Where do new genes come from? Duplication, divergence, and exon shuffling are the expected answers, so it is especially exciting when new genes are cobbled together from DNA of no related function (or no function at all). In this issue, Chen et al. (1) describe an antifreeze glycoprotein (AFGP) gene in an Antarctic fish that has arisen (in part) from noncoding DNA. Further, they show that a very similar AFGP from an Arctic fish is the product of some completely unrelated molecular processes (2). Together, these papers shed light on a number of key issues in molecular evolution.
In the late 1960s Arthur DeVries showed that freezing resistance in Antarctic fish was due to blood serum glycoproteins that lowered their freezing temperature below that of the subzero sea surrounding them (3, 4). The ensuing years have witnessed a great deal of work on AFPs (antifreeze proteins; not all are glycoproteins) in a number of phylogenetically diverse fish species, much of it by DeVries and his colleagues (5–7), revealing a number of types differing in their structure and amino-acid composition. These proteins, despite their diversity, function in similar ways to deter ice crystal growth (7, 8). But where did they come from, and how did they arise?
Birth of a Gene
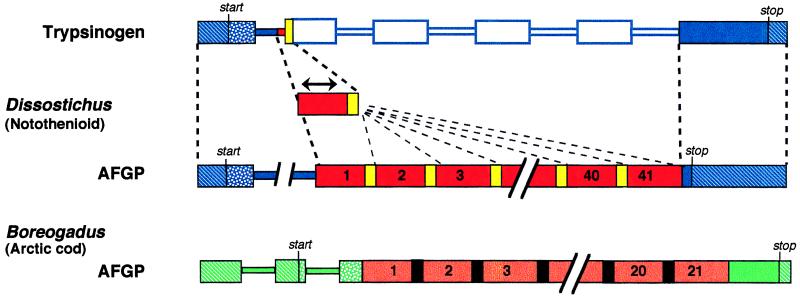
In the first of the two papers, Chen et al. (1) demonstrate that an AFGP gene from the Antarctic notothenioid Dissostichus mawsoni derives from a gene encoding a pancreatic trypsinogen. The relationship of these two genes is not simply one of duplication and divergence (9), co-option/recruitment (10), or exon shuffling (11), processes that have been appreciated by molecular evolutionists for some time now. Instead, the novel portion of the AFGP gene (encoding the ice-binding function) derives from the recruitment and iteration of a small region spanning the boundary between the first intron and second exon of the trypsinogen gene (Fig. 1). This newborn segment was expanded and then iteratively duplicated (perhaps by replication slippage or unequal crossing-over) to produce 41 tandemly repeated segments. Nonetheless, the contemporary AFGP gene retains, as its birthmark, sequences at both ends which are nearly identical to trypsinogen. Retention of the 5′ end of the trypsinogen gene may be significant, since this region encodes a signal peptide used for secretion from the pancreas into the digestive tract. Chen et al. (1) hypothesize that an early version of the notothenioid AFGP gene may have had its first function preventing freezing in the intestinal fluid, with this function later expanded into the circulatory system by way of its expression in the liver.
Figure 1.
Comparison of gene structures and their sequence similarities. The regions shown represent genomic regions encompassed by sequenced cDNAs, and are not to scale. Exons are shown as large boxes; introns are shown as thinner boxes; inferred initiation and termination codons are indicated. Untranslated regions are hatched, and regions encoding putative signal peptides are stippled. Regions in different genes that are the same color share sequence similarity, but only regions of the same color shade are homologous; dotted lines delineate regions of clear homology between Dissostichus trypsinogen and AFGP genes. The open region of the trypsinogen gene is absent in AFGP. The segment below the double-headed arrow represents expansion of a sequence element present in the Dissostichus trypsinogen gene that appears to have given rise to the canonical AFGP repeat; its subsequent tandem iteration is shown by thin dashed lines. AFGP repeats are numbered and discontinuities are indicated for presentation. Regions between the AFGP repeats (spacers; indicated as either yellow or black) are the presumed sites of posttranslational cleavage. A discontinuity in the intron Dissostichus AFGP gene is shown to represent an internal segment not present in the homologous trypsinogen gene intron.
So, what does this case tell us about the evolution of new genes? This AFGP gene is one of a very few newly invented genes that have arisen by processes other than duplication or exon-shuffling whose evolutionary history can be traced with confidence. One other notable case is the jingwei gene of Drosophila, a chimera of a processed alcohol dehydrogenase gene and another unrelated gene, which apparently arose by retrotransposition (12). Although naturally occurring sequence variation in jingwei strongly suggest that it is evolving under natural selection, its actual function is obscure. Other examples of innovations in gene function have been shown to be a result of exon shuffling, a process that has been especially important in animal gene evolution (11). Interestingly, the data from this AFGP suggests that iterative gene segments need not arise by exon-shuffling, as has also been noted in bacterial genes (13). Such repeating tandem duplication has been suggested as a source of protein novelty for some time (14). To consider the AFGP story as a special case of duplication and divergence would be oversimplifying; it is clear that the antifreeze function, or even a related function that could be converted to the purpose, was not present in trypsinogen. The molecular mechanisms involved in the formation of this gene were indeed more creative—making sense from nonsense—by calling into a functional coding capacity intronic DNA sequences.
Molecular Parallelism
Different AFPs likely arose as relatively recent adaptations to cooling during freezing of the Antarctic or Arctic Oceans. There are now known at least four types of fish AFPs, all apparently unrelated to each other (AFPs types I, II, III, and AFGPs; refs. 5 and 6). If each type arose only once, one would expect each to be characteristic of a single monophyletic group of fish. This has clearly turned out not to be the case for AFPs type II, as they have been found in three phylogenetically disparate fish species, Atlantic herring (Order Clupeiformes), smelt (Order Salmoniformes), and sea raven (Order Scorpaenoformes): the genes have evolved at least three times (15). Given this amount of parallel evolution, it is not surprising that these genes have apparently arisen by straightforward duplication and divergence, in this case from C-type lectin genes (15, 16).
AFGPs could also have evolved more than once, since they are found in both Antarctic notothenioids (Order Perciformes) and Arctic cod (Order Gadiformes) (5). This divergence time of these two orders is thought to be ≈40 million years ago (mya), long before the Antarctic freezing (≈14 mya). On the other hand, it had been postulated that AFGPs were present in the common ancestor of these two lineages (5). Still another possible case of parallelism has been suggested in the AFPs type I, since the northern fish that contain them, winter flounder (Order Perciformes) and sculpin (Order Scorpaenoformes), diverged long before the Arctic glaciation (5). Thus, AFPs may have a special propensity to arise by convergent and independent evolutionary means.
Parallelism in the case of AFGPs is quite nicely borne out in the second paper (2), in which Chen et al. describe the sequence of the AFGP gene from the Arctic cod, Boreogadus saida, and compare it to the notothenioid Dissostichus AFGP gene. Arctic cod and notothenioid AFGPs are nearly identical in amino acid composition and are comprised mainly of Thr-Ala-Ala repeats. In fact it was with a notothenioid gene probe (to the AFGP repeats) that Chen et al. isolated the Arctic cod gene, but the organization and sequence of the genes bespeaks their separate ancestries (Fig. 1). First, the coding regions flanking the AFGP repeats in the Arctic cod (including the signal peptide region) are not at all similar to notothenioid AFGP nor trypsinogen; indeed these regions in the Arctic cod AFGP are not identifiably similar to any known sequence. Second, the gene structure of the Arctic cod is quite different from the notothenioid AFGP, with each having different numbers and locations of introns, for example, in differing positions within the signal peptide. Since intron positions are highly conserved in vertebrate genes, they are reliable indicators of homology. Third, the repeating Thr-Ala-Ala of the AFGPs appears to be of different genetic origins. In the notothenioid, there is a strong bias for the specific codons aca-gct/g-gca, whereas many of the Arctic cod repeats are not this sequence, but instead use codons rarely if ever observed in the notothenioid AFGP gene. Finally, the spacers (which provide sites of posttranslational proteolytic cleavage) between AFGP repeats are clearly unrelated (having no sequence similarity) and are presumably processed by different proteases. Given all of this, Chen et al. (2) make a very strong case for the independent origins of AFGP genes.
It will be exciting to investigate the possible convergent evolution of AFPs type I in winter flounder and sculpin that is implied by their organismal phylogeny [as noted by Scott et al. (5)]. One clue is that the flounder and sculpin AFPs do have slightly different amino acid compositions; indeed, their protein sequences are notably dissimilar (17, 18). Although a sculpin AFP gene sequence has not yet been reported, sequences of AFP genes from flounder have been available for some time, and when used as a probe do not produce a signal with sculpin RNA or DNA (5). The isolation of a sculpin type I AFP gene will now be met with palpable evolutionary curiosity.
Caught in the Act: The Beauty of Recency
Nucleotide and amino acid substitutions often erase much, if not all, of the information necessary to draw strong conclusions about evolutionary molecular events, especially ancient ones. The fact that the evolutionary history of the Dissostichus AFGP gene can be pinpointed to trypsinogen and can strongly exclude common ancestry with Boreogadus AFGP is largely due the fact that these events have occurred so recently—there is 95% nucleotide identity between Dissostichus AFGP and trypsinogen genes when only the nucleotide sequences that are clearly homologous are taken together. There are no estimates of rates of substitutions in nuclear-encoded genes of these fish, so using the rate calculated from salmon mitochondrial DNA (probably an overestimate), Chen et al. (1) estimate the origin of the Dissostichus AFGP gene at 5–14 million years (myr). They argue that this correlates well with the presumed date of freezing of the Antarctic Ocean (10–14 myr), as well as molecular phylogenetic estimates of the emergence of AFGP-containing taxa (19). Clearly more data will be needed to precisely define the time of origin of these genes, but the recency of these events should make the job quite manageable
The fact that the Arctic cod AFGP gene arose independently from the notothenioid inevitably leads one to ask about its origin. Although Chen et al. (2) did not find any database matches to the sequence, the source of this gene and the evolutionary events that lead to its birth should be easily within reach. If the evolutionary impetus that created the Boreogadus AFGP gene was indeed Arctic glaciation, then the relevant time frame is only ≈2.5 myr. Given such a limited slice of time, and assuming an endogenous origin, one might isolate the source gene by sequence similarity (possibly even higher than that observed in the trypsinogen/AFGP comparison in Dissostichus). It will be quite interesting to compare the molecular mechanisms that have resulted in such a striking case of convergent evolution.
AFGPs are encoded by multigene families in both Dissostichus and Boreogadus (see figure 4B of ref. 2). Nonetheless, there is no reason to presume that the AFGP genes that have been sequenced from each species are not representative of the gene families as a whole. The two Dissostichus AFGP genes reported are very similar, differing mainly in the number of repeats (41 and 21). In fact, an AFGP gene had also been previously sequenced from another notothenioid species (Notothenia coriiceps neglectia; ref. 20); it is also very similar to the Dissostichus AFGP genes, except that it encodes 46 repeats. There is also promise that a further comparative study, both within gene families of individual species as well as with related species, would more clearly elucidate the pathway of evolution of these genes, possibly by providing examples of intermediates. This approach would be most fruitful in the notothenioids to test the scenario Chen et al. (1) propose in the context of the molecular phylogenetic framework developed by Bargelloni et al. (19).
The strong message from this work is the clear link between a new function that has arisen out of strong selective pressure and an abrupt shift in environmental conditions—adaptive molecular evolution. Demonstrations of this sort at the molecular level are rare and noteworthy. This case could be considered as one of macro-adaptation at the level of whole genes, in contrast to what might be termed micro-adaptation observed at individual sites within preexisting genes (21). With all of its interesting and diverse aspects, the story of AFGP genes will likely be cited as a textbook example of molecular evolution in the years to come.
Acknowledgments
We would like to thank S. L. Baldauf, L. Chen, C.-H. C. Cheng, A. L. DeVries, J. Eastman, and A. Stoltzfus for comments on the manuscript.
References
- 1.Chen L, DeVries A L, Cheng C-H C. Proc Natl Acad Sci USA. 1997;94:3811–3816. doi: 10.1073/pnas.94.8.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen L, DeVries A L, Cheng C-H C. Proc Natl Acad Sci USA. 1997;94:3817–3822. doi: 10.1073/pnas.94.8.3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeVries A L, Wohlschlag D E. Science. 1969;163:1073–1075. doi: 10.1126/science.163.3871.1073. [DOI] [PubMed] [Google Scholar]
- 4.DeVries A L. Science. 1971;172:1152–1155. doi: 10.1126/science.172.3988.1152. [DOI] [PubMed] [Google Scholar]
- 5.Scott G K, Fletcher G L, Davies P L. Can J Fish Aquat Sci. 1986;43:1028–1034. [Google Scholar]
- 6.Davies P L, Hew C L, Fletcher G L. Can J Zool. 1988;66:2611–2617. [Google Scholar]
- 7.Cheng C C, DeVries A L. In: Life Under Extreme Conditions. di Prisco G, editor. Berlin: Springer; 1991. pp. 1–14. [Google Scholar]
- 8.Davies P L, Hew C L. FASEB J. 1990;4:2460–2468. doi: 10.1096/fasebj.4.8.2185972. [DOI] [PubMed] [Google Scholar]
- 9.Ohta T. Genome. 1989;31:304–310. doi: 10.1139/g89-048. [DOI] [PubMed] [Google Scholar]
- 10.Piatagorsky J, Wistow G. Science. 1991;252:1078–1079. doi: 10.1126/science.252.5009.1078. [DOI] [PubMed] [Google Scholar]
- 11.Patthy L. Curr Opin Struct Biol. 1991;1:1351–1361. [Google Scholar]
- 12.Long M, Langley C H. Science. 1993;260:91–95. doi: 10.1126/science.7682012. [DOI] [PubMed] [Google Scholar]
- 13.Little E, Bork P, Doolittle R F. J Mol Evol. 1994;39:631–643. doi: 10.1007/BF00160409. [DOI] [PubMed] [Google Scholar]
- 14.Zuckerkandl E. J Mol Evol. 1975;7:1–57. doi: 10.1007/BF01732178. [DOI] [PubMed] [Google Scholar]
- 15.Ewart K V, Fletcher G L. Mol Mar Biol Biotechnol. 1993;2:20–27. [PubMed] [Google Scholar]
- 16.Ewart K V, Rubinsky B, Fletcher G L. Biochem Biophys Res Commun. 1992;185:335–340. doi: 10.1016/s0006-291x(05)90005-3. [DOI] [PubMed] [Google Scholar]
- 17.Ananthanarayanan V S. Life Chem Rep. 1989;7:1–32. [Google Scholar]
- 18.Knight C A, Cheng C C, DeVries A L. Biophys J. 1991;59:409–418. doi: 10.1016/S0006-3495(91)82234-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bargelloni L, Ritchie P A, Patarnello T, Battaglia B, Lambert D M, Meyer A. Mol Biol Evol. 1994;11:854–863. doi: 10.1093/oxfordjournals.molbev.a040168. [DOI] [PubMed] [Google Scholar]
- 20.Hsiao K C, Cheng C H, Fernandes I E, Detrich H W, DeVries A L. Proc Natl Acad Sci USA. 1990;87:9265–9269. doi: 10.1073/pnas.87.23.9265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Messier W, Stewart C-B. Nature (London) 1997;385:151–154. doi: 10.1038/385151a0. [DOI] [PubMed] [Google Scholar]