Abstract
Essential tremor (ET) is a common neurodegenerative disorder that is characterized by a postural or motion tremor. Despite a strong genetic basis, a gene with rare pathogenic mutations that cause ET has not yet been reported. We used exome sequencing to implement a simple approach to control for misdiagnosis of ET, as well as phenocopies involving sporadic and senile ET cases. We studied a large ET-affected family and identified a FUS p.Gln290∗ mutation as the cause of ET in this family. Further screening of 270 ET cases identified two additional rare missense FUS variants. Functional considerations suggest that the pathogenic effects of ET-specific FUS mutations are different from the effects observed when FUS is mutated in amyotrophic lateral sclerosis cases; we have shown that the ET FUS nonsense mutation is degraded by the nonsense-mediated-decay pathway, whereas amyotrophic lateral sclerosis FUS mutant transcripts are not.
Introduction
Essential tremor (ET [MIM 190300]) is a neurodegenerative disorder that is considered to be one of the most common adult-onset movement disorders.1,2 A recent meta-analysis that used population-based studies (n = 28) estimated that the pooled prevalence of ET (at all ages) was 0.9%2 and found that there is an increasing prevalence as age increases (the prevalence is 4.6% for individuals ≥ 65 years old).2 In 1998, the Movement Disorder Society created a consensus statement defining ET as a bilateral, mainly symmetrical postural or motion tremor that primarily affects the upper limbs.3 The heterogeneity of tremors (including their clinical expression, therapeutic response, pathology, and etiology) has, however, been suggested to underlie the common misdiagnosis of ET; 37%–50% of individuals previously diagnosed with ET are reportedly misdiagnosed.4,5
There are three subtypes of ET—hereditary, sporadic, and senile6—and most studies indicate that ET is a hereditary disorder in 50%–70% of affected individuals (and presumably has autosomal-dominant inheritance).7 Studies of large ET-affected families have shown that a family history of ET typically means an early age of onset, and the phenotype is usually fully penetrant by the age of 65 years. Thus, a hereditary-ET-affected family is defined as having at least two immediate family members affected with the disease and at least two family members diagnosed before 65 years of age.6
Linkage studies on families have identified three ET-associated loci (ETM1 [MIM 190300], ETM2 [MIM 602134], and ETM3 [MIM 611456]), but no gene with causative mutations has been reported.8–10 A common variation, c.312G>A, in dopamine D3 receptor (DRD3 [MIM 126451]) within the ETM1 locus has been suggested to be a susceptibility factor for ET.11 However, this association has not been consistently replicated. More recently, common sequence variants in LINGO1 (MIM 609791) have been associated with ET,12 but the significance of these findings remains unclear as well. Exome sequencing has recently been validated as a method of identifying rare coding variants that cause monogenic disease.13 This approach allows the use of only a few selected affected individuals and controls for the identification of disease-associated genes, which has been noted to be particularly beneficial when large families are not available for linkage analysis.13 Many large ET-affected families have been reported in the literature, but their usage in classical linkage-based gene-discovery efforts has been hindered by a number of factors. First, ET is a disorder that has been reported to be overdiagnosed;4,5 therefore, it is very likely that some unaffected members in large families are misdiagnosed as affected individuals, which would hinder linkage studies. Second, the high prevalence of ET increases the risk that a sporadic or senile case (or cases) exists in large families; such cases would be phenocopies that would also hinder linkage studies. Therefore, we hypothesized that exome sequencing could help identify rare penetrant variants that cause hereditary ET through the selection of a small number of “definitely” affected individuals (with an early age of onset) from ET-affected families; this would thus minimize the clinical barriers associated with ET.
Subjects and Methods
A detailed version of this study’s methodology is supplied in the Supplemental Data, available online. In brief, one ET-affected family, FET1, was chosen for study (Figure S1). During clinical assessment, ET was diagnosed as either “definite,” “probable,” or “possible”3,14 (Figure S2). The genomic DNA from four individuals with a “definite” ET diagnosis and an age of onset before 40 years and from a clinically unaffected married-in family member was captured with Agilent SureSelect all exome kits and sequenced with an Applied Biosystems SOLiD apparatus. Ethics approval for the recruitment and genetic analysis of ET-affected individuals and their families was granted by the following institutes: the Centre de recherche du Centre hospitalier de l’Université de Montréal (project ND043076), the Centre hospitalier affilié universitaire de Québec (project PEJ-280), and the Sainte-Justine University Hospital Center (project 2352).
Results
Exome Sequencing
After a list of exome-sequencing variants was generated for each family member, segregation analysis revealed a list of six exome-sequencing variants (three synonymous, two nonsynonymous, and one nonsense) that were shared exclusively by “definitely” affected individuals from family FET1 and that had a sequencing quality score greater than 50; capture efficiency for each individual was comparable between individuals (Figure S3 and Table S1). After Sanger sequencing, only one of those six variants proved to be legitimate—a nonsense mutation in FUS/TLS (fused in sarcoma/translocated in liposarcoma [MIM 137070]) (Figure 1 and Table 1). This variant, c.868C>T (in exon 9), corresponding to a stop mutation at p.Gln290∗ (RefSeq accession number NM_004960.3), had an average sequencing quality of 54.7, coverage of 133×, and mutation frequency of 36%. Furthermore, p.Gln290∗ was not detected in a cohort of 450 ethnically-matched control individuals (Table 1). Additionally, no truncating mutation has been identified in the ∼5,000 exomes for which variants are presently listed in the National Heart, Lung, and Blood Institute (NHLBI) Grand Opportunity (GO) Exome Sequencing Project (ESP) database. In fact, this mutation is only the third nonsense mutation ever reported in FUS; the other two (p.Arg495∗ and p.Gln519∗) cause amyotrophic lateral sclerosis (ALS [MIM 608030]).15,16 Amino acid 290 is located in the nuclear export signal (NES) motif of FUS (Figure 1).
Figure 1.
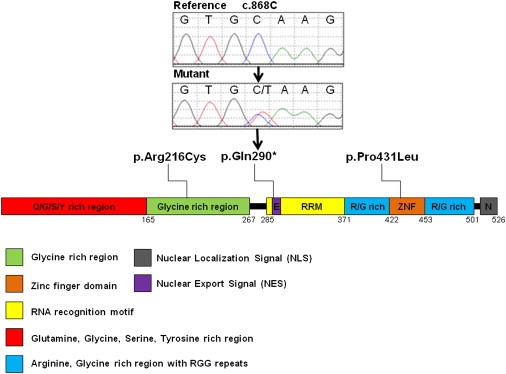
A Schematic of ET Variants within FUS
Table 1.
Rare FUS Variants that Have Been Identified in ET-Affected Individuals
| Identification | Exon |
Mutation Nomenclature |
Ethnically Matched Control Cohort |
||||
|---|---|---|---|---|---|---|---|
| Coding (NM_004960.3) | Genomic (hg19) | Protein | Individuals Screened | Alleles with Variant | Allele Frequency | ||
| Through exome sequencing | exon 9 | c.868C>T | chr16: g.31200479C>T | p.Gln290∗ | 450 | 0/900 | 0% |
| During the subsequent screening of an ET cohort | exon 6 | c.646C>T | chr16: g.31196382C>T | p.Arg216Cys | 450 | 1/900 | 0.1% |
| exon 12 | c.1292C>T | chr16: g.31201719C>T | p.Pro431Leu | 450 | 0/900 | 0% | |
Segregation Analysis of FUS c.868C>T in FET1 Family Members
All “definitely” and “probably” affected individuals from FET1 carried FUS c.868C>T (Figure S1 and Table S2). However, only 54% (7/13) of “possibly” affected individuals had the mutation. Of the seven “possibly” affected individuals with the mutation, three had an onset before 40 years of age and four had an unknown age of onset. Overall, the age of disease onset of individuals who harbored the mutation was variable and life expectancy was normal; notably, numerous individuals were over the average age of ALS onset and had no symptoms of the disease (Figure S1 and Table S2). Out of 13 clinically unaffected individuals, one (individual III:38, who was 24 years of age at the time of clinical observation; Figure S1) was mutation positive and might harbor a nonpenetrant variant (Figure S1 and Table S2).
FUS Screening in an ET Cohort
After screening of 270 ET cases for FUS coding variants, two rare missense variants were detected (Table 1). A c.1292C>T (p.Pro431Leu) variant was observed in exon 12 from a case with familial ET and was absent from our control cohort (Table 1). Additional family members were not available for segregation analysis. The proline at position 431 is a highly conserved amino acid (Figure S4) that is located in the zinc finger domain of FUS (Figure 1), and bioinformatics prediction software programs MutationTaster, SIFT, and Polyphen predict the substitution of a proline for a leucine to be disease causing, intolerable, and probably damaging, respectively. Interestingly, this variant, which is located in the last nucleotide of exon 12, is predicted to affect the splicing donor site of intron 12 (the donor-site score efficiency decreases from 0.90 to 0.61 with the variant). However, no splicing abnormality was detected with the use of cDNA prepared from lymphoblastoid cells derived from the affected individual with the c.1292C>T variant.
In exon 6, a c.646C>T (p.Arg216Cys) variant was detected in two ET cases, one with familial ET and the other a sporadic case (Table 1). With regard to the familial case, additional family members were not available for segregation analysis. A recent study reported this variant in a sporadic ALS case and not in any of their 500 control individuals; no comment was made regarding the presence of a tremor.17 The same report stated that three in silico prediction programs—PolyPhen, SNAP, and PMUT—predicted p.Arg216Cys to be damaging.17 SIFT also predicts this variant to be intolerable, and MutationTaster suggests that it is disease causing; the arginine at amino acid 216 is highly conserved (Figure S4) and is located in the glycine-rich domain (Figure 1). This variant was, however, detected in our control cohort (1/900 alleles) (Table 1). Additionally, it did not lead to splicing abnormalities in lymphoblastoid cells derived from an individual with this variant. Interestingly, this nucleotide variation disrupts a CpG site.
Functional Studies
Overall, the definitive mechanism by which FUS mutations cause ALS remains elusive; therefore, we used lymphoblastoid cell lines derived from ET- and ALS-affected individuals to compare mRNA expression in order to gain insights into the distinct mechanisms likely to be involved in the two diseases (Figure 2). Quantitative RT-PCR showed that the overall expression of FUS in the ET-affected individuals who carry FUS c.868C>T (p.Gln290∗) was lower than that of ALS-affected individuals with FUS mutations (Figure 2A); additionally, the expression of FUS in ET-affected individuals who carry c.868C>T increased 3.6-fold (p value = 0.02) upon treatment with the translation inhibitor puromycin, an antibiotic that suppresses nonsense-mediated decay (NMD) and facilitates the observation of mRNA with nonsense mutations18 (Figure 2A). These observations led to the hypothesis that mutant mRNA in cells with the ET FUS nonsense mutation is degraded by the NMD pathway and that ALS FUS mutant transcripts are not. Of note is that ALS mutations generally cluster at the 3′ end of FUS and would thus probably escape NMD; in fact, recent observations support the notion that the NMD pathway is not a major determinant of either toxicity or expression of ALS FUS mutants.19 To test our hypothesis, after the lymphoblastoid cells of five ET-affected individuals who carry the c.868C>T mutation and three ALS-affected individuals with three different FUS mutations were treated with puromycin, we prepared nonquantitative FUS RT-PCR of cDNA from treated and untreated cells and sequenced the products from each set. Upon examination of the various sequence traces, the mutated allele from the ET-affected individuals could only be observed in cells treated with puromycin (Figure 2B). This result suggests that a substantial fraction of ET mutant RNA might be degraded through NMD. In contrast, the mutated alleles of ALS-affected individuals were clearly observed in both untreated and puromycin-treated cells (Figure 2B). This observation prompted us to quantitatively measure the expression of the two separate c.868 FUS alleles in cells from the ET-affected individuals. In the absence of puromycin, the expression of the mutant RNA was below the automatic threshold of detection of the assay (Figure 2C and Figure S5). However, in the presence of puromycin, the mutant RNA was observed, specifically at approximately 25% of the level of expression of the wild-type allele; this level is comparable to the allelic ratios observed in the sequence traces (Figures 2B and 2C).
Figure 2.
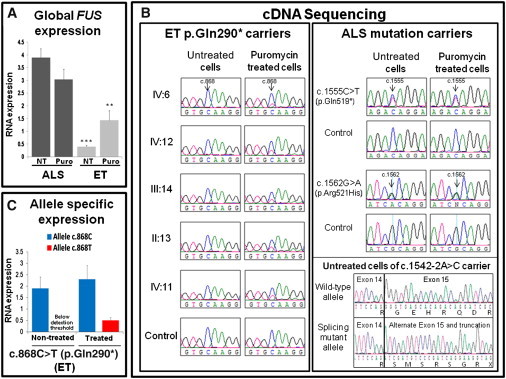
Expression of FUS mRNA Carrying ET and ALS Mutations
(A) Lymphoblastoid cells derived from five ET-affected individuals (individuals II:13, III:14, IV:6, IV:11, and IV:12 in Figure S1) who carry the FUS c.868C>T (p.Gln290∗) mutation and from ALS-affected individuals expressing three different FUS mutations (c.1555C>T [p.Gln519∗], c.1562G>A [p.Arg521His], and c.1542-2A>C) were treated with the protein-synthesis inhibitor puromycin (300 μg for 6 hr). After this, treated cells were harvested, and total RNA was prepared (Trizol extraction, Invitrogen) alongside the total RNA of untreated cells. Quantitative RT-PCR reactions were then performed with a Taqman probe specific to FUS (∗∗∗p value = 6.47 × 10−5 [comparing the difference in mRNA expression in ET and ALS untreated cells]; ∗∗p value = 0.03 [comparing the difference in mRNA expression in ET and ALS treated cells]). For each group of affected (ET and ALS) individuals, the average levels of expression are plotted. t tests were used for statistics, and error bars correspond to the standard error of the mean (SEM).
(B) Standard nonquantitative RT-PCR reactions were prepared with the RNA used in (A) and primers in the flanking exons (or UTRs) of each mutation. Amplified cDNA was sequenced, and whereas the ALS mutations were seen independently of a puromycin treatment, the ET mutation could only be seen in puromycin-treated cells.
(C) Quantitative allele-specific expression measurements of the same five FUS c.868C>T ET individuals from above were made with a set of Taqman custom-designed probes and primers for the wild-type (c.868C) and mutant (c.868T) alleles. After the puromycin treatment, the expression of the mutant allele became detectable. The specificity of the custom probe for the c.868C>T transcript is demonstrated in Figure S5. All expression levels were normalized with the human 18S ribosomal RNA (rRNA) gene and were calculated in comparison to the average level of expression of two healthy controls. t tests were used for statistics, and error bars correspond to SEM.
Discussion
Traditional gene-discovery approaches (linkage studies and homozygosity mapping) have successfully discovered causal variants of monogenic disorders over the past two decades.20 However, not all monogenic disorders accommodate well to such studies, and recently, exome sequencing has been recognized as a way of identifying rare causal variants for such disorders.13 The exome-sequencing approach has thus facilitated gene discovery of monogenic disorders, the majority of which, however, have an autosomal-recessive mode of inheritance.21 Nonetheless, the adoption of different variant identification strategies has allowed the identification of the causative variants for a smaller number of autosomal-dominant disorders.22–25 In this study, we report on a modified genetic approach for overcoming clinical barriers associated with ET, and this approach has allowed us to identify FUS mutations that cause ET.
We identified FUS c.868C>T (p.Gln290∗) as the pathogenic variant that causes ET in family FET1. Although we did not obtain ideal exome-sequence coverage by using the Applied Biosystems SOLiD technology, we were fortunate to identify this extremely rare nonsense FUS variant. FUS c.868C>T fully segregated with the disease in individuals with a “definite” and “probable” ET diagnosis. However, it was only present in 54% of the “possibly” affected individuals, supporting our initial belief that ET genetic studies still have to overcome many clinical barriers, such as distinguishing sporadic phenocopies from the hereditary cases within a family. In fact, past attempts at identifying the mutant gene in FET1 failed as a result of this manner. During a previous linkage study that was performed on FET1 family members, the highest two-point LOD score obtained was on chromosome 16 at marker D16S3034 (Z = 2.73), and haplotype construction showed segregation of a disease haplotype (Rouleau, unpublished data). FUS is within this disease region, but a recombination event in a “possibly” affected individual (individual IV:10, who does not have the FUS mutation [Figure S1]) was used for reducing the disease region and excluded FUS as a positional candidate gene.
Two additional rare variants, c.1292C>T (p.Pro431Leu) and c.646C>T (p.Arg216Cys), were identified in FUS after screening of a cohort of ET cases. Both variants are highly conserved and predicted to be pathogenic by bioinformatics software. The substitution of a proline for a leucine at position 431 was not detected in our control cohort. Despite the fact that this variant was predicted to have an effect on splicing, a splicing defect was not apparent in lymphoblastoid cells of the individual with the c.1292C>T variant. However, given that it was predicted to affect the efficiency of the intron 12 donor site by approximately 30%, perhaps such a marginal decrease was undetectable in our experiments in which lymphoblastoid cells were used and only a low level of aberrant messenger influences the disease onset. An alternative explanation might be that the predicted splicing defect is specific, or more frequent, in cells from the nervous system and can thus not be detected with the material that was currently available. The c.646C>T variant was detected in two ET probands but has also been reported in one sporadic ALS case17 and in one control in our study, corresponding to a frequency that is plausible for an ET-causing variant considering the prevalence of ET (0.9% for all ages). This variant is in exon 6 of FUS and in the glycine-rich region of FUS. Interestingly, this arginine residue is one of two FUS arginines that are physiologically dimethylated,17 which is a modification that can be important for protein shuttling and signal transduction, particularly in proteins involved in splicing.26 A splicing defect was not apparent in lymphoblastoid cells of the c.646C>T mutant, and, again, such an event might also be specific, or more frequent, in cells from the nervous system. Furthermore, this variant disrupts a CpG site, and an epigenetic effect on gene expression (through demethylation) could thus possibly explain disease pathogenesis. Also, methylation of cytosines at CpG sites and subsequent deamination is a common mechanism of recurrent mutations and could thus explain the slightly higher occurrence of c.646C>T in this cohort. Intriguingly, the lymphoblastoid cells of individuals with each ET missense variant had a significant lower overall expression of the FUS mRNA than did those of ALS-affected individuals with FUS mutations (without treatment, the c.646C>T p value = 8.88 × 10−4 and the c.1292C>T p value = 0.013). They also showed a significant increase in overall FUS mRNA expression after puromycin treatment: a 2.9-fold increase for c.646C>T (p value = 0.01) and a 3.2-fold increase for c.1292C>T (p value = 0.04). These observations are similar to the total expression of FUS in the c.868C>T mutant cells and need further exploration. Overall, FUS variants appear to be a rare cause of ET given that they explain, at most, 1.5% of our ET cohort.
Interestingly, FUS mutations were recognized as a cause of ALS in 2009;27,28 they explain approximately 4% of familial cases.29 ALS is an adult-onset neurodegenerative disorder characterized by the premature loss of upper and lower motor neurons and is usually fatal within the first 5 years of disease onset. Over 30 ALS FUS mutations have been identified; most are missense mutations that are inherited in an autosomal-dominant fashion and clustered in the extreme C terminus or the glycine-rich region of FUS.29
FUS is a 526 amino acid RNA binding protein and is involved in multiple steps of RNA processing.29 It is mainly a nuclear protein, but when it is mutated in ALS, it forms cytoplasmic inclusions in neurons and glial cells.30 Interestingly, the FUS nuclear localization signal (NLS), which normally targets a protein located in the cytoplasm for import to the nucleus, is predicted to be located in the conserved C terminus of the protein. This is precisely where the vast majority of ALS mutations cluster; thus, it is possible that these mutations disrupt the NLS, which explains the irregular distribution of FUS in the cytoplasm, a theory recently explored by Ito et al.31 Overall, the mechanism by which FUS mutations cause ALS is unknown—the mutations might gain an aberrant function or toxic property or lose a particular function. Nonetheless, several reports are now suggesting that the FUS proteinopathy underlying ALS is unlikely to involve a loss of the protein’s function but instead involve the gain of toxic effects.31,32 Only two FUS nonsense mutations have been reported in ALS, and both affect regions located in the C terminus (p.Arg495∗ and p.Gln519∗) of the protein and are associated with an extreme ALS phenotype.15,16 It is important to note that FUS truncation mutations have never been reported in control individuals.
The FUS mRNA with the nonsense mutation (c.868C>T) that segregates in FET1 appears to be mainly degraded by NMD, which suggests a loss-of-function disease mechanism. On the other hand, our data suggest that mRNA from FUS mutant ALS cells escapes NMD, which confirms that the NMD pathway is not a major determinant of either toxicity or expression of ALS FUS mutants.33 These preliminary expression data suggest differences in disease mechanism, but elucidating true disease mechanisms will require further investigation. ET variant p.Gln290∗ is located in the predicted NES of the protein, i.e., amino acids 289–298, which is a short amino acid sequence that targets the protein for export from the cell nucleus to the cytoplasm through the nuclear pore complex (the opposite effect of a NLS). No ALS mutations have been reported in the NES, as is the case for the zinc finger domain, where the p.Pro431Leu variation is located. Additionally, upon analysis of the RNA-Seq data from Illumina’s Human BodyMap 2.0 project (which was assembled by the Broad Institute with the program Scripture and the “Brain_R” library), all FUS isoforms appear to encompass the position of both ET and ALS mutations; thus, disease-specific isoforms are not likely to exist. Notably, Fus-knockout mice have been established and found to die shortly after birth.34 Heterozygous Fus+/− animals have been reported to be phenotypically undistinguishable from Fus+/+ animals;34 however, Fus+/− animals were not extensively studied because, in essence, they were primarily used for the generation of Fus−/− animals. Follow-up studies of Fus+/− mice are warranted. Interestingly, a motor phenotype has been observed in a recent publication of a fus-knockout zebrafish.35
Comorbidity of ALS with frontotemporal lobe dementia (FTD [MIM 600274]) and parkinsonism (PD [MIM 168601]) has suggested that these conditions might share a common pathogenesis; overlapping FUS mutations have now been found in individuals with ALS/FTD, FTD and ALS/PD;33,36 FUS inclusions have also been seen in FTD-affected individuals.29 Interestingly, individuals affected by ET have been reported to have an increased genetic risk of developing Parkinson disease;37 however, there is no direct link between ET and ALS, although it might be noteworthy that a rapid voice tremor has been reported as an extrapyramidal symptom in some cases of ALS.38 Overall, FUS seems to be a functionally important protein in neuronal cells.
Acknowledgments
N.D.M. is the recipient of a Claude Laberge fellowship from the Réseau de Médecine Génétique Appliqué (RMGA). The RMGA also supports the bioinformatics analytical team of G.A.R. G.A.R. holds the Canada Research Chair in Genetics of the Nervous System. This work was supported by the Chaire Jeanne-et-J.-Louis-Lévesque en Génétique des Maladies du Cerveau de l’Université de Montréal.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
NHLBI GO Exome Sequencing Project, https://esp.gs.washington.edu/drupal/
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org
References
- 1.Erickson-Davis C.R., Faust P.L., Vonsattel J.P., Gupta S., Honig L.S., Louis E.D. “Hairy baskets” associated with degenerative Purkinje cell changes in essential tremor. J. Neuropathol. Exp. Neurol. 2010;69:262–271. doi: 10.1097/NEN.0b013e3181d1ad04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louis E.D., Ferreira J.J. How common is the most common adult movement disorder? Update on the worldwide prevalence of essential tremor. Mov. Disord. 2010;25:534–541. doi: 10.1002/mds.22838. [DOI] [PubMed] [Google Scholar]
- 3.Deuschl G., Bain P., Brin M., Ad Hoc Scientific Committee Consensus statement of the Movement Disorder Society on Tremor. Mov. Disord. 1998;13(Suppl 3):2–23. doi: 10.1002/mds.870131303. [DOI] [PubMed] [Google Scholar]
- 4.Jain S., Lo S.E., Louis E.D. Common misdiagnosis of a common neurological disorder: How are we misdiagnosing essential tremor? Arch. Neurol. 2006;63:1100–1104. doi: 10.1001/archneur.63.8.1100. [DOI] [PubMed] [Google Scholar]
- 5.Schrag A., Münchau A., Bhatia K.P., Quinn N.P., Marsden C.D. Essential tremor: An overdiagnosed condition? J. Neurol. 2000;247:955–959. doi: 10.1007/s004150070053. [DOI] [PubMed] [Google Scholar]
- 6.Deuschl G., Elble R. Essential tremor—neurodegenerative or nondegenerative disease towards a working definition of ET. Mov. Disord. 2009;24:2033–2041. doi: 10.1002/mds.22755. [DOI] [PubMed] [Google Scholar]
- 7.Deng H., Le W., Jankovic J. Genetics of essential tremor. Brain. 2007;130:1456–1464. doi: 10.1093/brain/awm018. [DOI] [PubMed] [Google Scholar]
- 8.Gulcher J.R., Jónsson P., Kong A., Kristjánsson K., Frigge M.L., Kárason A., Einarsdóttir I.E., Stefánsson H., Einarsdóttir A.S., Sigurthoardóttir S. Mapping of a familial essential tremor gene, FET1, to chromosome 3q13. Nat. Genet. 1997;17:84–87. doi: 10.1038/ng0997-84. [DOI] [PubMed] [Google Scholar]
- 9.Higgins J.J., Pho L.T., Nee L.E. A gene (ETM) for essential tremor maps to chromosome 2p22-p25. Mov. Disord. 1997;12:859–864. doi: 10.1002/mds.870120605. [DOI] [PubMed] [Google Scholar]
- 10.Shatunov A., Sambuughin N., Jankovic J., Elble R., Lee H.S., Singleton A.B., Dagvadorj A., Ji J., Zhang Y., Kimonis V.E. Genomewide scans in North American families reveal genetic linkage of essential tremor to a region on chromosome 6p23. Brain. 2006;129:2318–2331. doi: 10.1093/brain/awl120. [DOI] [PubMed] [Google Scholar]
- 11.Jeanneteau F., Funalot B., Jankovic J., Deng H., Lagarde J.P., Lucotte G., Sokoloff P. A functional variant of the dopamine D3 receptor is associated with risk and age-at-onset of essential tremor. Proc. Natl. Acad. Sci. USA. 2006;103:10753–10758. doi: 10.1073/pnas.0508189103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stefansson H., Steinberg S., Petursson H., Gustafsson O., Gudjonsdottir I.H., Jonsdottir G.A., Palsson S.T., Jonsson T., Saemundsdottir J., Bjornsdottir G. Variant in the sequence of the LINGO1 gene confers risk of essential tremor. Nat. Genet. 2009;41:277–279. doi: 10.1038/ng.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ng S.B., Buckingham K.J., Lee C., Bigham A.W., Tabor H.K., Dent K.M., Huff C.D., Shannon P.T., Jabs E.W., Nickerson D.A. Exome sequencing identifies the cause of a mendelian disorder. Nat. Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Louis E.D., Ford B., Lee H., Andrews H., Cameron G. Diagnostic criteria for essential tremor: A population perspective. Arch. Neurol. 1998;55:823–828. doi: 10.1001/archneur.55.6.823. [DOI] [PubMed] [Google Scholar]
- 15.Waibel S., Neumann M., Rabe M., Meyer T., Ludolph A.C. Novel missense and truncating mutations in FUS/TLS in familial ALS. Neurology. 2010;75:815–817. doi: 10.1212/WNL.0b013e3181f07e26. [DOI] [PubMed] [Google Scholar]
- 16.Belzil V.V., Daoud H., St-Onge J., Desjarlais A., Bouchard J.P., Dupre N., Lacomblez L., Salachas F., Pradat P.F., Meininger V. Identification of novel FUS mutations in sporadic cases of amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. 2011;12:113–117. doi: 10.3109/17482968.2010.536840. [DOI] [PubMed] [Google Scholar]
- 17.Corrado L., Del Bo R., Castellotti B., Ratti A., Cereda C., Penco S., Sorarù G., Carlomagno Y., Ghezzi S., Pensato V. Mutations of FUS gene in sporadic amyotrophic lateral sclerosis. J. Med. Genet. 2010;47:190–194. doi: 10.1136/jmg.2009.071027. [DOI] [PubMed] [Google Scholar]
- 18.Carter M.S., Doskow J., Morris P., Li S., Nhim R.P., Sandstedt S., Wilkinson M.F. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J. Biol. Chem. 1995;270:28995–29003. doi: 10.1074/jbc.270.48.28995. [DOI] [PubMed] [Google Scholar]
- 19.Ju S., Tardiff D.F., Han H., Divya K., Zhong Q., Maquat L.E., Bosco D.A., Hayward L.J., Brown R.H., Jr., Lindquist S. A yeast model of FUS/TLS-dependent cytotoxicity. PLoS Biol. 2011;9:e1001052. doi: 10.1371/journal.pbio.1001052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Botstein D., Risch N. Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nat. Genet. 2003;33(Suppl):228–237. doi: 10.1038/ng1090. [DOI] [PubMed] [Google Scholar]
- 21.Ku C.S., Naidoo N., Pawitan Y. Revisiting Mendelian disorders through exome sequencing. Hum. Genet. 2011;129:351–370. doi: 10.1007/s00439-011-0964-2. [DOI] [PubMed] [Google Scholar]
- 22.Johnson J.O., Mandrioli J., Benatar M., Abramzon Y., Van Deerlin V.M., Trojanowski J.Q., Gibbs J.R., Brunetti M., Gronka S., Wuu J., ITALSGEN Consortium Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng S.B., Bigham A.W., Buckingham K.J., Hannibal M.C., McMillin M.J., Gildersleeve H.I., Beck A.E., Tabor H.K., Cooper G.M., Mefford H.C. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat. Genet. 2010;42:790–793. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Varela I., Tarpey P., Raine K., Huang D., Ong C.K., Stephens P., Davies H., Jones D., Lin M.L., Teague J. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J.L., Yang X., Xia K., Hu Z.M., Weng L., Jin X., Jiang H., Zhang P., Shen L., Guo J.F. TGM6 identified as a novel causative gene of spinocerebellar ataxias using exome sequencing. Brain. 2010;133:3510–3518. doi: 10.1093/brain/awq323. [DOI] [PubMed] [Google Scholar]
- 26.Brahms H., Meheus L., de Brabandere V., Fischer U., Lührmann R. Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B’ and the Sm-like protein LSm4, and their interaction with the SMN protein. RNA. 2001;7:1531–1542. doi: 10.1017/s135583820101442x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwiatkowski T.J., Jr., Bosco D.A., Leclerc A.L., Tamrazian E., Vanderburg C.R., Russ C., Davis A., Gilchrist J., Kasarskis E.J., Munsat T. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 28.Vance C., Rogelj B., Hortobágyi T., De Vos K.J., Nishimura A.L., Sreedharan J., Hu X., Smith B., Ruddy D., Wright P. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lagier-Tourenne C., Polymenidou M., Cleveland D.W. TDP-43 and FUS/TLS: Emerging roles in RNA processing and neurodegeneration. Hum. Mol. Genet. 2010;19(R1):R46–R64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tateishi T., Hokonohara T., Yamasaki R., Miura S., Kikuchi H., Iwaki A., Tashiro H., Furuya H., Nagara Y., Ohyagi Y. Multiple system degeneration with basophilic inclusions in Japanese ALS patients with FUS mutation. Acta Neuropathol. 2010;119:355–364. doi: 10.1007/s00401-009-0621-1. [DOI] [PubMed] [Google Scholar]
- 31.Ito D., Seki M., Tsunoda Y., Uchiyama H., Suzuki N. Nuclear transport impairment of amyotrophic lateral sclerosis-linked mutations in FUS/TLS. Ann. Neurol. 2011;69:152–162. doi: 10.1002/ana.22246. [DOI] [PubMed] [Google Scholar]
- 32.Murakami T., Yang S.P., Xie L., Kawano T., Fu D., Mukai A., Bohm C., Chen F., Robertson J., Suzuki H. ALS mutations in FUS cause neuronal dysfunction and death in Caenorhabditis elegans by a dominant gain-of-function mechanism. Hum. Mol. Genet. 2012;21:1–9. doi: 10.1093/hmg/ddr417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Langenhove T., van der Zee J., Sleegers K., Engelborghs S., Vandenberghe R., Gijselinck I., Van den Broeck M., Mattheijssens M., Peeters K., De Deyn P.P. Genetic contribution of FUS to frontotemporal lobar degeneration. Neurology. 2010;74:366–371. doi: 10.1212/WNL.0b013e3181ccc732. [DOI] [PubMed] [Google Scholar]
- 34.Kuroda M., Sok J., Webb L., Baechtold H., Urano F., Yin Y., Chung P., de Rooij D.G., Akhmedov A., Ashley T., Ron D. Male sterility and enhanced radiation sensitivity in TLS(-/-) mice. EMBO J. 2000;19:453–462. doi: 10.1093/emboj/19.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kabashi E., Bercier V., Lissouba A., Liao M., Brustein E., Rouleau G.A., Drapeau P. FUS and TARDBP but not SOD1 interact in genetic models of amyotrophic lateral sclerosis. PLoS Genet. 2011;7:e1002214. doi: 10.1371/journal.pgen.1002214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan J., Deng H.X., Siddique N., Fecto F., Chen W., Yang Y., Liu E., Donkervoort S., Zheng J.G., Shi Y. Frameshift and novel mutations in FUS in familial amyotrophic lateral sclerosis and ALS/dementia. Neurology. 2010;75:807–814. doi: 10.1212/WNL.0b013e3181f07e0c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spanaki C., Plaitakis A. Essential tremor in Parkinson’s disease kindreds from a population of similar genetic background. Mov. Disord. 2009;24:1662–1668. doi: 10.1002/mds.22655. [DOI] [PubMed] [Google Scholar]
- 38.Aronson A.E., Ramig L.O., Winholtz W.S., Silber S.R. Rapid voice tremor, or “flutter,” in amyotrophic lateral sclerosis. Ann. Otol. Rhinol. Laryngol. 1992;101:511–518. doi: 10.1177/000348949210100612. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


