Abstract
The number of T cell progenitors is significantly reduced in the involuted thymus, and the growth and developmental potential of the few cells that are present is severely attenuated. This review provides an overview of how aging affects T cell precursors before and following entry into the thymus and discusses the age related genetic changes that may occur in them. Finally, interventions that rejuvenate thymopoiesis in the elderly by targeting T cell progenitors are discussed.
Keywords: Thymus Involution, Early T Cell Progenitors, p16Ink4a, Rejuvenation
1. Introduction
Involution of the thymus is arguably the most well recognized, age-related change in the hematopoietic system [1]. The major consequence of thymic involution is the diminished production of naïve T lymphocytes. As a result, the peripheral immune repertoire is not replenished, and this is thought to contribute to impaired immunity in the elderly. In view of the critically important role of T cells in the immune response, rejuvenation of the involuted thymus has been proposed as an effective strategy to reverse the declines in immune function that accompany aging. This in turn has stimulated studies in multiple laboratories aimed at defining the causes of age-related thymic involution and using this information to reverse that process.
These efforts have identified multiple processes contributing to thymic involution that include altered production of various endocrine hormones [1, 2], deterioration of the thymic microenvironment [3, 4], and a reduction in the number and quality of T cell precursors [5–7]. While each may independently contribute to thymus involution, it is increasingly clear that changes in one parameter may in turn trigger changes in the others. For example, age-related changes in the thymus microenvironment significantly impact the growth and differentiation of immature thymocytes.
The goal of this review is to discuss thymus involution from the perspective of bone marrow and thymic T cell progenitors. In particular, we describe possible genetic changes that occur in these cells with age and strategies for reversing them in order to stimulate thymopoiesis in the elderly. In order to provide a background for how aging affects T cell development, we begin with a brief overview of the cellular stages of that process.
2. T cell development
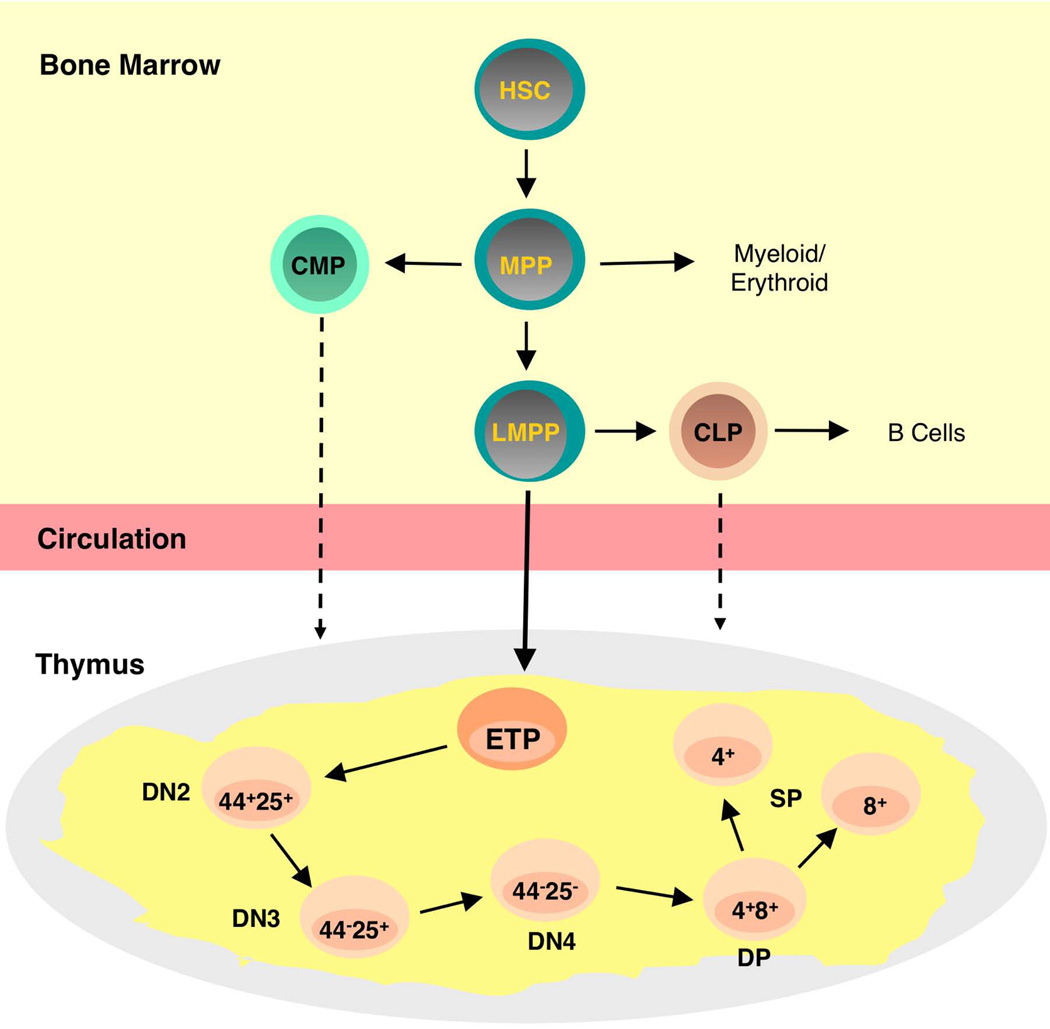
T cell development in the thymus is a highly regulated process in which immature progenitors progress through various intermediate stages before generating mature T lymphocytes [8–10]. The most immature T cell precursors in the thymus have been termed Early T cell Progenitors (ETP) and are defined by their lineage negative (Lin−) CD44+ CD25− CD117(c-kit)high phenotype [11, 12]. The requirements for resolution of these cells, which are present at a frequency of ~ 0.01%, have recently been discussed [9]. If these criteria are met, the thymus of a young mouse, which contains around 200 million thymocytes, includes approximately 20,000 ETP.
ETP mature into double negative (DN) thymocytes characterized by the differential expression of the CD44 and/or CD25 cell surface determinants (Figure 1). As development through the DN2, DN3, and DN4 stages of development occurs, these progenitors lose non-T lineage potential, and by the time the DN3 stage has been reached, commitment to the T cell lineage has occurred [13, 14]. DN3 thymocytes express the pre-T cell receptor (TCR), which consists of the TCR β-chain and the pre-Tα chain, and mature into DN4 cells. DN4 cells express low levels of the α and β TCR chains and mature into double positive (DP) thymocytes as they acquire the CD4 and CD8 determinants and up-regulate TCR expression levels. Finally, DP thymocytes generate CD4+ or CD8+ single positive cells. This process and a detailed discussion of the changes in gene expression that occur at each of these stages have recently been reviewed in depth [8, 15, 16] and in chapters by Sempowski and Manley and Dixit in this volume.
Figure 1.
T cell progenitors in the bone marrow and thymus. Hematopoietic stem cells (HSCs) in the bone marrow generate Multipotential Progenitors (MPP) capable of producing lymphoid and myeloid progeny. Lymphoid Primed Multipotential Progenitors (LMPP) have lost most myeloid and erythroid potential and are candidate thymus seeding cells as shown by the solid arrow. Early T Lineage Progenitors (ETP) are the most immature intrathymic progenitor and are defined by their lineage negative CD44high CD25− CD117+ phenotype. ETP then mature into double negative (DN) stages defined by differential expression of CD44 and/or CD25. DN4 cells express the T cell receptor at low levels and then mature into CD4+CD8+ double positive (DP) thymocytes from which CD4 or CD8 single positive (SP) thymocytes derive. Note that other cells in addition to LMPPs may under some circumstances seed the thymus. These could include HSCs, CLPs, and even CMPs. The latter cells have recently been demonstrated to have residual T cell potential.
The thymus does not contain an endogenous source of progenitors that can maintain long-term thymopoiesis. Instead, sustained cell production in that organ is dependent upon the migration of bone marrow precursors into it. The identification of these cells has been challenging. One reason why this is the case is that the entry of progenitors into the thymus is periodic rather than continuous and involves a very low number of cells [17]. In addition, while multiple bone marrow derived cells (Figure 1), including hematopoietic stem cells (HSCs), Multi-Potential Progenitors (MPP), Lymphoid Primed Multipotential Progenitors (LMPP), and common lymphoid progenitors (CLP), have T cell potential when tested in various in vitro culture systems or in vivo transplantation assays, not all of these populations necessarily migrate to the thymus under physiologic conditions. Further complicating this issue is that progenitors originally thought to have no thymopoietic potential, such as common myeloid progenitors (CMP) have now been shown to be able to generate T cells [18].
Because the thymus does not contain HSCs, it is generally accepted that they do not normally migrate to that organ [19]. However, which of their down-stream progeny sustains thymopoiesis remains a controversial issue. Various studies, using a range of cellular and genetic approaches that examined bone marrow, blood, and intrathymic populations, have suggested that LMPP like cells [20–22], T/myeloid progenitors [23, 24], and lymphoid specified precursors [25] may be thymus homing cells. Thus, when we discuss the effects of aging on pre-thymic cells in the bone marrow in the following section, we do so with the caveat that this is an evolving area of study.
3. Age-related changes in thymopoiesis
The tremendous advances in the resolution of stem and progenitor cells in the bone marrow and thymus have provided a foundation for delineating the effects of aging on T lymphopoiesis. This section will review how aging impacts cells at these stages of development.
3.1 Effects of aging on bone marrow T cell progenitors
In view of the dependence of thymopoiesis on continual input from the bone marrow, a logical prediction is that pre-thymic defects contribute to thymic involution. However, the extensive literature focused on this issue is contradictory and not easy to compare because of the different experimental approaches utilized. For example, peripheral T cell reconstitution, rather than thymus repopulation was analyzed in some experiments. In this case, the homeostatic expansion of mature T cells could obscure intrathymic deficiencies. In other instances, whole bone marrow rather than purified stem and progenitor cell populations was assessed. These issues have recently been discussed by Zediak et al. [6]. Those investigators compared the ability of purified HSCs and MPPs from young and old mice to generate thymocytes using in vitro and in vivo approaches. Even if these are not the precise thymus seeding cell(s), any age-related changes detected in these immature precursors would likely be reflected in their downstream, thymus seeding progeny. The results were clear-cut and demonstrated that the T lymphopoietic potential of old HSCs and MPPs was attenuated. Importantly, the latter cells were injected directly into the thymus of non-irradiated recipients in order to avoid artifacts that could be induced by the irradiation of recipients. Based on this approach, it was demonstrated that aged MPPs produced over 2-fold fewer thymocytes compared to their young counterparts.
These results are consistent with the conclusion that the lymphopoietic potential of the HSC compartment is significantly affected by aging. A paradigm shift in our thinking of how this occurs was formulated by Muller-Sieburg and colleagues who observed that HSCs are heterogeneous and include subsets that are lymphoid or myeloid biased or balanced in their developmental potential. Strikingly, aging was accompanied by an accumulation of myeloid biased HSCs and a loss of those which are lymphoid biased [26, 27]. A similar myeloid bias of HSCs was recently documented in humans [28].
The analysis of these HSC subpopulations has been greatly facilitated by three reports demonstrating that they can be prospectively isolated based on levels of expression of the signaling lymphocytic activation molecule (SLAM) 150 [29] cell surface determinant [30–32]. Thus, Lin– CD117high Sca-1+ cells that express CD150 at high levels were shown to be myeloid biased as demonstrated by their reconstituting potential following transplantation. In contrast, HSCs that express CD150 at low to undetectable levels were lymphoid biased. HSCs that express CD150 at intermediate levels exhibit no distinct pattern of reconstitution and presumably represent balanced HSCs. The enrichment in CD150high myeloid biased and decrease of CD150low lymphoid biased stem cells with age is consistent with previous reports showing increased myeloid potential of old HSCs [33] and provides an explanation for the reduced potential of stem cells from old bone marrow to generate T cell progeny as observed by Zediak et al. [6].
Even though there are changes in the proportion of lymphoid and myeloid stem cells with age, it has generally been assumed that the HSCs present, regardless of their bias, have normal developmental potential. However, this assumption has been questioned in a recent study demonstrating that myeloid biased HSCs from old donors have decreased self-renewal potential and generate smaller daughter cell clones than their young counterparts [34]. This observation is consistent with studies showing that common myeloid progenitors from old mice exhibit a similar age-related decline [35]. These findings raise the possibility that the few lymphoid biased HSCs present in old mice also would exhibit reduced lymphoid potential.
3.2 Effects of aging on ETP and intrathymic progenitors
After exiting the bone marrow, T cell progenitors enter the circulation and ultimately gain access to the thymus. Entry into the thymus is dependent upon expression of various receptors as recently reviewed [9]. For example, T cell progenitors express the CCR7 and CCR9 chemokine receptors as well as the PSGL1 glycoprotein that recognize ligands expressed on thymic stromal cells. Limited studies on the effects of aging on thymus entry have been performed, but the results suggest that aging does not impair this process [36].
However, the total number of ETP is reduced in the involuted thymus [7, 37] and this may occur for two reasons. First, as discussed in section 3.1, the potential of old bone marrow precursors to generate ETP is reduced significantly. A second factor is that those ETP that are produced proliferate at significantly lower levels and have higher rates of apoptosis than their young counterparts [7]. This decline in proliferation and increased cell death is a general feature of aging in multiple tissues and in the lymphoid lineage in particular. For example, B cell development in the bone marrow is significantly reduced with age, and pro-B cells from that tissue also exhibit reduced proliferation and increased apoptosis [38]. Furthermore, ETP harvested from the involuted thymus also exhibit developmental alterations. Thus, when the T cell potential of young and old ETP is compared in fetal thymic organ culture, few mature T cells are generated from the old progenitors [7].
In view of the above considerations, the conclusion that emerges is that aging results in intrinsic changes in bone marrow and intra-thymic T cell progenitors. However, what triggers these events is less clear. As mentioned in the introductory comments, thymus involution is a complex, multi-factorial process. Thus, it is difficult to estimate the degree to which the age-related changes occurring in hematopoietic stem and progenitor cells are intrinsically programmed or occur secondarily due to microenvironmental and systemic influences [39, 40].
4. Molecular Basis for Thymocyte Progenitor Aging
Because of the relative ease in isolating hematopoietic stem and progenitor cells in mice and humans and the wide availability of various platforms for global gene analysis, it has been possible to assess how aging affects patterns of gene expression in these populations. While most of these efforts have focused on HSCs and not more mature lymphoid progenitors, a discussion of the age-related genetic changes occurring in stem cells is relevant, as these will likely influence the T lymphopoietic potential of their more mature lymphoid specified progeny.
4.1 Changes in gene expression
The pattern of gene expression in young and old HSCs has been compared using various microarray platforms. One study that used this approach demonstrated that genes involved in specifying lymphoid fate are down-regulated in aging HSCs [41]. This approach has also revealed age-related changes in the expression of genes that regulate specific aspects of cellular physiology. For example, the Goodell laboratory examined more than 14,000 genes in HSCs from young and old mice and reported that 1,500 were up-regulated with age while 1,600 were repressed. Genes that were associated with stress responses, inflammation, and protein aggregation dominated the up-regulated genes while those involved in preservation of genomic integrity and chromatin remodeling were most frequently repressed [42].
Given the shift in the proportion of HSC subsets with age, it is not surprising, in retrospect, that different patterns of gene expression were observed in studies that compared the total HSC population from young and old mice. It will ultimately be important to compare gene expression patterns in young and old lymphoid biased HSCs and young and old myeloid biased stem cells. In view of the report that the quality of myeloid biased stem cells is attenuated with age [34], these studies are predicted to show differences between HSC subsets from young and old bone marrow. In addition, the data will likely reveal gene expression differences between lymphoid and myeloid biased HSCs, regardless of age. In this regard, Challen and colleagues compared the patterns of gene expression of myeloid and lymphoid biased HSCs from young mice and identified 785 genes that were differentially expressed. For example, the lymphoid biased HSCs preferentially expressed genes related to cell cycling and high metabolism [30]. It will also be important to extend these types of genetic analyses to young and old B and T cell progenitors [43].
If differences in patterns of gene expression between young and old myeloid and young and old lymphoid biased stem cells are observed, the question of how they are initiated will need to be addressed. On the one hand, they could be intrinsically programmed. However, given the fact that changes in the systemic and hematopoietic microenvironment occur with age, it is tempting to speculate that external signals are involved. In either case, changes in gene expression that occur in old stem cells may be the result of epigenetic modification of genes that regulate stem cell self-renewal, survival, and/or developmental potential [44, 45]. One of the most common epigenetic modifications is methylation of CpG dinucleotides, which is regulated by at least three different DNA methyltransferases (DNMTs). DNMT 3a and DNMT 3B are de novo methylators while DNMT1 is involved in the maintenance of the methylated state [46].
Changes in methylation status due to the effects of aging on DNMT expression could explain the well documented observation that some strains of old mice have an increased number of HSCs [33]. In this regard, it was recently demonstrated that loss of DNMT3a impairs HSC differentiation while expanding their number in the bone marrow [47], and DNMT3a levels have been reported to decline with age [48]. The age-related decline in expression of other DNMTs such as DNMT1 might also account for decreased production of lymphoid cells. Thus, mice with hypomorphic expression of DNMT1 exhibit relatively normal myeloerythroid potential but a significant block in lymphoid development [49]. While most of these studies have been performed in mice, changes in DNA methylation patterns have also been reported for aged human CD34+ cells [50].
While genome wide assessment strategies can identify changes in the pattern of gene expression in stem and progenitor cell during aging, the precise role played by specific genes will ultimately need to be tested in biological systems. We provide an example of this in the following section using the genes encoded by the Cdkn2a locus as an example.
4.2 The Cdkn2a locus
Declines in proliferation and survival are hallmarks of stem and progenitor cell aging in multiple tissues, and changes in expression of genes that regulate these processes are likely to contribute to the aging phenotype. Particular focus in this regard has been placed on the INK4a and Arf proteins encoded by the Cdkn2a locus [51]. INK4a activates retinoblastoma (Rb) and inhibits cell cycle progression while Arf activates p53 leading to cell cycle arrest and/or apoptosis [52, 53]. The expression of both INK4a and Arf increases in multiple tissues with age [52]. A role for INK4a in aging was substantiated in a study that showed that deletion of p16Ink4a expressing cells in vivo delayed the onset of aging in multiple tissues in a progeroid mouse model [54]. As discussed below, the delay in thymic involution observed following p16Ink4a deletion in thymocytes is consistent with this observation. However, whether this will hold true for the normal aging of other tissues remains to be determined.
INK4a and Arf are tumor suppressor proteins, raising the question as to why tumor suppression and aging would be linked. One view is that self-renewing stem and progenitor cells accumulate DNA damage that could predispose to malignancy. Thus, it is advantageous to remove them from the organism, and proteins such as INK4a and Arf that promote cell cycle arrest and apoptosis are able to do so. The ultimate result, however, is a reduction in the number of stem and progenitor cells, resulting in tissue aging, which would be the price to be paid for protection against cancer [55–59].
There has been controversy as to whether INK4a and Arf are expressed in HSCs with age, although it appears that if this occurs [60], it is at very low levels [61]. On the other hand, there is strong evidence to support a role for these tumor suppressors in aging of the lymphoid lineage. We recently reported that levels of both proteins increase with age in B cell progenitors in the bone marrow and that down-regulating their expression increased the survival and proliferation of these cells [38].
There is evidence from gain and loss of function studies that INK4a is an important regulator of thymopoiesis. For example, thymus size is increased in p16Ink4a deficient mice [62, 63], and mice engineered to express p16Ink4a under the control of an lck promoter have a block in T cell differentiation at the DN3 stage [64]. In order to test the effects of p16Ink4a deletion in an aging model, Liu and colleagues conditionally deleted p16Ink4a in thymocytes by mating mice with a floxed p16Ink4a allele to lck-Cre mice and measured thymic size in groups of mice up to 25 months of age. The striking result of this study was that thymic involution was significantly delayed [65], providing clear evidence that p16Ink4a plays a role in thymic involution.
However, the thymus of p16Ink4a deficient mice ultimately underwent involution. One reason this occurred is that genes other than p16Ink4a are likely involved in the regulation of that process. In addition, because expression of lck is most robust starting at the DN2 stage of development [13], p16Ink4a might not have been deleted in earlier stages of development. In fact, our preliminary results indicate that p16Ink4a is expressed in old but not young ETP and plays a major role in the decline in the number and developmental potential of these immature intrathymic progenitors. In view of this, it will ultimately be interesting to develop models in which p16Ink4a is conditionally deleted beginning at the ETP stage of development. Additional studies to define the role of Arf in the aging of ETP are also needed [66].
It will also be of interest to examine the regulation of p16Ink4a in the aging thymus (Figure 2). p16Ink4a expression is strongly suppressed by the polycomb group protein Bmi-1 [67, 68]. Consistent with this, the thymus of Bmi-1 deficient mice is atrophied [68], while this is not the case in Bmi-1/p16Ink4a deficient animals [69]. However, it is important to note that the loss of Bmi-1 has effects beyond increased expression of INK4a. For example, Bmi-1 deficient thymocytes exhibit significant mitochondrial dysfunction and increased reactive oxygen species [70].
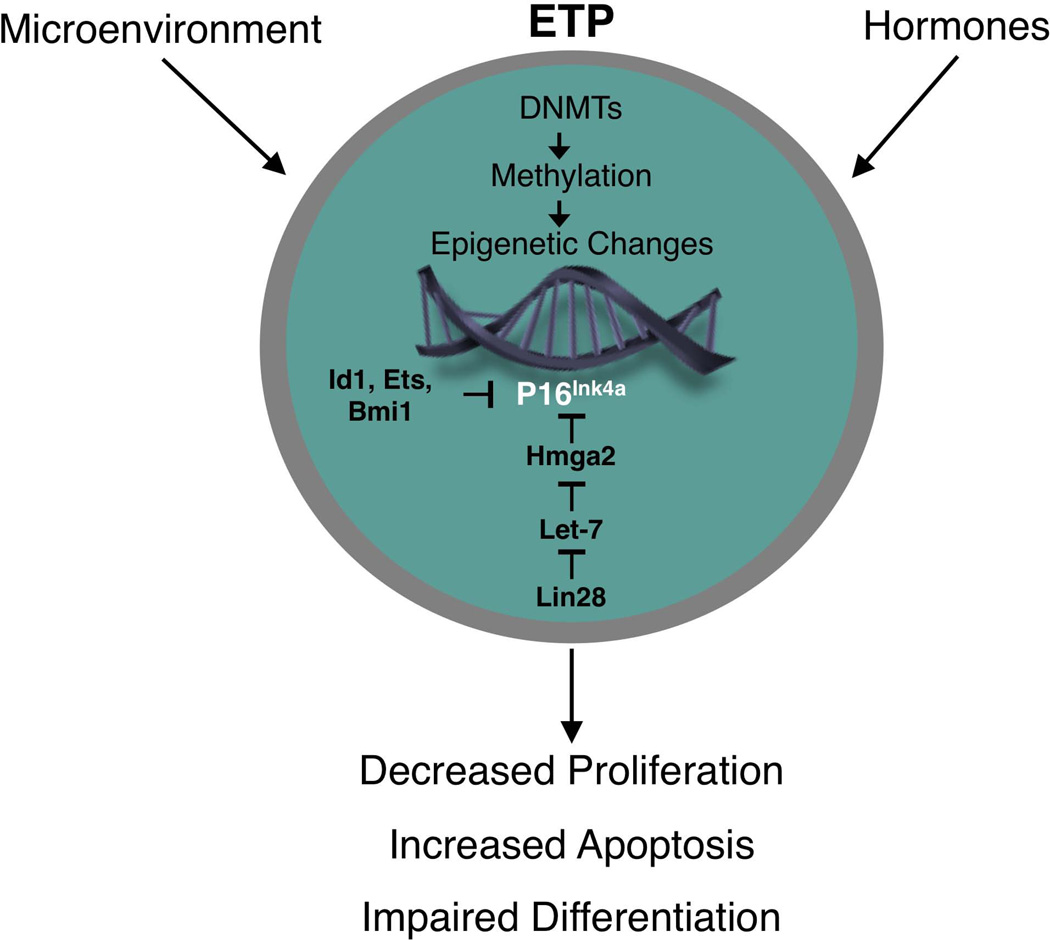
Figure 2.
Proposed changes in gene expression in aging ETP. Changes in expression of one or more DNA methyltransferases (DNMTs), as a result of cell intrinsic or extracellular influences, may cause epigenetic modifications that affect the expression of genes that regulate ETP growth, differentiation, and/or survival. Our preliminary data indicate that expression of p16INK4a is increased in ETP with age and is a factor that contributes to their reduced proliferation. A complex network regulates expression of p16INK4a in cells. For example, various factors that include Id1, Ets1, Ets2, Bmi1, and Hmga2 can suppress p16INK4a expression. Hmga2 in turn is under control of multiple upstream factors that include Lin 28 and the Let-7b microRNA. The precise factors that regulate p16INK4a expression in ETP have not been defined.
In addition to Bmi-1, p16Ink4a expression is also regulated by factors that include Id-1, Ets1, Ets2, and the chromatin associated Hmga-2 protein [71, 72], and understanding how their expression is altered in the aging thymus is needed. These regulatory circuits are complex and include multiple layers. For example, the direct regulators of p16Ink4a expression are themselves controlled by a hierarchy of factors. Thus, Hmga-2 expression is blocked by let-7b micro RNA, and let-7b expression in turn is regulated by the Lin-28 RNA binding protein [73, 74]. A prominent role for miRNAs as key modulators of cellular senescence is emerging [75], but almost nothing is known about the role they play in thymic involution and how they regulate T lineage associated genes.
5. Rejuvenation of the involuted thymus
While a significant body of information regarding why the thymus involutes now exists, the goal is to use this information to stimulate thymopoiesis in the elderly. As we recently discussed, such interventions may not need to restore the thymus to levels in a young child [76]. Instead, even a modest, transient stimulation of T cell production may be sufficient to generate a wave mature, naïve T cells that will migrate to the periphery and improve immune function.
There is increasing evidence from various systems that the aging phenotype is at least partially reversible. This is particularly well illustrated by studies of muscle cell aging. Rando and colleagues demonstrated that aging of muscle satellite cells occurs due to systemic/environmental changes and that exposure of old satellite cells to a young environment reversed the effects of aging [77]. It has also been suggested that exposure of old hematopoietic stem and progenitor cells to a young microenvironment can at least partially reverse the effects of aging [39, 40].
As noted in section 1, there is a decline in the production of various endocrine hormones with age, and this has been implicated in the process of thymus involution. For example, it has been recognized for some time that reduced production of growth hormone (GH) occurs with age. Many of its effects are mediated via induction of insulin like growth factor-1 (IGF-1) which also declines with age [78]. Numerous murine studies have shown that administration of GH or IGF-I to old mice can increase the number of cells in the thymus [79], and this information has in turn been the basis for a clinical trial in which GH was administered to HIV-1 patients whose disease was in remission [80]. This resulted in increased thymic mass, enhanced thymic output as demonstrated by an increased frequency of T cell receptor rearrangement excision circles in circulating T cells, and an increased number of naïve CD4+ T cells. Taken together, these results provide evidence that T cell production in the involuted human thymus can be stimulated.
The potential of numerous additonal agents that include IL-7 [81], androgen inhibitors [82], and fibroblast growth factor (Fgf) 7, also referred to as keratinocyte growth factor [83–85], to stimulate thymopoiesis has been evaluated in pre-clinical and/or clinical trials as recently discussed in depth [86]. Fgf7 has been evaluated in pre-clinical studies and exhibits remarkable effects on increasing the number of cells in the involuted thymus [84]. Strikingly, thymus cell number in old mice can be restored to levels in young animals following several rounds of Fgf7 treatment. The use of hormones and cytokines to stimulate thymopoiesis is particularly attractive in contrast to cell-based therapies, because they can be easily administered.
These factors may have differential effects on thymocytes and/or thymic stromal cells. For example, IGF-I receptors are expressed on both thymocytes and thymic epithelial cells, although a recent study indicates that its effects on increasing thymus cellularity are primarily mediated via effects on the latter population [87]. The effects of Fgf7 on thymocytes are indirectly mediated, as Fgf7 receptors are found on thymic epithelial cells but not thymocytes [85]
Regardless of whether the effects of a particular agent are direct or indirect on ETP and other immature thymocytes, they must somehow be reversing the effects of aging in these cells. How this occurs at the genetic and molecular levels remains to be determined. For example, administration of a particular factor could directly or indirectly down-regulate p16Ink4a expression in ETP, possibly through effects on its upstream regulators.
6. Concluding remarks
As a result of efforts in multiple laboratories over the past decade, it is no longer necessary to start reviews by stating that the causes of thymic involution are unknown. In fact, as reviewed herein and in other chapters in this volume, a tremendous amount of information about the causes of thymus involution is now available. The fact that clinical trials that have tested the potential of various compounds to rejuvenate the involuted thymus have been conducted is further testament to how far the field has progressed.
Nevertheless, there is still much to be learned. In particular, how aging affects the patterns of gene expression in thymic epithelium [88] and in thymocyte progenitors need further definition. The hope is that by doing so, key targets that can be therapeutically manipulated to rejuvenate the aging thymus will be identified.
Highlights.
Thymic involution is a multifactorial process
The proliferative potential of T cell progenitors declines with age
Genes encoded by the Cdkn2a locus may be involved in T cell progenitor aging
Pharmacological rejuvenation of the thymus may be possible
Acknowldegements
This work was supported by NIH grant AG 034875.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Linton PJ, Dorshkind K. Age-related changes in lymphocyte development and function. Nat Immunol. 2004;5:133–139. doi: 10.1038/ni1033. [DOI] [PubMed] [Google Scholar]
- 2.Taub D, Murphy W, Longo D. Rejuvenation of the aging thymus: growth hormone-mediated pathways and ghrelin-mediated signaling pathways. Curr Opin Pharmacol. 2010;10:408–424. doi: 10.1016/j.coph.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrew D, Aspinall R. Thymic atrophy in the mouse is a soluble problem of the thymic environment. Vaccine. 2000;18:1629–1637. doi: 10.1016/s0264-410x(99)00498-3. [DOI] [PubMed] [Google Scholar]
- 4.Mackall CL, Punt JA, Morgan P, Farr AG, Gress RE. Thymic function in young/old chimeras: substantial thymic T cell regenerative capacity despite irreversible age-associated thymic involution. Eur J Immunol. 1998;28:1886–1893. doi: 10.1002/(SICI)1521-4141(199806)28:06<1886::AID-IMMU1886>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 5.Sharp A, Kukulansky T, Globerson A. In vitro analysis of age-related changes in the developmental potential of bone marrow thymocyte progenitors. Eur J Immunol. 1990;20:2541–2546. doi: 10.1002/eji.1830201203. [DOI] [PubMed] [Google Scholar]
- 6.Zediak VP, Maillard I, Bhandoola A. Multiple prethymic defects underlie age-related loss of T progenitor competence. Blood. 2007;110:1161–1167. doi: 10.1182/blood-2007-01-071605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Min H, Montecino-Rodriguez E, Dorshkind K. Reduction in the developmental potential of intrathymic T cell progenitors with age. J Immunol. 2004;173:245–250. doi: 10.4049/jimmunol.173.1.245. [DOI] [PubMed] [Google Scholar]
- 8.Rothenberg E. T cell lineage commitment: identity and renunciation. J Immunol. 2011;186:6649–6655. doi: 10.4049/jimmunol.1003703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Love P, Bhandoola A. Signal integration and crosstalk during thymocyte migration and emigration. Nat Rev Immunol. 2011;11:469–477. doi: 10.1038/nri2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petrie HT, Zuniga-Pflucker JC. Zoned out: functional mapping of stromal signaling microenvironments in the thymus. Annu Rev Immunol. 2007;25:649–679. doi: 10.1146/annurev.immunol.23.021704.115715. [DOI] [PubMed] [Google Scholar]
- 11.Allman D, Sambandam A, Kim S, Miller JP, Pagan A, Well D, Meraz A, Bhandoola R. Thymopoiesis independent of common lymphoid progenitors. Nat Immunol. 2003;4:168–174. doi: 10.1038/ni878. [DOI] [PubMed] [Google Scholar]
- 12.Porritt HE, Rumfelt LL, Tabrizifard S, Schmitt TM, Zuniga-Pflucker JC, Petri HT. Heterogeneity among DN1 prothymocytes reveals multiple progenitors with different capacities to generate T cell and non-T cell lineages. Immunity. 2003;20:735–745. doi: 10.1016/j.immuni.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 13.Rothenberg E, Zhang J, Li L. Multilayered specification of the T-cell lineage fate. Immunol Rev. 2010;238:150–168. doi: 10.1111/j.1600-065X.2010.00964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhandoola A, von Boehmer H, Petrie HT, Zuniga-Flucker JC. Commitment and developmental potential of extrathymic and intrathymic T cell precursors: plenty to choose from. Immunity. 2007;26:678–689. doi: 10.1016/j.immuni.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 15.Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rothenberg E. Transcriptional drivers of the T-cell lineage program. Curr Opin Immunol. 2012 doi: 10.1016/j.coi.2011.12.012. PMID: 22264928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldschneider I. Cyclical mobilization and gated importation of thymocyte progenitors in the adult mouse: evidence for a thymus-bone marrow feedback loop. Immunol Rev. 2006;209:58–75. doi: 10.1111/j.0105-2896.2006.00354.x. [DOI] [PubMed] [Google Scholar]
- 18.Chi A, Chavez A, Xu L, Weber B, Shestova O, Schaffer A, et al. Identification of Flt3+CD150− myeloid progenitors in adult mouse bone marrow that harbor T lymphoid developmental potential. Blood. 2011;118:2723–2732. doi: 10.1182/blood-2010-09-309989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mori S, Shortman K, Wu L. Characterization of thymus-seeding precursor cells from mouse bone marrow. Blood. 2001;98:696–704. doi: 10.1182/blood.v98.3.696. [DOI] [PubMed] [Google Scholar]
- 20.Schwarz A, Bhandoola A. Circulating hematopoietic progenitors with T lineage potential. Nat Immunol. 2004;5:953–960. doi: 10.1038/ni1101. [DOI] [PubMed] [Google Scholar]
- 21.Perry SS, Welner RS, Kouro T, Kincade PW, Sun XH. Primitive lymphoid progenitors in bone marrow with T lineage reconstituting potential. J Immunol. 2006;177:2880–2887. doi: 10.4049/jimmunol.177.5.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benz C, Martins VC, Radtke F, Bleul CC. The stream of precursors that colonizes the thymus proceeds selectively through the early T lineage precursor stage of T cell development. J Exp Med. 2008;205:1187–1189. doi: 10.1084/jem.20072168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bell JJ, Bhandoola A. The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature. 2008;452:764–767. doi: 10.1038/nature06840. [DOI] [PubMed] [Google Scholar]
- 24.Wada H, Masuda K, Satoh R, Kakugawa K, Ikawa T, Katsura Y, Kawamoto H. Adult T-cell progenitors retain myeloid potential. Nature. 2008;452:768–772. doi: 10.1038/nature06839. [DOI] [PubMed] [Google Scholar]
- 25.Schlenner S, Madan V, Busch K, Tietz A, Laufle C, Costa C, et al. Fate mapping reveals separate origins of T cells and myeloid lineages in the thymus. Immunity. 2010;32:426–436. doi: 10.1016/j.immuni.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 26.Cho R, Sieburg H, Muller-Sieburg C. A new mechanism for the aging of hematopoietic stem cells: aging changes the clonal composition of the stem cell compartment but not the individual stem cells. Blood. 2008;111:5553–5561. doi: 10.1182/blood-2007-11-123547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muller-Sieburg C, Cho R, Karlsson L, Huang J, Sieburg H. Myeloid-biased hematopoietic stem cells have extensive self-renewal capacity but generate diminished lymphoid progeny with impaired IL-7 responsiveness. Blood. 2004;103:4111–4118. doi: 10.1182/blood-2003-10-3448. [DOI] [PubMed] [Google Scholar]
- 28.Pang W, Price E, Sahoo D, Beerman I, Maloney W, Rossi D, et al. Human bone marrow hematopoietic stem cells are increased in frequency and myeloid-biased with age. Proc Natl Acad Sci U S A. 2011;108:20012–20017. doi: 10.1073/pnas.1116110108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 30.Challen G, Boles N, Chambers S, Goodel M. Distinct hematopoietic stem cells subtypes are differentially regulated by TGF-β1. Cell Stem Cell. 2010;6:265–278. doi: 10.1016/j.stem.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beerman I, Bhattacharya D, Zandi S, Sigvardsson M, Weissman I, Bryder D, et al. Functionally distinct hematopoietic stem cells modulate hematopoietic lineage potential during aging by a mechanism of clonal expansion. Proc Natl Acad Sci U S A. 2010;107:5465–5470. doi: 10.1073/pnas.1000834107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morita Y, Ema H, Nakauchi H. Heterogeneity and hierarchy within the most primitive hematopoietic stem cell compartment. J Exp Med. 2010;207:1173–1182. doi: 10.1084/jem.20091318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sudo K, Ema H, Morita Y, Nakauchi H. Age-associated characteristics of murine hematopoietic stem cells. J Exp Med. 2000;192:1273–1280. doi: 10.1084/jem.192.9.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dykstra B, Olthof S, Schreuder J, Ritsema M, De Haan G. Clonal analysis reveals multiple functional defects of aged murine hematopoietic stem cells. J Exp Med. 2011;208:2691–2703. doi: 10.1084/jem.20111490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Signer RAJ, Montecino-Rodriguez E, Witte ON, McLaughlin J, Dorshkind K. Age-related defects in B lymphopoiesis underlie the myeloid dominance of adult leukemia. Blood. 2007;110:1831–1839. doi: 10.1182/blood-2007-01-069401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gui J, Zhu X, Dohkan J, Cheng L, Barnes P, Su D. The aged thymus shows normal recruitment of lymphohematopoietic progenitors but has defects in thymic epithelial cells. Int Immunol. 2007;19:1201–1211. doi: 10.1093/intimm/dxm095. [DOI] [PubMed] [Google Scholar]
- 37.Heng TSP, Goldberg GL, Gray DHD, Sutherland JS, Chidgey AP, Boyd RL. Effects of castration on thymocyte development in two different models of thymic involution. J Immunol. 2005;175:2982–2993. doi: 10.4049/jimmunol.175.5.2982. [DOI] [PubMed] [Google Scholar]
- 38.Signer RAJ, Montecino-Rodriguez E, Witte ON, Dorshkind K. Aging and cancer resistance in lymphoid progenitors are linked processes conferred by p16Ink4a and Arf. Genes Develop. 2008;22:3115–3120. doi: 10.1101/gad.1715808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song Z, Wang J, Guachalla L, Terszowski G, Rodewald H, Ju Z, et al. Alterations of the systemic environment are the primary cause of impaired B and T lymphopoiesis in telomere-dysfunctional mice. Blood. 2010;115:1481–1489. doi: 10.1182/blood-2009-08-237230. [DOI] [PubMed] [Google Scholar]
- 40.Ju Z, Jiang H, Jaworski M, Rathinam C, Gompf A, Klein C, et al. Telomere dysfunction induces environmental alterations limiting hematopoietic stem cell function and engraftment. Nat Med. 2007;13:742–747. doi: 10.1038/nm1578. [DOI] [PubMed] [Google Scholar]
- 41.Rossi DJ, Bryder D, Zahn JM, Ahlenius H, Sonu R, Wagers AJ, et al. Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc Natl Acad Sci USA. 2005;102 doi: 10.1073/pnas.0503280102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chambers S, Shaw C, Gatza C, Fisk C, Donehower L, Goodel M. Aging hematopoietic stem cells decline in function and exhibit epigentic dysregulation. PLoS Biol. 2007;5:e201. doi: 10.1371/journal.pbio.0050201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lustig A, Carter A, Bertak D, Enika D, Vandanmagsar B, Wood W, et al. Transcriptome analysys of murine thymocytes reveals age-associated changes in thymic gene expression. Int J Med. 2009;6:51–64. doi: 10.7150/ijms.6.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fraga MF, Esteller M. Epigenetics and aging: the targets and the marks. Trends Genet. 2007;23:413–418. doi: 10.1016/j.tig.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 45.Rando T, Chang H. Aging, rejuvenation, and epigenetic reprogramming: resetting the aging clock. Cell. 2012;148:46–57. doi: 10.1016/j.cell.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;35:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 47.Challen G, Sun D, Jeong M, Luo M, Jelinek J, Berg J, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet. 2011;44:23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Casillas M, Lopatina N, Andrews L, Tollefsbol T. Transcriptional control of the DNA methyltransferases is altered in aging and neoplastically-transformed human fibroblasts. Mol Cell Biochem. 2003;252:33–43. doi: 10.1023/a:1025548623524. [DOI] [PubMed] [Google Scholar]
- 49.Broske A, Vockentanz L, Kharazi S, Huska M, Mancini E, Scheller M, et al. DNA methylation protects hematopoietic stem cell multipotency from myeloerythorid restriction. Nat Genet. 2009;41:1207–1215. doi: 10.1038/ng.463. [DOI] [PubMed] [Google Scholar]
- 50.Bocker M, Hellwig I, Breiling A, Eckstein V, Ho A, Lyko F. Genome-wide promoter DNA methylation dynamics of human hematopoietic progentors cells during differentiation and aging. Blood. 2011;117:3182–3189. doi: 10.1182/blood-2011-01-331926. [DOI] [PubMed] [Google Scholar]
- 51.Sharpless NE. Ink4a/Arf links senescence and aging. Exp Gerontol. 2004;39:1751–1759. doi: 10.1016/j.exger.2004.06.025. [DOI] [PubMed] [Google Scholar]
- 52.Krishnamurthy J, Torrice C, Ramsey MR, Kovalev GI, Al-Regaiey K, Su L, et al. Ink4a/Arf expression is a biomarker of aging. J Clin Invest. 2004;114:1299–1307. doi: 10.1172/JCI22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Randle DH, Zindy F, Sherr CJ, Roussel MF. Differential effects of p19Arf and p16Ink4a loss on senescence of murine bone marrow-derived preB cells and macrophages. Proc Natl Acad Sci USA. 2001;98:9654–9659. doi: 10.1073/pnas.171217498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baker D, Wijshake T, Tchkonia T, LeBrasseur N, Childs B, van de Sluis B, et al. Clearance of p16Ink4a positive sensescent cells delays ageing-associated disorders. Nature. 2011;479:232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sharpless NE, DePinho RA. How stem cells age and why this makes us grow old. Nature Rev Mol Biol. 2007;8:703–713. doi: 10.1038/nrm2241. [DOI] [PubMed] [Google Scholar]
- 56.Mooi WJ, Peeper DS. Oncogene-induced cell senescence-halting on the road to cancer. New Eng J Med. 2006;355:1037–1046. doi: 10.1056/NEJMra062285. [DOI] [PubMed] [Google Scholar]
- 57.Collado M, Blasco MA, Serrano M. Cellular senescence in cancer and aging. Cell. 2007;130:223–233. doi: 10.1016/j.cell.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 58.Campisi J. Cancer and ageing: rival demons? Nat Rev Cancer. 2003;3:339–349. doi: 10.1038/nrc1073. [DOI] [PubMed] [Google Scholar]
- 59.Finkel T, Serrano M, Blasco M. The common biology of cancer and ageing. Nature. 2007;448:767–774. doi: 10.1038/nature05985. [DOI] [PubMed] [Google Scholar]
- 60.Janzen V, Forkert R, Fleming H, Saito Y, Waring MT, Dombkowski, Cheng T, DePinho RA, Sharpless NE, Scadden DT. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature. 2006;443:421–426. doi: 10.1038/nature05159. [DOI] [PubMed] [Google Scholar]
- 61.Attema J, Pronk C, Norddahl G, Nygren J, Bryder D. Hematopoietic stem cell aging is uncoupled from p16INK4a-mediated senescence. Oncogene. 2009;28 doi: 10.1038/onc.2009.94. 22-38-2243. [DOI] [PubMed] [Google Scholar]
- 62.Sharpless NE, Bardeesy N, Lee KH, Carrasco D, Castrillon DH, Aguirre AJ, Wu EA, Horner JW, DePinho RA. Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature. 2001;413:86–91. doi: 10.1038/35092592. [DOI] [PubMed] [Google Scholar]
- 63.Bianchi T, Rufer N, MacDonald H, Migliaccio M. The tumor suppressor p16Ink4a regulates T lymphocyte survival. Oncogene. 2006;25:4110–4115. doi: 10.1038/sj.onc.1209437. [DOI] [PubMed] [Google Scholar]
- 64.Lagresle C, Gardie B, Eyquem S, Fasseu M, Vieville J-C, Pla M, et al. Transgenic expression of the p16INK4a cyclin-dpendent kinase inhibitor leads to enhanced apoptosis and differentiation arrest of CD4−CD8− immature thymocytes. J Immunol. 2002;168:2325–2331. doi: 10.4049/jimmunol.168.5.2325. [DOI] [PubMed] [Google Scholar]
- 65.Liu Y, Johnson S, Fedoriw Y, Rogers A, Yuan H, Krishnamurthy J, et al. Expression of p16Ink4a prevents cancer and promotes aging in lymphocytes. Blood. 2011;117:3257–3267. doi: 10.1182/blood-2010-09-304402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miyazaki M, Miyazaki K, Itoi M, Katoh Y, Guo Y, Kanno R, Katoh-Fukui Y, Honda H, Amagai T, van Lohuizen M, Kawamoto H, Kanno M. Thymocyte proliferation induced by pre-T cell receptor signaling is maintained through polycomb gene product Bmi-1-mediated Cdkn2a repression. Immunity. 2008;28:231–245. doi: 10.1016/j.immuni.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 67.Jacobs JJL, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and polycomb group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 68.van der Lugt DJ, Linders K, van Roon M, Robanus-Maandag E, te Riele H, van der Valk M, Deschamps J, Sofroniew M, van Lohuizen M. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted dletion of the bmi-1 protooncogene. Genes Develop. 1994;8:757–769. doi: 10.1101/gad.8.7.757. [DOI] [PubMed] [Google Scholar]
- 69.Bruggeman SWM, Valk-Lingbeek ME, van der Stoop PPM, Jacobs JJL, Kieboom K, Tanger E, et al. Ink4a and Arf differentially affect cell proliferation and neural stem cell self-renewal in Bmi1-deficient mice. Genes Develop. 2005;19:1438–1443. doi: 10.1101/gad.1299305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu J, Cao L, Chen J, Song S, Lee I, Quijano C, et al. Bmi1 regulates mitochondrial function and the DNA damage response pathway. Nature. 2009;459:387–392. doi: 10.1038/nature08040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ohtani N, Z Z, Huot T, Stinson J, Sugimoto M, Ohashi Y, et al. Opposing effects of Ets and Id proteins on p16Ink4a expression during cellular senescence. Nature. 2001;409:1067–1070. doi: 10.1038/35059131. [DOI] [PubMed] [Google Scholar]
- 72.Alani R, Young A, Shifflett C. Id1 regulation of cellular senescence through transcriptional repression of p16/Ink4a. Proc Natl Acad Sci U S A. 2001;98:7812–7816. doi: 10.1073/pnas.141235398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nishino J, Kim I, Chada K, Morrison SJ. Hmga2 promotes neural sem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf expression. Cell. 2008;135:227–239. doi: 10.1016/j.cell.2008.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hammond S, Sharpless N. HMGA2, microRNAs, and stem cell aging. Cell. 2008;135:0113–1016. doi: 10.1016/j.cell.2008.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gorospe M, Abdelmohsen K. MicroRegulators come of age in senescence. Trends Genet. 2011;27:233–241. doi: 10.1016/j.tig.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Signer RAJ, Montecino-Rodriguez E, Dorshkind K. The ageing immune system: is it ever too old to become oung again? Nat Rev Immunol. 2009;9:57–62. doi: 10.1038/nri2471. [DOI] [PubMed] [Google Scholar]
- 77.Conboy I, Conboy M, Wagers A, Girma E, Weissman I, Rando T. Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature. 2005;433:760–764. doi: 10.1038/nature03260. [DOI] [PubMed] [Google Scholar]
- 78.Clark R. The somatogenic hormones and insulin-like growth factor-1: stimulators of lymphopoiesis and immune function. Endocrine Rev. 1997;18:157–179. doi: 10.1210/edrv.18.2.0296. [DOI] [PubMed] [Google Scholar]
- 79.Dorshkind K, Horseman ND. The roles of prolactin, growth hormone, insulin-like growth factor-I, and thyroid hormones in lymphocyte development and function: insights from genetic models of hormone and hormone receptor deficiency. Endocr Rev. 2000;21:292–312. doi: 10.1210/edrv.21.3.0397. [DOI] [PubMed] [Google Scholar]
- 80.Napolitano LA, Schmidt D, Gotway MB, Ameli N, Filbert EL, Ng MM, et al. Growth hormone enhances thymic function in HIV-1-infected adults. J Clin Invest. 2008;118:1085–1098. doi: 10.1172/JCI32830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bolotin E, Smogorzewska M, Smith S, Widmer M, Weinberg K. Enhancement of thymopoiesis after bone marrow transplant by in vivo interleukin-7. Blood. 1996;88:1887–1894. [PubMed] [Google Scholar]
- 82.Sutherland JS, Spyroglou L, Muirhead JL, Heng TS, Prieto-Hinojosa A, Prince HM, et al. Enhanced immune system regeneration in humans following allogeneic or autologous hemopoietic stem cell transplantation by temporary sex steroid blockade. Clin Cancer Res. 2008;14:1138–1149. doi: 10.1158/1078-0432.CCR-07-1784. [DOI] [PubMed] [Google Scholar]
- 83.Alpdogan O, Hubbard V, Smith O, Patel N, Lu S, Goldberg G, et al. Keratinocyte growth factor (KGF) is required for postnatal thymic regeneration. Blood. 2006;107:2453–2460. doi: 10.1182/blood-2005-07-2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Min D, Panoskaltsis-Motari A, Juro-o M, Hollander GA, Blazar B, Weinberg KI. Sustained thymopoiesis and improvement in functional immunity induced by exogenous KGF administration in murine models of aging. Blood. 2007;109:2529–2537. doi: 10.1182/blood-2006-08-043794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rossi SW, Jeker LT, Ueno T, Kuse S, Keller MP, Zuklys S, et al. Keratinocyte growth factor (KGF) enhances postnatal T-cell development via enhancements in proliferation and function of thymic epithelial cells. Blood. 2007;109:3803–3811. doi: 10.1182/blood-2006-10-049767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hollander GA, Krenger W, Blazar B. Emerging strategies to boost thymic function. Curr Opin Pharmacol. 2010;10:1–11. doi: 10.1016/j.coph.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chu Y, Schmitz S, Choudhury B, Telford W, Kapoor V, Garfield S, et al. Exogenous insulin-like growth factor 1 enhances thymopoiesis predominantly through thymic epithelial cell expansion. Blood. 2008;112:2836–2846. doi: 10.1182/blood-2008-04-149435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Griffith A, Fallahi M, Nakase H, Gosink M, Young B, Petrie H. Spatial mapping of thymic stromal microenvironments reveals unique features influencing T lymphoid differentiation. Immunity. 2009;31:999–1009. doi: 10.1016/j.immuni.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]