A., Hamosh A., Biery N.J., Montgomery R.A., Duke M., Elkins R., Dietz H.C. Familial Tetralogy of Fallot caused by mutation in the jagged1 gene. Hum. Mol. Genet. 2001;10:163–169. doi: 10.1093/hmg/10.2.163. [DOI] [PubMed] [Google Scholar]
- 4.Adams F.H., Oliver C.P. Hereditary deformities in man due to arrested development. J. Hered. 1945;36:3–7. [Google Scholar]
- 5.Whitley C.B., Gorlin R.J. Adams-Oliver syndrome revisited. Am. J. Med. Genet. 1991;40:319–326. doi: 10.1002/ajmg.1320400315. [DOI] [PubMed] [Google Scholar]
- 6.Bork K., Pfeifle J. Multifocal aplasia cutis congenita, distal limb hemimelia, and cutis marmorata telangiectatica in a patient with Adams-Oliver syndrome. Br. J. Dermatol. 1992;127:160–163. doi: 10.1111/j.1365-2133.1992.tb08049.x. [DOI] [PubMed] [Google Scholar]
- 7.Bamforth J.S., Kaurah P., Byrne J., Ferreira P. Adams Oliver syndrome: A family with extreme variability in clinical expression. Am. J. Med. Genet. 1994;49:393–396. doi: 10.1002/ajmg.1320490408. [DOI] [PubMed] [Google Scholar]
- 8.Martínez-Frías M.L., Arroyo Carrera I., Muñoz-Delgado N.J., Nieto Conde C., Rodríguez-Pinilla E., Urioste Azcorra M., Omeñaca Teres F., García Alix A. [The Adams-Oliver syndrome in Spain: The epidemiological aspects] An. Esp. Pediatr. 1996;45:57–61. [PubMed] [Google Scholar]
- 9.Southgate L., Machado R.D., Snape K.M., Primeau M., Dafou D., Ruddy D.M., Branney P.A., Fisher M., Lee G.J., Simpson M.A. Gain-of-function mutations of ARHGAP31, a Cdc42/Rac1 GTPase regulator, cause syndromic cutis aplasia and limb anomalies. Am. J. Hum. Genet. 2011;88:574–585. doi: 10.1016/j.ajhg.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shaheen R., Faqeih E., Sunker A., Morsy H., Al-Sheddi T., Shamseldin H.E., Adly N., Hashem M., Alkuraya F.S. Recessive mutations in DOCK6, encoding the guanidine nucleotide exchange factor DOCK6, lead to abnormal actin cytoskeleton organization and Adams-Oliver syndrome. Am. J. Hum. Genet. 2011;89:328–333. doi: 10.1016/j.ajhg.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tekin M., Bodurtha J., Ciftçi E., Arsan S. Further family with possible autosomal recessive inheritance of Adams-Oliver syndrome. Am. J. Med. Genet. 1999;86:90–91. doi: 10.1002/(sici)1096-8628(19990903)86:1<90::aid-ajmg20>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 12.Unay B., Sarici S.U., Gül D., Akin R., Gökçay E. Adams-Oliver syndrome: Further evidence for autosomal recessive inheritance. Clin. Dysmorphol. 2001;10:223–225. doi: 10.1097/00019605-200107000-00014. [DOI] [PubMed] [Google Scholar]
- 13.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang K., Li M., Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kovall R.A., Hendrickson W.A. Crystal structure of the nuclear effector of Notch signaling, CSL, bound to DNA. EMBO J. 2004;23:3441–3451. doi: 10.1038/sj.emboj.7600349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 19.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chung C.N., Hamaguchi Y., Honjo T., Kawaichi M. Site-directed mutagenesis study on DNA binding regions of the mouse homologue of Suppressor of Hairless, RBP-J kappa. Nucleic Acids Res. 1994;22:2938–2944. doi: 10.1093/nar/22.15.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Digilio M.C., Marino B., Dallapiccola B. Autosomal dominant inheritance of aplasia cutis congenita and congenital heart defect: A possible link to the Adams-Oliver syndrome. Am. J. Med. Genet. A. 2008;146A:2842–2844. doi: 10.1002/ajmg.a.32526. [DOI] [PubMed] [Google Scholar]
- 22.Dong Y., Jesse A.M., Kohn A., Gunnell L.M., Honjo T., Zuscik M.J., O'Keefe R.J., Hilton M.J. RBPjkappa-dependent Notch signaling regulates mesenchymal progenitor cell proliferation and differentiation during skeletal development. Development. 2010;137:1461–1471. doi: 10.1242/dev.042911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vauclair S., Nicolas M., Barrandon Y., Radtke F. Notch1 is essential for postnatal hair follicle development and homeostasis. Dev. Biol. 2005;284:184–193. doi: 10.1016/j.ydbio.2005.05.018. [DOI] [PubMed] [Google Scholar]
- 24.Dou G.R., Wang Y.C., Hu X.B., Hou L.H., Wang C.M., Xu J.F., Wang Y.S., Liang Y.M., Yao L.B., Yang A.G., Han H. RBP-J, the transcription factor downstream of Notch receptors, is essential for the maintenance of vascular homeostasis in adult mice. FASEB J. 2008;22:1606–1617. doi: 10.1096/fj.07-9998com. [DOI] [PubMed] [Google Scholar]
- 25.Mead T.J., Yutzey K.E. Notch signaling and the developing skeleton. Adv. Exp. Med. Biol. 2012;727:114–130. doi: 10.1007/978-1-4614-0899-4_9. [DOI] [PubMed] [Google Scholar]
- 26.Krebs L.T., Shutter J.R., Tanigaki K., Honjo T., Stark K.L., Gridley T. Haploinsufficient lethality and formation of arteriovenous malformations in Notch pathway mutants. Genes Dev. 2004;18:2469–2473. doi: 10.1101/gad.1239204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


