Abstract
Mitogen-activated protein kinase-activated protein kinase 2 (MAPKAPK2) may promote cancer development and progression by inducing tumorigenesis and drug resistance. To assess whether the copy-number variation g.CNV-30450 located in the MAPKAPK2 promoter has any effect on lung cancer risk or prognosis, we investigated the association between g.CNV-30450 and cancer risk in three independent case-control studies of 2,332 individuals with lung cancer and 2,457 controls and the effects of g.CNV-30450 on cancer prognosis in 1,137 individuals with lung cancer with survival data in southern and eastern Chinese populations. We found that those subjects who had four copies of g.CNV-30450 had an increased cancer risk (odds ratio = 1.94, 95% confidence interval [CI] = 1.61–2.35) and a worse prognosis for individuals with lung cancer (with a median survival time of only 9 months) (hazard ratio = 1.47, 95% CI = 1.22–1.78) compared with those with two or three copies (with a median survival time of 14 months). Meanwhile, four copies of g.CNV-30450 significantly increased MAPKAPK2 expression, both in vitro and in vivo, compared with two or three copies. Our study establishes a robust association between the functional g.CNV-30450 in MAPKAPK2 and risk as well as prognosis of lung cancer, and it presents this functional copy-number variation as a potential biomarker for susceptibility to and prognosis for lung cancer.
Main Text
Lung cancer (MIM 211980) ranks as the leading cause of cancer death in China and worldwide.1 Epidemiological studies have established many environmental risk factors for lung cancer, including smoking, air pollution, and radiation.2 All of these factors are stimuli to cells and could activate the mitogen-activated protein kinase (MAPK) pathways (Kyoto Encyclopedia of Genes and Genomes [KEGG] pathway: hsa04010), which are known to regulate apoptosis, inflammation, and tumorigenesis.3 Therefore, genetic variation in genes of the MAPK pathway may influence cancer susceptibility. The MAPK-activated protein kinase 2 (also known as MAPKAPK2 or MK2 [MIM 602006]) belongs to the p38 MAPK pathway,4 which is involved in regulating a variety of biological responses to extracellular signals that include oxidative stress, radiation, genotoxicity, inflammation, and infection.5 MAPKAPK2 has been identified as oncogenic and involved in tumor growth and invasion.6,7 Therefore, genetic variants in MAPKAPK2 may contribute to cancer susceptibility and prognosis.
In the present study, the procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation and approved by the institutional review boards of Guangzhou Medical University and Soochow University. We performed three independent case-control studies that included a total of 2,332 individuals with lung cancer and 2,457 controls in southern and eastern Chinese populations to investigate the association between copy-number variations (CNVs) in MAPKAPK2 and cancer risk. After their provision of written informed consent, as described previously,8–10 1,056 individuals with primary lung cancer and 1,056 age- (±5 years) and sex-frequency-matched healthy controls who were recruited between March 2007 and March 2009 in the Guangzhou area were used as a discovery set, and 503 individuals with lung cancer and 623 frequency-matched controls who were enrolled between March 2008 and May 2010 in the Suzhou area were used as the validation set I. Then, another 773 individuals with lung cancer and 778 frequency-matched controls who were continuously recruited from the Guangzhou area between April 2009 and June 2011 were employed as the validation set II. All the individuals were genetically unrelated Han Chinese. Each participant was scheduled for an interview for collection of data on smoking status, drinking status, preexisting chronic obstructive pulmonary disease (COPD) conditions, and other risk factors with a structured questionnaire (Table S1 available online) and for donation of a one-time 5 ml peripheral-blood sample.8–10
By searching the Database of Genomic Variants (DGV), we found that there were three CNVs (g.CNV-2091, g.CNV-30450, and g.CNV-71062) in MAPKAPK2. Considering the low frequencies of g.CNV-2091 and g.CNV-71062 (1/39 = 0.026 and 15/269 = 0.055, respectively), we selected only g.CNV-30450 (frequency: 6/30 = 0.20), which spans over the MAPKAPK2 promoter region and has 1.7 kb sequences (−1098∼+668 nt to initiation transcription code ATG) (Figure S1). Genomic DNA was extracted from 2 ml whole blood of each participant and normalized to 20 ng/μl with a good purity (optical density 260 [OD260]/OD280 = 1.8∼2.0). We genotyped the g.CNV-30450 for all 4,789 subjects by using the TaqMan real-time quantitative PCR (qPCR) method according to the protocol of Applied BioSystems (catalog no. Hs01173160, Applied BioSystems, Foster City, CA, USA)11 and validated the results with the AccuCopy assay (a multiple competitive real-time PCR) by Genesky Bio-Tech (Shanghai, China) and Affymetrix Genome-Wide Human SNP Array 6.0 by Bio Miao Biological Technology (Beijing) in 200 randomly selected samples (Figures S1 and S2);12 the results were 97.5% or 98.0% concordant.
We detected three kinds of the g.CNV-30450 (i.e., two, three, and four copies) in blood samples, and found a significant difference in the distributions of the g.CNV-30450 genotypes between individuals with lung cancer and unaffected controls (p = 2.26 × 10−6) (Table 1). After adjustment for possible confounding factors including age, sex, smoking status, drinking status, and family history of cancer in logistic-regression models, we found that compared with the common two copies, individuals who carried four copies had a 2.03-fold increased risk of lung cancer (odds ratio [OR] = 2.03, 95% confidence interval [CI] = 1.54–2.65; p = 0.001), but individuals with three copies did not (p = 0.371), which best fitted the recessive-genetic model according to the smallest Akaike information criterion (AIC) value.13 The four copies conferred a 2.01-fold increased risk of lung cancer compared with two or three copies (OR = 2.01, 95% CI = 1.54–2.63; p = 3.1 × 10−7). We subsequently replicated the association in the two validation sets, in which the four copies of g.CNV-30450 contributed to a 2.04-fold increased cancer risk (validation set I: OR = 2.04, 95% CI = 1.36–3.06; p = 0.001) and a 1.75-fold increased lung cancer risk (validation set II: OR = 1.75, 95% CI = 1.23–2.51; p = 0.002). Because the risk associations observed in the three independent case-control studies were homogeneous (p = 0.870), we further combined the three sets to increase the study power for stratification analysis. We found that the adverse effect of the four-copy genotype was significant in most subgroups, except for individuals with a family history of lung cancer, ever drinkers, and other histological types, which may be due to limited sample size in these subgroups; however, there was no significant heterogeneity between the strata-OR as suggested by the Breslow-Day test (p > 0.05 for all), and no significant interaction between the MAPKAPK2 adverse genotypes and selected factors (p > 0.05 for all, Figure S3). Interestingly, in those individuals with a family history of cancer, there was an intuitively higher OR than that found in individuals without a family history of cancer, suggesting that the g.CNV-30450 explains some of the genetic roles in cancer risk.
Table 1.
Distribution of g.CNV-30450 Genotypes of MAPKAPK2 and Associations with the Risk of Lung Cancer
| Genotype/Allele Copy Number |
Discovery Set (Southern Chinese) |
Validation Set I (Eastern Chinese) |
Validation Set II (Southern Chinese) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Cases n (%) | Controls n (%) | Adjusted OR (95% CI)a | Cases n (%) | Controls n (%) | Adjusted OR (95% CI)a | Cases n (%) | Controls n (%) | Adjusted OR (95% CI)a | |
| Total n of subjects | 1,056 | 1,056 | 503 | 623 | 773 | 778 | |||
| Codominant Modelb | 2,751.80 | 1,526.86 | 2,122.54 | ||||||
| 2 | 709 (67.1) | 787 (74.5) | 1.00 (ref.) | 359 (71.4) | 485 (77.8) | 1.00 (ref.) | 563 (72.8) | 591 (76.0) | 1.00 (ref.) |
| 3 | 161 (15.3) | 163 (15.4) | 1.04 (0.81–1.33) | 78 (15.5) | 94 (15.1) | 1.11 (0.80–1.55) | 118 (15.3) | 131 (16.8) | 0.96 (0.73–1.26) |
| 4 | 186 (17.6) | 106 (10.1) | 2.03 (1.54–2.65) | 66 (13.1) | 44 (7.1) | 2.07 (1.38–3.13) | 92 (11.9) | 56 (7.2) | 1.74 (1.22–2.48) |
| Additive Modelb | 2755.25 | 1523.66 | 2122.75 | ||||||
| 4 versus 3 versus 2 | 1.34 (1.19–1.52) | 1.40 (1.14–1.66) | 1.22 (1.04–1.43) | ||||||
| Dominant Modelb | 2,765.03 | 1,531.12 | 2,128.443 | ||||||
| 2 | 709 (67.1) | 787 (74.5) | 1.00 (ref.) | 359 (71.4) | 485 (77.8) | 1.00 (ref.) | 563 (72.8) | 591 (76.0) | 1.00 (ref.) |
| 3 + 4 | 347 (32.9) | 269 (25.5) | 1.42 (1.16–1.72) | 144 (28.6) | 138 (22.2) | 1.42 (1.08–1.86) | 210 (27.2) | 187 (24.0) | 1.19 (0.94–1.50) |
| Recessive Modelb | 2,749.89 | 1,525.24 | 2,120.65 | ||||||
| 2 + 3 | 870 (82.4) | 950 (90.0) | 1.00 (ref.) | 437 (86.9) | 579 (92.9) | 1.00 (ref.) | 681 (88.1) | 722 (92.8) | 1.00 (ref.) |
| 4 | 186 (17.6) | 106 (10.0) | 2.01 (1.54–2.63) | 66 (13.1) | 44 (7.1) | 2.04 (1.36–3.06) | 92 (11.9) | 56 (7.2) | 1.75 (1.23–2.51) |
The following abbreviations are used: OR, odds ratio; CI, confidence interval; n, number; and ref., reference.
Adjusted in a logistic-regression model that included age, sex, smoking status, alcohol use, and family history of cancer.
Akaike information criterion (AIC) value.
Additionally, we found association of the g.CNV-30450 with poor prognosis of lung cancer in 1,137 individuals, using complete survival data on deaths caused by lung cancer obtained from the follow-ups conducted every 3 months via telephone from medical records, or from individuals’ families (Table S2). In the discovery set of 510 individuals with lung cancer, we found that the median survival time (MST) of individuals who carried the four-copy-variant genotype in the discovery set was significantly shorter (9 months) than those with the two- or three-copy genotype (13 months, log-rank test: p = 0.001). Similarly, cancer-affected individuals with the four-copy genotype had an increased hazard of death (Cox proportional hazards model: hazard ratio [HR] = 1.57, 95% CI = 1.22–2.01; p = 4.48 × 10−5; Figure 1A). We then confirmed the above results in the validation set I of 296 individuals and the validation set II of 331 individuals. As shown in Figures 1B and 1C, those carrying four copies of g.CNV-30450 had a decreased MST by 7 months compared with the MST for those with two or three copies (8 months versus 15 months, log-rank test: p = 0.045) and also had a significant hazard as shown by the Cox model (HR = 1.48, 95% CI = 1.01–2.17; p = 0.046; Table 2) in the validation set I, and those with the four-copy genotype exhibited a shorter MST (7 months) than did those with the two- or three-copy genotype (15 months), with a borderline significance (log-rank test: p = 0.051) and poor survival rate (Cox model: HR = 1.53, 95% CI = 0.98–2.39; p = 0.061; Table 2) in the validation set II. When we merged all individuals with lung cancer from the three studies, the four-copy adverse genotype conferred an MST decreased by 5 months compared with the two- or three-copy genotype (9 months and 14 months, respectively; log-rank test: p = 9.30 × 10−6) and had a 47% higher death risk (Cox model: HR = 1.47, 95% CI = 1.22–1.78; p = 5.12 × 10−5; Table 2). Furthermore, the multivariate Cox model showed that age, smoking status, stage, surgery, chemotherapy, radiotherapy, and MAPKAPK2 g.CNV-30450 genotypes remained significantly associated with lung cancer survival (Table S3). In a stratified survival analysis, we found that the association between four copies of g.CNV-30450 and poor lung cancer survival was significant in most subgroups, except for individuals with the histological types of large-cell carcinoma and small-cell lung cancer. Furthermore, we did not observe any significant interactions between confounding factors or clinical characteristics and g.CNV-30450 genotypes in lung cancer survival (p > 0.05 for all; Figure S4).
Figure 1.
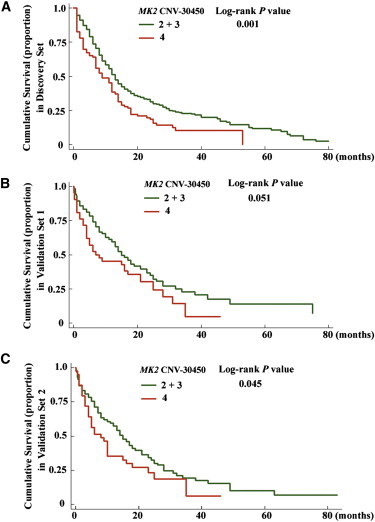
Kaplan-Meier Survival Curve for Individuals with Lung Cancer by MAPKAPK2 g.CNV-30450 Genotype
(A) The discovery set.
(B) The validation set I.
(C) The validation set II.
Table 2.
Analysis of MAPKAPK2 g.CNV-30450 and Lung Cancer Survival
| g.CNV-30450 | n Individuals (%) | n Deaths | MST (months) | Log-Rank p Value | HR (95% CI)a | Cox model p Valuea |
|---|---|---|---|---|---|---|
| Discovery Set | 510 | 413 | 12 | |||
| 2 | 352 (69.0) | 273 | 13 | 0.001 | 1.00 (ref.) | |
| 3 | 72 (14.1) | 63 | 12 | 1.17 (0.89–1.55) | 0.258 | |
| 4 | 86 (16.9) | 77 | 9 | 1.60 (1.24–2.07) | 3.57 × 10−4 | |
| Trend test p value | 3.06 × 10−4 | |||||
| 2 + 3 | 424 (83.1) | 336 | 13 | 0.001 | 1.00 (ref.) | |
| 4 | 86 (16.9) | 77 | 9 | 1.57 (1.22–2.01) | 4.48 × 10−5 | |
| Validation Set I | 296 | 203 | 13 | |||
| 2 | 204 (68.9) | 133 | 15 | 0.105 | 1.00 (ref.) | |
| 3 | 53 (17.9) | 38 | 13 | 1.23 (0.84–1.80) | 0.278 | |
| 4 | 39 (13.2) | 32 | 8 | 1.58 (1.06–2.33) | 0.024 | |
| Trend test p value | 0.027 | |||||
| 2+3 | 257 (86.8) | 171 | 15 | 0.045 | 1.00 (ref.) | |
| 4 | 39 (13.2) | 32 | 8 | 1.48 (1.01–2.17) | 0.046 | |
| Validation Set II | 331 | 208 | 12 | |||
| 2 | 228 (68.9) | 132 | 15 | 0.083 | 1.00 (ref.) | |
| 3 | 61 (18.4) | 41 | 13 | 1.28 (0.81–2.03) | 0.282 | |
| 4 | 42 (12.7) | 35 | 7 | 1.63 (1.02-2.59) | 0.039 | |
| Trend test p value | 0.028 | |||||
| 2 + 3 | 289 (87.3) | 173 | 15 | 0.051 | 1.00 (ref.) | |
| 4 | 42 (12.7) | 35 | 7 | 1.53 (0.98–2.39) | 0.061 | |
| Merged Set | 1137 | 824 | 12 | |||
| 2 | 784 (69.0) | 538 | 14 | 7.07 × 10−6 | 1.00 (ref.) | |
| 3 | 186 (16.3) | 142 | 12 | 1.19 (0.99–1.46) | 0.069 | |
| 4 | 167 (14.7) | 144 | 9 | 1.53 (1.26–1.85) | 1.56 × 10−5 | |
| Trend test p value | 1.08 × 10−5 | |||||
| 2 + 3 | 970 (85.3) | 680 | 14 | 9.30 × 10−6 | 1.00 (ref.) | |
| 4 | 167 (14.7) | 144 | 9 | 1.47 (1.22–1.78) | 5.12 × 10−5 | |
The following abbreviations are used: MST, median survival time; HR, hazard ratio; ref., reference.
Cox regression analysis was adjusted for age, sex, preexisting chronic obstructive pulmonary disease, and smoking, drinking, histology, stages, surgery, chemotherapy, and radiotherapy status.
Previous studies have shown that copy-number aberrations at 1q32, a region that covers human MAPKAPK2, are associated with increased cancer risk and poor prognosis.14,15 The gain of human chromosome 1q32 conferred adverse prognosis for atypical meningioma.14,15 With a large population included and three independent case-control studies, our results consistently showed that the four-copy genotype of g.CNV-30450 in MAPKAPK2 was associated with an increased risk of lung cancer compared with the genotype of two or three copies, and the four copies also conferred a worse prognosis for individuals with lung cancer. Therefore, the g.CNV-30450 may be a potential biomarker for risk and prognosis of lung cancer in Chinese populations.
Because the g.CNV-30450 is located in the promoter of MAPKAPK2, we therefore performed luciferase assays to show whether the CNV may influence the promoter activity. Using similar methods to those described previously,8–10 we constructed two reporter genes integrated with the single copy or duplicated copies of g.CNV-30450 into pGL3 luciferase reporter (Figure 2A). Three human cell lines 16HBE, A549, and NC1-520 were cotransfected with 0.5 μg reporter and 10 ng pRL-TK plasmids. At 16 hr after transfection, we measured the luciferase activity with the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA). The transcription activities of the reporter genes with dual copies were consistently higher in 16HBE, A549, and NC1-520 (p < 0.001 for all; Figure 2B) compared with the reporter gene with a single copy of the MAPKAPK2 promoter region, suggesting that the dual copy of g.CNV-30450 may increase the transcriptional activity of MAPKAPK2.
Figure 2.
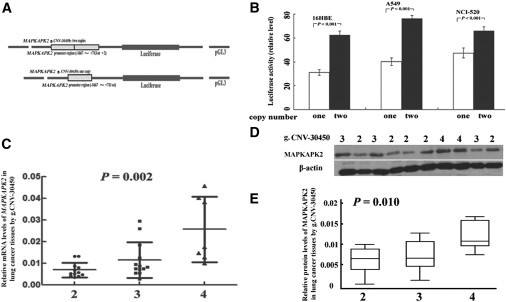
Functional Analysis of MAPKAPK2 g.CNV-30450 In Vivo and In Vitro
(A) Schematic of the two reporter-gene constructs containing the single or dual g.CNV-30450.
(B) Luciferase expression of the two constructs in lung cells (16HBE, A549, and NCI-520). Student’s t test was used to test the differences in the expression levels of different constructs.
(C) The relative mRNA level of MAPKAPK2 by g.CNV-30450 genotype.
(D) The MAPKAPK2 and β-actin protein band in lung cancer tissues.
(E) MAPKAPK2 expressions by g.CNV-30450 genotype. Columns, mean from three independent experiments; bars, SD. The copy number of g.CNV-30450 was from blood DNA. The dual copies of g.CNV-30450 had significantly higher luciferase activity than the single copy; thus, the four copies of MAPKAPK2 g.CNV-30450 had significantly increased expressions (mean ± SD) compared with the two or three copies.
We next detected the MAPKAPK2 expression in 32 primary lung cancer tissues at both mRNA and protein levels with SYBR-Green real-time PCR and western blotting, respectively, as described previously.10 The mRNA expression levels of MAPKAPK2 and an internal reference β-actin were detected with the ABI Prism 7500 sequence detection system (Applied BioSystems) with self-designed primers (Table S4). The MAPKAPK2 protein-expression levels normalized to that of β-actin were detected with the Phototope - Horseradish Peroxidase Western Blot detection kit (Cell Signaling Technology, Danvers, MA, USA) and were semiquantified with the GeneTools software (version 4.01, Syngene, Cambridge). Genotyping of the g.CNV-30450 in samples of blood and tissue DNA were detected with the TaqMan qPCR method and validated with the AccuCopy assay. Expression levels of both mRNA and protein of MAPKAPK2 increased as the copy numbers of g.CNV-30450 increased, and the four-copy genotype was associated with significantly increased MAPKAPK2 mRNA levels (ANOVA test: p = 0.002; linear-regression test: p < 0.001; Figure 2C) and protein levels (ANOVA test: p = 0.010; linear-regression test: p = 0.005; Figures 2D and 2E) in these 32 individuals with lung cancer. Additionally, we used another 62 paraffin sections of primary lung cancer tissues from the Second Affiliated Hospital of Guangzhou Medical University for immunohistochemistry to detect the MAPKAPK2 expressions in situ by using the Image-Pro Plus software (Media Cybernetics, Bethesda, MD, USA) to score the MAPKAPK2 cytoplasmic expressions. Consistently, individuals carrying the four-copy genotype had a significantly higher MAPKAPK2-expression score than that of those with two- and three- copy genotypes (ANOVA test: p = 0.016; linear-regression test: p = 0.004; Figure S5). These findings further suggested that the g.CNV-30450 is functional by inducing MAPKAPK2 expression. Furthermore, it appeared that the four-copy genotype may be responsible for the observed CNV in the somatic genome (linear-regression test: p = 9.95 × 10−5; Figure S6).
Previous studies showed that overexpression of P38/MAPKAPK2 pathway signals was common in various cancers.7,16 When phosphorylated and activated by the P38 pathway, the active MAPKAPK2 participates in multiple roles in cell apoptosis,17 cell cycle,18 adhesion,19 movement,20 and response to oxidative stress.6 The MAPKAPK2 could phosphorylate several other important cancer-related proteins, such as Cdc25B/C,21 tuberin (TSC2 [MIM 191092]),18,19 Polo-like kinase 1 (Plk1 [MIM 602098]),22 HSP27,6,20,23 and especially the mRNA-AU-rich-element (ARE)-binding proteins (i.e., TTP and hnRNP A0 [MIM 609409]), which further regulate mRNA stability of multiple genes, such as TNF-α [MIM 191160], CCND1[MIM 168461], Plk3 [MIM 602913], c-Fos [MIM 164810], c-Myc [MIM 190080], and MMP-1 [MIM 120353],24,25 affecting cell metabolism, differentiation, and carcinogenesis. Meanwhile, MAPKAPK2 can promote tumor growth and metastasis through the activation of LIM in VEGF-induced angiogenesis progress.26 MAPKAPK2 also helps tumor cells in resisting chemotherapy by regulating DNA repair through operating checkpoint kinase 1 (Chk1 [MIM 603078]) and Chk2 (MIM 603078) in response to cisplatin, camptothecin, and doxorubicin.27 Genetic variations in MAPKAPK2, especially in its promoter region, may influence the gene’s function; therefore, it is biologically plausible that g.CNV-30450 could increase risk and cause poor prognosis in individuals with lung cancer by enhancing the expression of MAPKAPK2, as demonstrated in the present study.
At present, several CNVs have been reported to be associated with human diseases,28 including some systemic autoimmune diseases29,30 and cancers.31–33 CNVs in the genome may cause a greater adverse effect on the host genes’ function than other genetic variants, such as SNPs, and thus may provide a more effective tool to predict the risk and prognosis of diseases. In the present study, we found that the four copies of g.CNV-30450 in MAPKAPK2 are associated with an increased risk and poor prognosis for lung cancer in Chinese populations, which is probably caused by an underlying mechanism that increases the expression of MAPKAPK2 in vivo, and thus promotes tumorigenesis. In conclusion, our findings suggest that the g.CNV-30450 in MAPKAPK2 may be a biomarker for susceptibility to and prognosis for lung cancer.
Acknowledgments
We thank Qingyi Wei (Department of Epidemiology, University of Texas MD Anderson Cancer Center) for careful scientific editing. This study was supported by the National Natural Scientific Foundation of China grants 30671813, 30872178, 81072366 (J.L.), and 30972540 (B.L.), and partly by 30872142 (W.J.) and 81001278 (Y.Z.); Guangdong Provincial Scientific Research grants 8251018201000005 (J.L.) and 2008B060600008 (B.L.); Guangdong Provincial High Level Experts grant 2010-79; Changjiang Scholars and Innovative Research Team in University grant IRT0961; and Guangdong Natural Science Foundation team grant 10351012003000000 (J.L.). We thank Zhanhong Xie, Wanmin Zeng, and Ling Liu for their assistance in recruiting the subjects and Fuman Qiu and Yehua Liu for their laboratory assistance.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
Applied BioSystems, http://www.appliedbiosystems.com/absite/us/en/home.html
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org
Database of Genomic Variants, http://projects.tcag.ca/variation/?source=hg18
References
- 1.Siegel R., Ward E., Brawley O., Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J. Clin. 2011;61:212–236. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 2.Spitz M.R., Hong W.K., Amos C.I., Wu X., Schabath M.B., Dong Q., Shete S., Etzel C.J. A risk model for prediction of lung cancer. J. Natl. Cancer Inst. 2007;99:715–726. doi: 10.1093/jnci/djk153. [DOI] [PubMed] [Google Scholar]
- 3.Maruyama J., Naguro I., Takeda K., Ichijo H. Stress-activated MAP kinase cascades in cellular senescence. Curr. Med. Chem. 2009;16:1229–1235. doi: 10.2174/092986709787846613. [DOI] [PubMed] [Google Scholar]
- 4.Ronkina N., Kotlyarov A., Gaestel M. MK2 and MK3—a pair of isoenzymes? Front. Biosci. 2008;13:5511–5521. doi: 10.2741/3095. [DOI] [PubMed] [Google Scholar]
- 5.ten Hove W., Houben L.A., Raaijmakers J.A., Bracke M., Koenderman L. Differential regulation of TNFalpha and GM-CSF induced activation of P38 MAPK in neutrophils and eosinophils. Mol. Immunol. 2007;44:2492–2496. doi: 10.1016/j.molimm.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 6.Jackson R.M., Garcia-Rojas R. Kinase activity, heat shock protein 27 phosphorylation, and lung epithelial cell glutathione. Exp. Lung Res. 2008;34:245–262. doi: 10.1080/01902140802022500. [DOI] [PubMed] [Google Scholar]
- 7.Stearman R.S., Dwyer-Nield L., Zerbe L., Blaine S.A., Chan Z., Bunn P.A., Jr., Johnson G.L., Hirsch F.R., Merrick D.T., Franklin W.A. Analysis of orthologous gene expression between human pulmonary adenocarcinoma and a carcinogen-induced murine model. Am. J. Pathol. 2005;167:1763–1775. doi: 10.1016/S0002-9440(10)61257-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang L., Li Y., Cheng M., Huang D., Zheng J., Liu B., Ling X., Li Q., Zhang X., Ji W. A functional polymorphism at microRNA-629-binding site in the 3′-untranslated region of NBS1 gene confers an increased risk of lung cancer in Southern and Eastern Chinese population. Carcinogenesis. 2012;33:338–347. doi: 10.1093/carcin/bgr272. [DOI] [PubMed] [Google Scholar]
- 9.Huang B., Liu B., Yang L., Li Y., Cheng M., Huang D., Wang H., Zhang X., Zheng J., Li Q. Functional genetic variants of c-Jun and their interaction with smoking and drinking increase the susceptibility to lung cancer in southern and eastern chinese. Int. J. Cancer. 2012;131:E744–E758. doi: 10.1002/ijc.27407. [DOI] [PubMed] [Google Scholar]
- 10.Lu J., Yang L., Zhao H., Liu B., Li Y., Wu H., Li Q., Zeng B., Wang Y., Ji W., Zhou Y. The polymorphism and haplotypes of PIN1 gene are associated with the risk of lung cancer in Southern and Eastern Chinese populations. Hum. Mutat. 2011;32:1299–1308. doi: 10.1002/humu.21574. [DOI] [PubMed] [Google Scholar]
- 11.Mayo P., Hartshorne T., Li K., McMunn-Gibson C., Spencer K., Schnetz-Boutaud N. CNV analysis using TaqMan copy number assays. Curr. Protoc. Hum. Genet. 2010;67 doi: 10.1002/0471142905.hg0213s67. 2.13.1–2.13.10. [DOI] [PubMed] [Google Scholar]
- 12.Eckel-Passow J.E., Atkinson E.J., Maharjan S., Kardia S.L., de Andrade M. Software comparison for evaluating genomic copy number variation for Affymetrix 6.0 SNP array platform. BMC Bioinformatics. 2011;12:220. doi: 10.1186/1471-2105-12-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akaike H. Information theory as an extension of the maximum likelihood principle. In: Petrov B.N., Csaki F., editors. Second International Symposium on Information Theory. Akademiai Kiado; Budapest: 1973. pp. 267–281. [Google Scholar]
- 14.Gabeau-Lacet D., Engler D., Gupta S., Scangas G.A., Betensky R.A., Barker F.G., 2nd, Loeffler J.S., Louis D.N., Mohapatra G. Genomic profiling of atypical meningiomas associates gain of 1q with poor clinical outcome. J. Neuropathol. Exp. Neurol. 2009;68:1155–1165. doi: 10.1097/NEN.0b013e3181ba3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jansen M., Mohapatra G., Betensky R.A., Keohane C., Louis D.N. Gain of chromosome arm 1q in atypical meningioma correlates with shorter progression-free survival. Neuropathol. Appl. Neurobiol. 2012;38:213–219. doi: 10.1111/j.1365-2990.2011.01222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pomérance M., Quillard J., Chantoux F., Young J., Blondeau J.P. High-level expression, activation, and subcellular localization of p38-MAP kinase in thyroid neoplasms. J. Pathol. 2006;209:298–306. doi: 10.1002/path.1975. [DOI] [PubMed] [Google Scholar]
- 17.Seisenbacher G., Hafen E., Stocker H. MK2-dependent p38b signalling protects Drosophila hindgut enterocytes against JNK-induced apoptosis under chronic stress. PLoS Genet. 2011;7:e1002168. doi: 10.1371/journal.pgen.1002168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y., Inoki K., Vacratsis P., Guan K.L. The p38 and MK2 kinase cascade phosphorylates tuberin, the tuberous sclerosis 2 gene product, and enhances its interaction with 14-3-3. J. Biol. Chem. 2003;278:13663–13671. doi: 10.1074/jbc.M300862200. [DOI] [PubMed] [Google Scholar]
- 19.Chakraborty S., Mohiyuddin S.M., Gopinath K.S., Kumar A. Involvement of TSC genes and differential expression of other members of the mTOR signaling pathway in oral squamous cell carcinoma. BMC Cancer. 2008;8:163. doi: 10.1186/1471-2407-8-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu R., Kausar H., Johnson P., Montoya-Durango D.E., Merchant M., Rane M.J. Hsp27 regulates Akt activation and polymorphonuclear leukocyte apoptosis by scaffolding MK2 to Akt signal complex. J. Biol. Chem. 2007;282:21598–21608. doi: 10.1074/jbc.M611316200. [DOI] [PubMed] [Google Scholar]
- 21.Manke I.A., Nguyen A., Lim D., Stewart M.Q., Elia A.E., Yaffe M.B. MAPKAP kinase-2 is a cell cycle checkpoint kinase that regulates the G2/M transition and S phase progression in response to UV irradiation. Mol. Cell. 2005;17:37–48. doi: 10.1016/j.molcel.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 22.Tang J., Yang X., Liu X. Phosphorylation of Plk1 at Ser326 regulates its functions during mitotic progression. Oncogene. 2008;27:6635–6645. doi: 10.1038/onc.2008.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo H., Bai Y., Xu P., Hu Z., Liu L., Wang F., Jin G., Wang F., Deng Q., Tu Y. Functional promoter -1271G>C variant of HSPB1 predicts lung cancer risk and survival. J. Clin. Oncol. 2010;28:1928–1935. doi: 10.1200/JCO.2009.24.4954. [DOI] [PubMed] [Google Scholar]
- 24.Maitra S., Chou C.F., Luber C.A., Lee K.Y., Mann M., Chen C.Y. The AU-rich element mRNA decay-promoting activity of BRF1 is regulated by mitogen-activated protein kinase-activated protein kinase 2. RNA. 2008;14:950–959. doi: 10.1261/rna.983708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ronkina N., Menon M.B., Schwermann J., Tiedje C., Hitti E., Kotlyarov A., Gaestel M. MAPKAP kinases MK2 and MK3 in inflammation: complex regulation of TNF biosynthesis via expression and phosphorylation of tristetraprolin. Biochem. Pharmacol. 2010;80:1915–1920. doi: 10.1016/j.bcp.2010.06.021. [DOI] [PubMed] [Google Scholar]
- 26.Kobayashi M., Nishita M., Mishima T., Ohashi K., Mizuno K. MAPKAPK-2-mediated LIM-kinase activation is critical for VEGF-induced actin remodeling and cell migration. EMBO J. 2006;25:713–726. doi: 10.1038/sj.emboj.7600973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reinhardt H.C., Aslanian A.S., Lees J.A., Yaffe M.B. p53-deficient cells rely on ATM- and ATR-mediated checkpoint signaling through the p38MAPK/MK2 pathway for survival after DNA damage. Cancer Cell. 2007;11:175–189. doi: 10.1016/j.ccr.2006.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCarroll S.A., Altshuler D.M. Copy-number variation and association studies of human disease. Nat. Genet. 2007;39(7, Suppl):S37–S42. doi: 10.1038/ng2080. [DOI] [PubMed] [Google Scholar]
- 29.Chen J.M., Masson E., Le Maréchal C., Férec C. Copy number variations in chronic pancreatitis. Cytogenet. Genome Res. 2008;123:102–107. doi: 10.1159/000184697. [DOI] [PubMed] [Google Scholar]
- 30.Colobran R., Pedrosa E., Carretero-Iglesia L., Juan M. Copy number variation in chemokine superfamily: the complex scene of CCL3L-CCL4L genes in health and disease. Clin. Exp. Immunol. 2010;162:41–52. doi: 10.1111/j.1365-2249.2010.04224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Diskin S.J., Hou C., Glessner J.T., Attiyeh E.F., Laudenslager M., Bosse K., Cole K., Mossé Y.P., Wood A., Lynch J.E. Copy number variation at 1q21.1 associated with neuroblastoma. Nature. 2009;459:987–991. doi: 10.1038/nature08035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang L., Yu D., Wu C., Zhai K., Jiang G., Cao G., Wang C., Liu Y., Sun M., Li Z. Copy number variation at 6q13 functions as a long-range regulator and is associated with pancreatic cancer risk. Carcinogenesis. 2012;33:94–100. doi: 10.1093/carcin/bgr228. [DOI] [PubMed] [Google Scholar]
- 33.Shlien A., Malkin D. Copy number variations and cancer susceptibility. Curr. Opin. Oncol. 2010;22:55–63. doi: 10.1097/CCO.0b013e328333dca4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


