Abstract
Thioamide modifications of the peptide backbone are used to perturb secondary structure, to inhibit proteolysis, as photoswitches, and as spectroscopic labels. Thus far, their incorporation has been confined to single peptides synthesized on solid phase. We have generated thioamides in C-terminal thioesters or N-terminal Cys fragments and examined their compatibility with native chemical ligation conditions. Most sequence variants can be coupled in good yields with either TCEP or DTT as the reductant, though some byproducts are observed with prolonged TCEP incubations. Furthermore, we find that thioamides are compatible with thiazolidine protection of an N-terminal Cys, so that multiple ligations can be used to construct larger proteins. Since the acid-lability of the thioamide prohibits on-resin thioester synthesis using Boc chemistry, we devised a method for the synthesis of thioamide peptides with a masked C-terminal thioester that is revealed in situ. Finally, we have shown that thioamidous peptides can be coupled to expressed protein fragments to generate large proteins with backbone thioamide labels by synthesizing labeled versions of the amyloid protein α-synuclein for protein folding studies. In a proof-of-principle experiment, we demonstrated that quenching of fluorescence by thioamides can be used to track conformational changes during aggregation of labeled α-synuclein.
INTRODUCTION
The complex folding and function of proteins is dictated by the interplay of a large number of intra- and intermolecular noncovalent interactions. The interactions of amino acid sidechains are probed through mutation of a given residue to one of the other 19 natural amino acids, or by replacement with an unnatural amino acid through ribosomal incorporation or semi-synthesis.1,2 The role of the protein backbone can only be rationally probed by synthetic modification, as the natural amino acids all possess an oxoamide. A great number of backbone analogs have been developed, including isosteric ester, thioester, and thioamide substitutions.3–10 All of these modifications can be used to probe backbone electrostatic and hydrogen-bonding interactions, but thioamides can also be used to confer proteolytic stability, to photoisomerize the backbone, and as a spectroscopic label in circular dichroism studies.11–13 Very recently, we have demonstrated that they also function as fluorescence quenching probes to study protein dynamics.14,15
Thioamides are nearly isosteric with the natural oxoamides found in the peptide backbone, but there are some subtle differences. Sulfur has a larger van der Waals radius than oxygen (1.85 Å vs. 1.40 Å), and the thiocarbonyl bond is somewhat longer than the oxocarbonyl bond (~ 1.60 Å vs. ~ 1.25 Å).16–19 The thioamide NH is a stronger hydrogen bond donor than the oxoamide NH, while the sulfur is a slightly weaker hydrogen bond acceptor than the corresponding oxygen.20–23 The thioamide sulfur is also more reactive as a nucleophile, reactivity commonly observed in cyclization during Edman degradation.24,25 Despite these differences, the thioamide is generally stable at physiological pH and can be incorporated in an α-helix or a β-turn without grossly perturbing secondary structure.26–28
Thioamide substitutions can also be used as unique photochemical probes because the O-to-S substitution in the carbonyl causes a red-shift of the absorption spectrum. This is manifested as circular dichroism bands at 280 and 330 nm that can be used to monitor local conformational changes at the thioamide.13 The shift of the π → π* absorption from 200 nm for the oxoamide to 260 nm for the thioamide allows one to specifically excite the thiopeptide unit. Irradiation with 260 nm light drives the thiopeptide bond from a predominantly trans conformation to one with a significant cis population, making the thiopeptide a photoswitch.12,29 Our own research has shown that the thioamide bond can act as a quencher of p-cyanophenylalanine (Cnf, F*), tyrosine, and tryptophan fluorescence, and that this quenching can be used to monitor protein dynamics.14,15 Since analogs of the 20 natural amino acids can conceivably be inserted into a peptide sequence, photochemical applications of the thioamide should have no inherent positional restrictions.
One particularly exciting area of application for thioamides is the study of amyloid protein misfolding in neurological disease, such as the Aβ peptide in Alzheimer’s disease or α-synuclein (αS) in Parkinson’s disease.30–36 It is clear that these proteins exert their neurotoxic effect through oligomerization and fibrillization, but the chemical scale details of the misfolding process and the precise identity of the toxic species remain unclear. Thioamides could be used to perturb hydrogen bonding interactions important to misfolding or as minimally-perturbing spectroscopic probes to monitor the misfolding process itself. Kelly and coworkers have shown that some amyloid proteins are tolerant of backbone substitution, making oxoester and olefin isostere replacements of the amide bond to study Aβ.37–39 The short (40 – 42 residue) length of Aβ makes it a viable target for direct peptide synthesis, but other amyloid proteins are too large to be accessed by direct solid phase peptide synthesis (SPPS). Brik, Lashuel, and coworkers have shown that ubiquitin-modified αS can be made through native chemical ligation (NCL), the most common and robust segment condensation reaction for unprotected peptides.40,41 We sought to build off of their work in order to use αS as a test case for the incorporation of backbone thioamides into full-sized proteins. If successful, this would provide us with labeled αS for folding studies and test the general compatibility of thioamides with NCL for protein semi-synthesis. Fluorescence quenching studies with thioamidous αS can be used to make models of structural changes during aggregation.
In spite of their potential utility, the synthesis of thioamide-containing proteins has been limited to single chains assembled on solid phase. The longest fully synthetic thiopeptides are the 35 amino acid leucine zippers of Miwa and coworkers and the Villin headpiece variant HP35 described in Goldberg et al.14,26 Fischer and coworkers were able to reconstitute a functional Ribonuclease S (RNase S) through the noncovalent association of a proteolytic fragment of RNase S and a synthetic thiopeptide.8,42 However, the methods employed with RNase S clearly cannot be used for the general construction of thioamide-containing proteins. There have been, to our knowledge, no systematic studies of the compatibility of thioamides with NCL reactions.
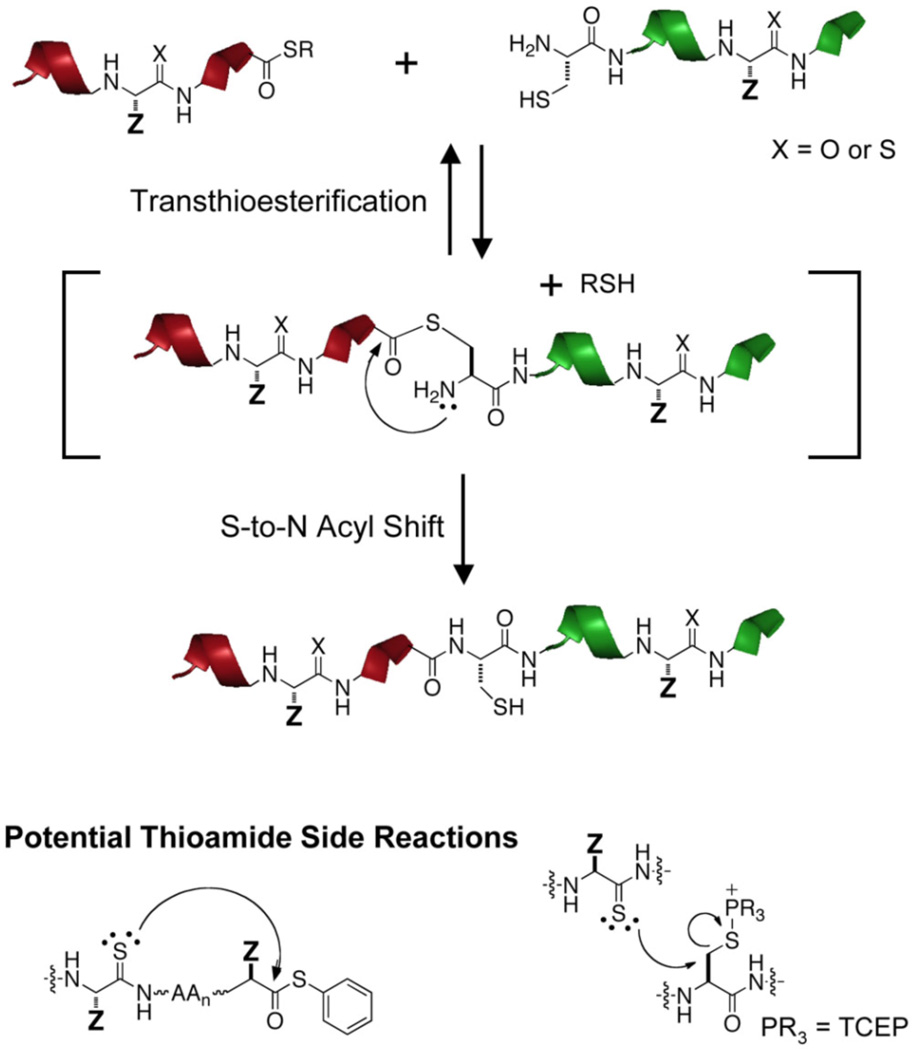
NCL reactions require that one peptide must bear an N-terminal Cys (or Cys surrogate) and that the other peptide be activated as a C-terminal thioester. The two fragments initially engage via reversible transthioesterification, followed by an irreversible S-to-N acyl shift to generate a “native” amide bond. (See Scheme 1, Top)
Scheme 1.
Native Chemical Ligation Mechanism and Potential Thioamide Side Reactions
NCL reactions have been shown to be tolerant of protein functional groups as well as a variety of non-native functional groups.43 Thus, we expected thioamides to be compatible with NCL conditions, although we had some concerns regarding Edman-type side reactions or Cys desulfurization through attack of the nucleophilic thioamide. (See Scheme 1, Bottom) Another potential concern was desulfurization of the thioamide via transient water attack at the thiocarbonyl carbon followed by expulsion of H2S from the tetrahedral intermediate.44,45
We began by synthesizing thioamide-containing C-terminal thioesters or N-terminal Cys peptides and testing their compatibility with NCL. Since the HF cleavage step of Boc-based peptide synthesis degrades thioamides, we used an Fmoc-based method. We also tested several tools that would allow us to make larger proteins: protecting groups for multiple ligations, in situ thioester formation from masked precursors, and finally ligation to expressed proteins. We applied the considerations from these methodological experiments to the successful synthesis of a thioamide-labeled α-synuclein, which can be used in folding experiments using our thioamide fluorescence quenching methods. Note: Thioamide residues are represented by the one or three letter codes of the equivalent oxoamide amino acids with a prime symbol (e.g. L' or Leu' represent thioleucine).
RESULTS AND DISCUSSION
Our initial tests of the stability of thioamides toward NCL reaction conditions used thioesters formed by solution phase thioesterification for ease of synthesis. We have previously shown that Fmoc-protected thiocarboxybenzotriazoles can be used with conventional Fmoc-protected amino acids in the solid-phase synthesis of thioamide-containing peptides. These syntheses are typically carried out on 2-chlorotrityl (Cl-Trt) resin so that deprotection and cleavage can be carried out with moderate (60 – 80%) TFA concentrations. This method required no modification for the synthesis of the N-terminal Cys fragments, and only minor modification to generate C-terminal thioesters. For a thioester, the peptide was synthesized and cleaved with an 8:1:1 CH2Cl2/AcOH/TFE solution that afforded the peptide with its N-terminus and sidechain protecting groups intact. The C-terminus was converted to a thioester using PyBOP and thiophenol, after which the acid-labile protecting groups were removed with TFA. The products were HPLC purified to give thioamide-containing C-terminal thioesters.
Solution-phase thioesterification of protected peptides can lead to epimerization of the α-carbon of the C-terminal residue. However, studies have shown that a judicious choice of activating agent and short reaction times can reduce epimerization.46,47 We find that using PyBOP and thiophenol for reaction times of less than one hour is effective. For example, the activation of Ac-XVA (X = 7-methoxycoumarin-4-yl alanine) was analyzed by HPLC through comparison to an authentic sample of the epimer containing d-Ala, and the yield of epimerized thioester was found to be 1 – 8%. (See Supporting Information) Since the focus of this study is on the reactivity of the thioamide, our initial test peptides featured a C-terminal Gly to avoid concerns about stereochemical integrity.
We generated a series of peptides with strategically placed thioamides and explored their reactivity under standard NCL conditions with either DTT or TCEP as the reducing agent. Reactions were monitored by HPLC (quantified by LC area percentage, LCAP) and MALDI MS or MS/MS. The trial sequences were selected to determine the NCL compatibility of thioamides placed in either the thioester (Table 1, C) or Cys-peptide (Table 1, B) fragment. We also wished to search for possible side reactions of thioamides directly adjacent to the thioester (Table 1, D) or Cys residue (Table 1, A). The peptide sequences are given in Table 1. Peptides A – C were synthesized successfully, but peptide D could not be synthesized because the thioester could not be prepared in pure form. For reaction D, although epimerization could be avoided, the Edman-type cyclization shown in Scheme 1 and subsequent side reactions prohibited the preparation of the thioester 6. In order to understand these side reactions, we subjected Boc-Ala'Gly to our thioesterification conditions. MS and NMR characterization of the intermediates were consistent with phosphonium-mediated deoxygenation followed by reaction with thiophenol. (See Supporting Information)
Table 1.
Thioamide Peptide Test Ligations
| Peptidesa | Red. | Conversionb | Purityb |
|---|---|---|---|
| A. Ac-AKXAGCOSPh (1) | DTT | 86% (18 h) | 74% (4 h) |
| + C𝕃'AKWAA (2) | TCEP | 77% (19 h) | 71% (4 h) |
| B. Ac-AKXAGCOSPh (1) | DTT | 99% (19 h) | 81% (4 h) |
| + CAG𝕃'KXAG (3) | TCEP | 62% (4 h) | 54% (4 h) |
| C. Ac-G𝕃'KXAGCOSPh (4) | DTT | 63% (19 h) | 83% (8 h) |
| + CAGLKXAG (5) | TCEP | 55% (3 h) | 54% (3 h) |
| D. Ac-GAKX𝕃'GCOSPh (6) | n/a | n/a | |
| + CAGLKXAG (5) |
Reactions carried out with 1 mM thioester peptide and 1 mM Cys-peptide in phosphate buffer, pH 7, with 20 mM reductant (Red.) under Ar.
Conversion and purity determined at indicated time as described in Supporting Information.
NCL reactions among the test peptides shown in Table 1 were carried out at 1 mM concentrations in Ar-sparged pH 7 phosphate buffer in the presence of 20 mM DTT or TCEP. For thiophenyl esters, no additional thiophenol was used. Reactions were monitored by HPLC; product and intermediate peaks were identified by MALDI MS. Peak assignments were confirmed by comparison to authentic samples synthesized directly on solid phase. In order to ascertain that no desulfurization occurred, oxoamide versions of some peptides were also prepared and their HPLC retention times compared to ligation HPLC traces. HPLC analysis of peptide C synthesis is shown in Figure 1, others are shown in Supporting Information.
Figure 1.
HPLC Analysis of Ligation to Form Peptide C. The top three panels show aliquots taken at different time points from a reaction using DTT. The bottom panel shows an aliquot taken from a reaction under otherwise identical conditions using TCEP as reductant. Absorbances recorded at 325 nm.
Reactions of these short peptides were typically rapid, with greater than 50% conversion observed after only a few minutes. The rate of conversion of the remaining thioester starting material depended on the addition of excess thiophenol or 4-mercaptophenyl acetic acid (MPAA). Similar results were observed at early timepoints when reactions were carried out in 20 mM TCEP. However, overnight incubation with TCEP resulted in a number of degradation products unless a rigorously oxygen-free atmosphere was maintained. (Figure 1, Bottom) In some cases, maximal yield (conversion) of the ligated products was achieved at different timepoints than maximal purity, and are therefore listed separately in Table 1.
When the preformed disulfide-bonded thiopeptide dimer (L'AF*LCKAXG)2 or its oxoamide equivalent (LAF*LCKAXG)2 were incubated with TCEP, side products unqiue to the thioamide sequence were observed. Cysteine-free thiopeptide L'AF*LAKAXG or oxopeptide monomer LAF*LAKAXG were unaffected by overnight incubation in TCEP buffer. Since no significant side products are observed in DTT ligations, and the TCEP-based side reactions occur only when disulfides are present, we hypothesize that they are initiated by attack of the thioamide sulfur on the thiophosphonium byproduct of disulfide cleavage.48–50 HPLC analyses of these TCEP reactions and our mechanistic hypothesis are included in Supporting Information. Importantly for synthetic considerations, these side reactions can be avoided by limiting initial disulfide bond formation through careful Ar sparging.
While these initial reactions established the fundamental compatibility of thioamides with NCL, several aspects needed further development in order to incorporate thioamides into proteins such as αS. First, we were still limited in size. A single NCL reaction can, at best, produce an 80mer product from two 40mers (lengths are more limited for Fmoc-based SPPS, 70mers have been reported for NCL from Boc-based SPPS).40,51 Second, the manner in which we originally formed thioesters was limited to C-terminal Gly or low thioesterification yields of other amino acids (from short reaction times to prevent epimerization). Third, we had not tested ligation under “protein” conditions, especially in the presence of denaturants. Finally, to access full-sized proteins, it is ideal if one can ligate the synthetic peptide to an expressed protein fragment in order to minimize synthesis of unmodified portions of the protein. To expand the potential application of thioamides through NCL, we began by testing conditions for multiple ligations using N-terminal thiazolidine (Thz, Cz) protection in one peptide fragment.
Kent and others have shown that protection as Thz can be used to block an N-terminal Cys that can then be revealed by treatment with methoxylamine hydrochloride.52,53 We wanted to ensure that MeONH2•HCl incubation would not degrade the thioamide. Therefore, we synthesized three peptide fragments (Fig. 2; 7: CZKKLAQXGG-COSR, 2: H2N-CL′AKWAA-CO2H, and 10: Ac-AKXAG-COSR). First, peptides 7 and 2 were ligated under standard conditions to form 8(Thz). (Fig. 2, Top Right) Then the product was deprotected with MeONH2•HCl to give peptide 9(Cys)Red. HPLC and MALDI analysis showed that the expected product was formed without significant byproducts. Although intramolecular disulfide-bonded 9(Cys)Ox macrocycles formed upon standing, these could be reduced by TCEP for subsequent ligations. (Fig. 2 Middle Right) Peptide 9 was then reacted with a large excess of peptide 10. The product, peptide 11, was confirmed by MADLI MS. This set of experiments showed that thioamides are inert toward methoxylamine treatment and confirmed the functional group compatibility observed in the test reactions.
Figure 2.
Multiple Ligations Using Thz (CZ) Protection of N-Terminal Cys. Left: Reaction scheme with exact masses of reactants and products. Sequences; 7: CZKKLAQXGG-COSR, 2: H2N-CL′AKWAA-CO2H, 9: H2N-CKKLAQXGGCL′AKWAA-CO2H, 10: Ac-AKXAG-COSR, 11: Ac-AKXAGCKKLAQXGGCL′AKWAA-CO2H. Right: HPLC chromatograms and MALDI mass spectra of indicated products. *Contaminant. Yields based on peak integration of HPLC traces shown in Supporting Information.
Thioesterification by activation with PyBOP was useful for the initial test reactions, but the possibility of epimerization and the difficulty of solubilizing fully-protected peptides for C-terminal activation make this an undesirable long term solution. Therefore, we examined on-resin thioesterification methods. “Safety-catch” linkers and related hindered amides require alkylation of the sulfonamide/amide nitrogen by electrophiles, which could react with the thioamide sulfur.54–57 On the other hand, thioesters masked as oxoesters or even as amides can be synthesized on solid phase, then rearrangement to the thioester can be initiated by reduction of a disulfide. Two methods using masked thioesters have been published in the literature. One strategy, used by Muir, Botti, and others, uses a peptide connected as an oxoester to a t-Bu-protected Cys hydroxy acid.58–61 Another strategy, described by Kawakami and MacMillan, uses a peptide connected to a Cys-Pro-Gly/Gla linker, where Gla (Go) is glycolic acid.62,63 Our method uses a Csb-Pro-Gla linker, where Csb (Cb) is a t-Bu-protected Cys; the linker as a whole is hereafter referred to as CbPGo. We focused on the CbPGo linker because of the ease of synthesis (protected Cys hydroxy acid synthesis requires 5 steps).60
We used a slightly different method than the published route, beginning by attaching bromoacetic acid to Rink amide resin. Pro and then a dipeptide of the C-terminal amino acid and Csb are attached and the rest of the peptide is elongated by standard Fmoc-peptide synthesis. After acidic cleavage from the resin, the Csb t-Bu group (inert to TFA) is removed by treatment with TCEP, initiating a cascade of reactions resulting in a peptide thioester attached to the diketopiperazine (Dkp) product of CbPGo rearrangement. (Scheme 2). The Dkp thioester can then either react directly with an N-terminal Cys or undergo transesterification with a thiol additive like MPAA.
Scheme 2.
Thioester Generation by N-to-S Rearrangment
We first tested this strategy by synthesizing thioester 4 from Table 1 (Ac-GL'KXAG-COSR) with the CbPGo linker. The rearrangement, transthioesterification, and ligation can be carried out in one pot, where a peptide such as Ac-GL'KXAGCbPGo (12a) is added to a solution with TCEP and a ligation partner, in this case Cys. TCEP reduction of the disulfide is rapid and efficient. (See Supporting Information) Cyclization is difficult to monitor directly, since the Dkp thioester exchanges readily and is rapidly converted to ligated product. If the reaction is carried out at a low concentration of Ac-GL'KXAGCbPGo in the absence of Cys, intermediates can be observed, such as the Dkp thioester, the branched Ac-GL'KXAG thioester dimer, and the hydrolyzed Ac-GL'KXAG carboxylic acid. (See Supporting Information)
To demonstrate the compatibility of thioamides with NCL conditions typical of full-sized proteins, we synthesized the Villin headpiece HP35 protein: a small 35 amino acid protein that we have previously studied using thioamide FRET.14 Although 35mers are synthetically-accessible without ligation, their convergent synthesis from two fragments would allow us to prepare versions with thioamides at different locations in a combinatorial fashion. More importantly, ligation for proteins of this size allows us to use protein ligation conditions, but the Villin product is small enough that we can monitor desulfurization by HPLC retention and MALDI MS. Here, we ligated a 17mer thioester fragment and a 18mer Cys-peptide. The thioester fragment was synthesized by activation of the protected peptide with PyBOP and thiophenol in DMF. Thioesterified peptide was recovered by precipitation through addition of water, and deprotected with TFA for ligation. Ligation reactions, carried out in phosphate buffer with 20 mM TCEP, saturating thiophenol, and 6 M Gdn•HCl, reached completion with respect to the limiting VillinN peptide in about 4 hours with no TCEP side reactions or desulfurization observed. This laid the ground work for using NCL reactions to attach thioamides to expressed protein fragments.
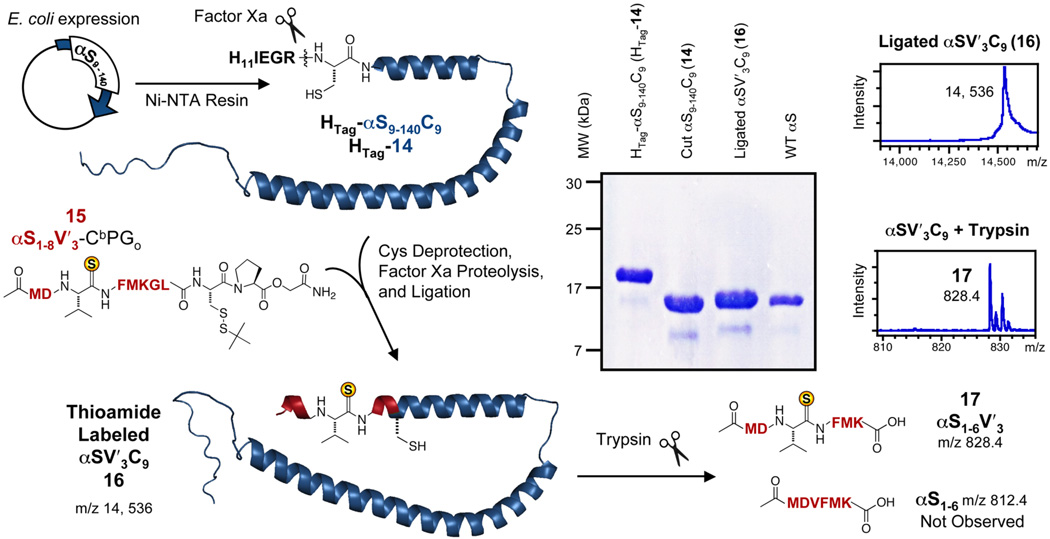
In order to make labeled αS as a first test of thioamide incorporation into proteins through semi-synthesis, we generated constructs for producing a version of αS with an N-terminal His tag which could be removed by proteolysis to reveal an N-terminal Cys for ligation. We chose to use commercially-available Factor Xa protease, which cleaves an IEGR sequence, and had been used previously in expressed protein ligation.1,64 Since αS has no native Cys, we introduced one by altering the expression construct to remove the first eight residues and leave an N-terminal Cys (αS9–140C9) after proteolysis. We chose position 9 because Trexler et al. had previously studied αS with a Ser-to-Cys mutation at this position in FRET experiments.65 The scheme for the semi-synthesis of Val3-labeled αS is shown in Figure 5.
Figure 5.
Synthesis of Labeled αS by Ligation of a Thioamide Peptide to an Expressed Protein Fragment. Left: C-Terminal fragment of αS9–140 with an N-terminal His tag, an S9C mutation, and IEGR cleavage sequence (HTag-14) expressed and purified on Ni-NTA resin. After Factor Xa cleavage to generate an N-terminal Cys (αS9–140C9, 14), the protein fragment is ligated to synthetic αS1–8 with a thioamide at position 3 (αS1–8V′3), generated in situ from precursor αS1–8V′3-CbPGo (15). Top Right: PAGE gel analysis of ligation. Far Right: MALDI MS of full length αSV′3C9 (16, calcd m/z 14, 536) or trypsinized αSV′3C9, giving αS1–6V′3 (17, calcd m/z 828.4). There is no peak corresponding to desulfurized oxopeptide αS1–6 (calcd m/z 812.4).
The C-terminal fragment, HTag-αS9–140C9 (HTag-14), was expressed in E. coli, purified by Ni-NTA chromatography, and proteolyzed with Factor Xa to give αS9–140C9 (14). PAGE gel analysis of these steps (Fig. 5, Top Right) shows that αS9–140C9 was pure and fully cleaved at this point. Additional cleanup could be carried out by HPLC purification using a C4 reverse phase column as necessary. The N-terminal fragment was synthesized on solid phase using the CbPGo linker, with a thioamide inserted at Val3 during SPPS (αS1–8V′3-CbPGo, 15). Activation of the peptide by TCEP yielded αS1–8V′3 Dkp thioester, which reacted with αS9–140C9 in situ to give full-length αSV′3C9 (16). Although ligations reached only 50% completion, ligated αSV′3C9 could be purified by HPLC for analysis. (See Supporting Information)
The integrity of the thioamide bond after ligation was assessed by MALDI MS of the full-length protein and trypsinized products, as well as by UV absorption spectroscopy. Edman-type degradation from thiocarbonyl attack at the F4/M5 amide would result in a loss of the four N-terminal residues, a loss of 552.2 Da. This was not observed in MALDI MS of the ligated αSV′3C9. (Fig. 5) However, this does not address sulfur exchange with oxygen or some more subtle modification. Trypsin digestion releases a peptide corresponding to residues 1 to 5 with a mass of 828.4 Da for the V′3 thiopeptide and 812.4 Da for the oxoamide peptide. MALDI MS of trypsinized αS showed only the mass of the intact thiopeptide, αS1–6V′3 (17). (Fig. 5) Finally, comparison of the UV absorption spectra of αSV′3C9 and αS9–140C9 indicate that 99% of the thioamide is intact after ligation. (See Supporting Information) By all of these measures, the thioamide appears to have been incorporated without degradation.
Finally, in order to demonstrate the value of incorporating thioamide probes into a protein such as αS, we carried out an aggregation experiment in which intramolecular conformational changes were monitored in an αS construct. αS, like other amyloidogenic proteins, is known to aggregate first into soluble oligomers of two to hundereds of subunits, and then into longer fibrils which can tangle to form Lewy bodies.30,31,68,69 Since there is a great deal of uncertainty as to the structures of the oligomers and the structural changes that lead to fibrillization, a method for site-selectively monitoring conformational change could be very valuable to understanding αS pathology. Indeed, many fluorescence-based studies have attempted to determine structures for aggregation intermediates, but they often employ fluorophores that can be perturbing to structure, particularly when substituted at sites that do not natively contain an aromatic amino acid.70–81 Our thioamide probe, which has been shown to quench Trp, Tyr, and the unnatural amino acid Cnf, should offer much greater freedom, particularly in the placement of the acceptor (i.e. the thioamide) in a donor/acceptor pair and can be used to access the stretch from residues 40 to 93, which contains no aromatic amino acids.
We employ an αS construct with a single Trp donor fluorophore (W94) and a single thioamide quencher (V′3). As noted above, our previous work has shown that Trp fluorescence is quenched by thioamides in a distant-dependent manner.15 The native αS sequence has no Trp residues, so selective excitation of an introduced Trp can be achieved with 295 nm light. Comparison to an oxoamide control (i.e. αSW94) allows us to isolate thioamide quenching effects from other environmental effects on Trp fluorescence.82 Since through-space quenching does not occur over distances longer than 30 Å, observation of Trp fluorescence quenching normalized to oxoamide controls shows that the Trp must be within 30 Å of the thioamide residue.15 However, in an oligomer or fibril, quenching can arise through either inter- or intramolecular interactions. In order to isolate intramolecular misfolding events, we carry out aggregation experiments with our αSV′3C9W94 construct present in a 1:30 ratio with WT αS. A ratio of less than one Trp/thioamide-labeled αS per 26 WT αS should ensure that, on average, no two labeled αS are directly next to each other in aggregates. This derives from a simple statistical packing model in which each monomer unit would be at the center of a cube with 26 partners along the centers of faces and vertices, or at corners.
The construct used in aggregation studies (18) was synthesized by ligation of the same synthetic fragment (αS1–8V′3) with an expressed C-terminal fragment (αS9–140C9W94). For an aggregation experiment, the presence of non-native Cys in the sequence could be problematic as the formation of disulfide-bonded dimers would alter the aggregation mechanism.66,67 Therefore, we incubated αS with β-mercaptoethanol (BME) to prevent disulfide bond formation. No significant increase in aggregation rate was observed in the Cys mutants.
Aggregation experiments were carried out by shaking the 1:30 αSV′3C9W94/αS mixtures at 37 °C in phosphate buffer with BME. Although overall Trp fluorescence increased over 4 days during protein aggregation, the relative fluorescence of the thioamide (FThio/FOxo) decreased. (See Fig. 6 for FThio/FOxo data, and see Supporting Information for primary fluorescence data.) This indicates that positions 3 and 94 approach each other to within 30 Å in the oligomeric and fibrillar states, but that they are more separated in the monomeric state. Since we used a 1:30 αSV′3C9W94/αS ratio, we interpret the quenching as arising primarily from intramolecular interactions within the labeled molecule. We normalize our data to an equivalent oxoamide protein (1:30 αSW94/αS) in order to account for other local effects on fluorescence, which are necessarily complex in an aggregate. We monitored overall fibrillization by ThT fluorescence, as well as the independent metrics of Congo Red staining and sedimentation PAGE gel analysis. (See Supporting Information)
Figure 6.
Monitoring Intramolecular Misfolding During αS Aggregation Using Thioamide Quenching. Left: Monomeric αSV′3C9W94 (18) mixed in a 1:30 ratio with WT αS. Aggregation assay carried out by shaking at 37 °C for several days. Samples taken daily and Trp fluorescence measured at 350 nm. ThT added and fluorescence at 490 nm measured after 2 min incubation. Trp fluorescence quenching is observed in oligomers and fibrils. ThT fluorescence is only observed for fibril-bound ThT. Top Right: Schematic representation of αSV′3C9W94 construct showing Trp, thiovaline, and Cys. Bottom Right: Normalized Trp (squares) and ThT (circles) fluorescence data for four aggregation experiments with αSV′3C9W94 (Thio) and control experiments with αSW94 (Oxo). Both Trplabeled proteins were used in 1:30 mixtures with WT αS. See Supporting Information for examples of primary fluorescence data and a table containing normalized fluorescence values used in generating this plot.
Comparing the ThT data to the Trp fluorescence data clearly indicates that we can observe quenching in oligomers, which occur at early timepoints (< 24 h), before fibrillization and ThT fluorescence. (Fig. 6) It is unlikely that multiple labeled αS molecules are present in oligomers of less than 30 monomer units. Therefore, quenching of Trp fluorescence provides evidence of close intramolecular approach of residues 3 and 94 when bound in oligomers, consistent with some previous studies showing folding of the N- and C-termini before fibrillization.81 Our experiments show that thioamide fluorescence quenching should be useful in monitoring intermediates in the aggregation process, which are silent in the conventional ThT protocol. Conducting many such experiments in conjunction with positional scanning of the thioamide probe should provide mechanistic insight into the aggregation process and the mode of toxicity of these meta-stable intermediates.
CONCLUSIONS
Thioamides can be valuable biophysical tools for perturbing backbone hydrogen bonds, tracking protein motions, or inducing cis/trans isomerization. Previously, their uses have been restricted to peptides that can be synthesized on solid phase. Here, we have shown that the thioamide functional group is compatible with native chemical ligation conditions. Furthermore, we have seen that thioamides are compatible with thiazolidine protection of an N-terminal Cys, so that multiple ligations can be used to construct larger proteins. To enable facile syntheses of the requisite thioesters using Fmoc chemistry, we adapted methods for the generation of a masked thioester using the CbPGo linker. Deprotection of the t-Bu sidechain protecting group can be carried out in situ so that the thioester is generated and reacts rapidly with an N-terminal Cys. Finally, we have shown that thioamidous peptides synthesized with the CbPGo linker can be coupled to expressed protein fragments by synthesizing a labeled version of αS.
We have recently shown that thioamide quenching of the unnatural amino acid Cnf can be used to monitor protein folding.14 In addition, thioamide quenching of Trp or Tyr could be used to monitor interactions for native proteins.15 Most significantly, we have found that visible wavelength fluorophores can be quenched by thioamides through electron transfer, so that folding experiments could be carried out with site-selective fluorophore excitation in the presence of biological chromophores.83 However, all of these experiments would be limited to thiopeptides synthesized on solid phase without the ability to ligate thioamide-containing peptide fragments. Thus, the methods developed here will be used to synthesize thioamide-labeled proteins for use in the study of protein folding and protein-protein interactions. Misfolded or aggregating proteins are particularly attractive as they are often refractory to higher resolution structural techniques such as crystallography or NMR. We envision generating a large number of labeled αS constructs and using thioamide fluorescence quenching to monitor conformational changes during aggregation and misfolding. Our proof-of-principle experiment indicates that these studies are feasible and should reveal information that cannot be accessed with common methods such as ThT binding. Of course, our methods are potentially applicable to a great number of protein dynamics questions, and we will continue to explore thioamide compatibility with NCL reactions for the synthesis and study of these proteins.
EXPERIMENTAL PROCEDURES
General Information
Fmoc-l-4-cyanophenylalanine (Fmoc-Cnf-OH) was purchased from Peptech (Burlington, MA). Boc-l-thionoleucine-1-(6-nitro)benzotriazolide, Fmoc-Gln(Trt)-OH, Fmoc-Asn(Trt)-OH, and Fmoc-β-(7-methoxy-coumarin-4-yl)-Ala-OH (denoted X) were purchased from Bachem (Torrance, CA) or EMD Chemicals (Philadelphia, PA). Benzotriazol-1-yl-oxy-trispyrrolidino-phosphonium hexafluorophosphate (PyBOP), Fmoc-Ala-OH, Fmoc-Leu-OH, Fmoc-Glu(OtBu)-OH, Fmoc-Lys(Boc)-OH, Fmoc-Trp(Boc)-OH, Fmoc-Arg(Pbf)-OH, Fmoc-Pro-OH, Fmoc-Thr(tBu)-OH, Fmoc-Met-OH, Fmoc-Gly-OH, Fmoc-Val-OH, Fmoc-Phe-OH, Fmoc-Asp(OtBu)-OH, Fmoc-Ser(tBu)-OH, Fmoc-Cys(Trt)-OH, 2-chlorotrityl chloride resin, and Rink amide resin were purchased from Novabiochem (San Diego, CA). Piperidine and 2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU) were purchased from American Bioanalytical (Natick, MA). Sigmacote, N,N-diisopropylethylamine (DIPEA), thiophenol, benzyl mercaptan, trifluoroacetic acid (TFA), tris(2-carboxyethyl)phosphine hydrochloride (TCEP), and Boc-4-thiazolidinecarboxylic acid (Boc-Thz-OH) were purchased from Sigma-Aldrich (St. Louis, MO). All deuterated solvents were purchased from Cambridge Isotopes Laboratories, Inc. (Andover, MA).
Ni-NTA resin was from Qiagen (Valencia, CA). E. coli BL21(DE3) cells were purchased from Stratagene (La Jolla, CA). Sequencing-grade trypsin was purchased from Promega (Madison, WI). Restriction Grade Factor Xa protease was purchased from Novagen (San Diego, CA). All other reagents were purchased from Fisher Scientific (Pittsburgh, PA).
Milli-Q filtered (18 MΩ) water was used for all solutions (Millipore; Billerica, MA). Matrix-assisted laser desorption ionization (MALDI) mass spectra were collected with a Bruker Ultraflex III MALDI-TOF-TOF mass spectrometer (Billerica, MA). Electrospray ionization (ESI) mass spectra were collected with a Waters LCT Premier XE liquid chromatograph/mass spectrometer (Milford, MA). UV/Vis absorbance spectra were obtained with a Hewlett-Packard 8452A diode array spectrophotometer (Agilent Technologies, Santa Clara, CA). Fluorescence spectra were collected with a Varian Cary Eclipse fluorescence spectrophotometer fitted with a Peltier multicell holder (currently Agilent Technologies). Nuclear Magnetic Resonance (NMR) spectra were obtained with either a Bruker DRX 500 MHz or DMX 500 MHz instrument.
Peptide Synthesis
All peptides were synthesized using a manual, Fmoc-based solid-phase procedure described previously.14 Thioamide monomers were synthesized using the general procedure previously described for thioleucine.14 Explicit procedures for the synthesis of thiovaline are given in Supporting Information. Off-resin peptide thioesterfications are also described in Supporting Information. The thiazolidine group was introduced as a Boc-4-thiazolidine carboxylic acid and followed the general SPPS procedure.
Ligation of Model Thioamide Peptides
The N-terminal peptide thioester 1 or 4 (1 equiv, 0.5 µmol) and the C-terminal peptide fragment 2, 3, or 5 (1 equiv, 0.5 µmol) were dissolved in a freshly made, argon-sparged ligation buffer (100 mM Na2HPO4, 20 mM DTT or TCEP, pH 7.0 – 7.2). A typical ligation began with the combination of the two peptide fragments in a 5 mL, septum-capped, and argon-purged round-bottom flask. The final volume of the reaction was 500 µL, giving a 1 mM final concentration of each peptide. The reaction mixture was sparged with argon for 15 min and stirred under an argon atmosphere at room temperature. To monitor the ligation progress, aliquots (50 µL) were taken periodically at regular intervals and diluted to 800 µL with 0.1 % TFA in water. Each sample was analyzed by analytical HPLC on a YMC-Pack Pro C18 analytical column (Kyoto, Japan) using gradient 2 (Table S2). HPLC chromatograms for the ligations in Table 1 are given in Figure S2 in Supporting Information.
Ligation of Thiazolidine-Protected Cys Peptides
CzKKLAQXGG-S(CH2)2CO2Me 7 (1 equiv, 0.2 µmol, 1 mM) and CL'AKWAA 2 (1 equiv, 0.2 µmol, 1 mM) were incubated in 200 µL of a freshly made, argon-sparged ligation buffer (6 M Gdn•HCl, 100 mM Na2HPO4, 25 mM TCEP, pH 7.2). The reaction was initiated by the addition of thiophenol (1 % v/v, 2 µL). After 22 h of stirring, the reaction was quenched with 0.1 % TFA in water. The isolated product, CzKKLAQXGGCL'AKWAA 8, was purified by HPLC on a Vydac 218 TP C18 semiprep column using gradient 2 and characterized by MALDI MS (Supporting Information Table S2 and Table S3). The ligated peptide was treated with 100 µL of a deprotection solution (6 M Gdn•HCl, 100 mM Na2HPO4, 200 mM MeONH2•HCl, pH 4) for 8 h. The conversion from Thz-thiopeptide to Cys-thiopeptide 4 was confirmed by MALDI MS (Fig. 2). The deprotected Cys-thiopeptide was purified by HPLC on a Vydac 218 TP C18 semiprep column using gradient 2 (Table S2). For the second ligation, the Cys-thiopeptide 9 (1 equiv, 0.009 µmol, 0.2 mM) and Ac-AKXAG-SPh 10 (2.5 equiv, 0.022 µmol, 0.5 mM) were dissolved in the ligation buffer (same as above). The reaction was initiated by the addition of thiophenol (4% v/v, 2 µL). The reaction was purged with argon for 15 min and stirred under an argon atmosphere at room temperature for 8 h. To monitor the ligation progress, aliquots (10 µL) of the reaction solution were taken periodically, diluted to 800 µL, and analyzed by analytical HPLC on a YMC-Pack Pro C18 analytical column using gradient 2 (Supporting Information Table S2). The final ligated product was confirmed by MALDI MS (Supporting Information Table S3).
Synthesis of CbPGo Peptides
Ac-GL'KXAGCbPGo 12a and Ac-MDV′3FMKGLCbPGo (αS1–8V'3-CbPGo, 15) were synthesized on Rink amide resin, each at 100 µmol scale. A synthetic scheme is given in Supporting Information. The resin was first deprotected with 20% piperidine in DMF and rinsed with DMF. Bromoacetic acid (0.0695 g, 500 µmol, 5 equiv) was pre-activated with DCC (0.0516 g, 250 µmol, 2.5 equiv) for 30 min in dry DMF (6 mL), and then coupled to the resin over 30 min. Subsequently, Fmoc-Pro-OH (0.1687 g, 500 µmol, 5 equiv) was dissolved in dry DMF (6 mL) with DIPEA (174.3 µL, 1 mmol, 10 equiv), and coupled to the resin for 30 min.
The next two residues were introduced as a dipeptide to avoid unwanted Dkp formation. For Ac-GL'KXAGCbPGo as an initial test, Fmoc-Gly-Cys(S-t-Bu)-OH was synthesized on 2-chlorotrityl resin at 125 µmol scale with the general SPPS protocol, and cleaved with CH2Cl2/AcOH/TFE (8:1:1 v/v). The crude product (0.0537 g, 110 µmol, 1.1 equiv) was dissolved in DMF (6 mL), with PyBOP (0.0572 g, 110 µmol, 1.1 equiv) and DIPEA (174.3 µL, 1 mmol, 10 equiv), and then coupled to the Pro residue for 1 h. For αS1–8V'3-CbPGo, an improved procedure was adopted, where Fmoc-Leu-Cys(S-t-Bu)-OH was synthesized at 2 mmol scale in solution phase (a detailed experimental procedure is given in Supporting Information). The purified dipeptide (0.1634 g, 300 µmol, 3 equiv) was dissolved in DMF (6 mL), with PyBOP (0.1561 g, 300 µmol, 3 equiv) and DIPEA (174.3 µL, 1 mmol, 10 equiv), and then coupled to the Pro residue for 90 min. A second coupling was carried out to maximize the coupling efficiency. The rest of both peptides were elongated and acetylated with the standard protocol.
For Ac-GL'KXAGCbPGo 12a, the product was cleaved from the resin with two treatments of CH2Cl2/TFA (7:3 v/v, 6 mL) on a rotisserie, each for 30 min. The resulting solutions were combined, dried by rotatory evaporation, and purified by HPLC on a Vydac 218 TP C18 semiprep column using gradient 12 (Supporting Information, Tables S1 and S2). For αS1–8V'3-CbPGo 15, thioanisole (500 µL) and TIPS (250 µL) were added to the resin at 0 °C, followed by TFA (5 mL). The system was stirred at 0 °C for 20 min, then allowed to warm up to room temperature over 30 min. The resulting solution was concentrated by rotatory evaporation, from which the crude product was precipitated with cold diethyl ether (10 mL) and purified by HPLC on a Vydac 218 TP C18 analytical column using gradient 10 (Supporting Information, Tables S1 and S2).
Ligation of Ac-GL'KXAGCbPGo with Cysteine
A 50 mM cysteine stock in water was freshly prepared. Peptide Ac-GL'KXAGCbPGo 15 (0.1 µmol, ε325 = 12, 000 M−1 cm−1) was dissolved in H2O/CH3CN (11:4 v/v, 31.2 µL), and mixed with the cysteine stock (20.8 µL).84 A 2 µL aliquot of the above solution was taken, and diluted with 0.1% TFA in H2O (798 µL) as a t0 HPLC standard. 50 µL of a freshly prepared ligation buffer stock (400 mM Na2HPO4, 100 mM TCEP, pH 8.4) was added to initiate the reaction. The reaction was placed in an incubator at 37 °C, shaking at 250 RPM. At appropriate time points, a 4 µL sample of the reaction mixture was taken, quenched with 0.1% TFA in H2O (796 µL), and analyzed by analytical HPLC on a YMC-Pack Pro C18 analytical column using gradient 11 (Supporting Information Table S2 and Table S3).
Native Chemical Ligation of Villin
The VillinN thioester (1 equiv, 0.2 µmol, 1 mM) and the VillinC fragment (2 equiv, 0.4 µmol, 2 mM) were dissolved in a freshly made, argon-sparged ligation buffer (6 M Gdn•HCl, 100 mM Na2HPO4, 20 mM TCEP, pH 7.2) to give a final volume of 200 µL. The ligation was initiated by the addition of thiophenol (1% v/v, 2 µL) and was stirred under an argon atmosphere at room temperature. Aliquots (20 µL) of the reaction solution were taken periodically, diluted to 800 µL, and analyzed by analytical HPLC on a YMC-Pack Pro C18 analytical column using gradient 6 (Supporting Information, Table S2). In this method, VillinC, VillinN thioester and ligated Villin eluted at 14.8 min, 25.1 min, and 24.1 min, respectively (Fig. 4). Ligated Villin was isolated, dried in vacuo, and characterized by UV/Vis spectroscopy and MALDI MS (Supporting Information, Table S3, Fig. S9, and Fig. S10).
Figure 4.
HPLC Analysis of Villin HP35 Synthesis by NCL. VillinN thiophenyl ester synthesized by PyBOP activation. Aliquots taken at different timepoints from a reaction using TCEP as reductant. Absorbance recorded at 215 nm.
Overexpression and Purification of Recombinant αS9–140C9 and αS9–140C9W94 Mutants
A plasmid encoding HTag-αS9–140C9 (pSB6159) or HTag-αS9–140C9W94 (pSB7021) was generated as described in Supporting Information. This plasmid was transformed into E. coli BL21-(DE3) cells grown on agar plates. A starter culture of 4 mL LB media was inoculated with a single colony and was grown at 37 °C in the presence of ampicillin (100 mg/L) for approximately 6 h. A secondary culture of 500 mL LB was inoculated with 1 mL of the starter culture and grown at 37 °C in the presence of ampicillin. The cultures were induced with 1 mM of isopropyl-β-d-thiogalactopyranoside (IPTG) at OD600 of 0.7 and allowed to grow at 25 °C overnight. Cells were pelleted at 6, 000 RPM using a GS3 rotor and Sorvall RC-5 centrifuge for 15 min at 4 °C. After discarding the supernatant, the pellet was resuspended in 10 mL of resuspension buffer (50 mM Tris, 150 mM NaCl, 50 µM ethylenediaminetetraacetic acid (EDTA), pH 8.0, protease inhibitor cocktail, 1 mM phenylmethanesulfonyl fluoride (PMSF), and 10 units/mL DNAse1–Grade II). The cells were lysed by sonication, and the lysate was centrifuged at 13, 200 RPM for 15 min at 4 °C. The supernatant was then incubated with Ni-NTA resin for 1 h on ice. This slurry was loaded into a column and the liquid allowed to flow through. The resin was washed extensively, first with 15 mL of Wash Buffer A (50 mM Tris, 150 mM NaCl, 20 mM TCEP, pH 8.0), then twice with 10 mL of Wash Buffer B (50 mM Tris, 150 mM NaCl, 20 mM TCEP, 5 mM imidazole, pH 8.0), and twice with 10 mL of Wash Buffer C (50 mM Tris, 150 mM NaCl, 20 mM TCEP, 30 mM imidazole, pH 8.0). The protein was eluted with six 2 mL portions of elution buffer (50 mM Tris, 150 mM NaCl, 20 mM TCEP, 250 mM imidazole, pH 8.0). The eluted fractions were combined and dialyzed against cleavage buffer (50 mM Tris, 150 mM NaCl, 5 mM CaCl2, pH 8.0) overnight. The protein that would not immediately be proteolyzed was stored at − 80 °C until further use.
Purification of αS9–140C9 or αS9–140C9W94
Ni-NTA-purified HTag-αS9–140C9 (1 mg/mL) was proteolyzed with Factor Xa protease (20 units per 1 mg of HTag-αS9–140C9) for 14 h at 37 °C without shaking. The completion of proteolysis was confirmed by MALDI MS. The reaction was quenched by the addition of 1 mM PMSF and 5 mM TCEP and incubated for 30 min at room temperature. Subsequently, the protein solution was incubated with 1 mL of Ni-NTA resin for 1 h at room temperature to remove the N-terminal fusion peptide. The resin was loaded into a column, and the flow-through was collected. The resin was washed with 3 × 2 mL of cleavage buffer, and the resulting solutions were combined with the flow-through. The combined solution was dialyzed against Milli-Q water, and further purified by HPLC on a Vydac 214 TP C4 prep. column using gradient 8 (Supporting Information Table S2). Purification of αS9–140C9W94 followed an identical procedure.
Native Chemical Ligation of αS1–8V'3-CbPGo and αS9–140C9 or αS9–140C9W94
Masked thioester αS1–8V'3-CbPGo 15 (1.5 equiv, 0.09 µmol, 1.5 mM) and expressed protein fragment αS9–140C9 (1 equiv, 0.06 µmol, 1 mM) were dissolved in 60 µL of a freshly made, degassed ligation buffer (6 M Gdn•HCl, 200 mM Na2HPO4, 20 mM TCEP, 1% v/v thiophenol, and 1% v/v benzylmercaptan, pH 8.0). The reaction solution was placed in an incubator at 37 °C, shaking at 250 RPM. An aliquot (30 µL) was removed from the solution at reaction time of 8 h and quenched with 0.1 % TFA in water (470 µL). The quenched aliquot was dialyzed against Milli-Q water, and analyzed by HPLC on a Vydac 214 TP C4 analytical column using gradient 12 (Table S2). The rest of the ligation solution was quenched at reaction time of 25 h, dialyzed, and analyzed by HPLC. The product fractions were pooled, dried in vacuo, and characterized by MALDI MS (Table S3).
Ligation of αS9–140C9W94 was carried out in a nearly identical fashion with the following changes: Protein buffer was exchanged using an Amicon (Millipore) Ultra 0.5 mL 10 kDa spin column prior to set up of the ligation reaction and 2 % v/v thiophenol was used.
Trypsin Digest Analysis of αSV'3C9
αSV'3C9 16 (20 µg) was dissolved in 18 µL of freshly prepared 25 mM NH4HCO3 (pH 7.5), and digested with 2 µL of sequencing grade modified trypsin (0.1 µg/µL). The digestion was allowed to proceed at 37 °C for 4 h. An aliquot (1.0 µL) of the digest was taken and analyzed by MALDI MS (Fig. 5).
αS Aggregation Assays
Aggregation experiments were performed according to literature precedent.73,79 350 µL samples for aggregation assays were prepared (97 µM WT αS with 3 µM αSW94 or 3 µM αSV′3C9W94) in phosphate buffered saline (pH 7.0) containing 1 mM β-mercaptoethanol. Aggregation was seeded by the addition of approximately 10% (wt/wt) preformed WT αS fibrils. The samples were incubated at 37 °C for 4 to 6 days with continuous shaking at 1100 rpm. 40 µL aliquots were periodically removed to monitor changes in tryptophan fluorescence and ThT or Congo Red binding. Protocols and examples of primary fluorescence or absorbance data are given in Supporting Information.
Supplementary Material
Figure 3.
HPLC and MALDI MS Analysis of Thioester Formation from Csb-Pro-Gla (CbPGo) Linker. Cleaved, purified thioester precursor Ac-GL'KXAGCbPGo-NH2 (12a) was incubated with TCEP and Cys at pH 8.4. Top: Representative HPLC chromatogram at t = 120 min showing deprotected reactant peptide Ac-GL'KXAGCPGo-NH2 (12, red circles), Cysligated product Ac-GL'KXAGC-CO2H (13, blue squares), and side products (green triangles). Absorbance recorded at 325 nm. Inset: MALDI MS of product HPLC peak (23.7 min retention time, m/z calcd 873.3). Bottom: Relative concentrations of reactant, product, and side products determined from HPLC chromatograms. Labels are as above. Reactant decay and product growth (sum of product and side products, orange diamonds) fit to single exponential functions to obtain half lives. See Supporting Information for HPLC data, MS characterization, and fitting procedures.
ACKNOWLEDGMENT
The authors thank Elizabeth Rhoades for the gift of the pT7 plasmid containing α-synuclein. We thank Virginia Lee, John Trojanowski, and members of the Center for Neurodegenerative Disease Research for the gift of the pRK172 plasmid and for helpful advice. We also thank Cheryl McCullough for assistance with peptide synthesis. SB thanks John Warner for guidance with molecular biology.
Funding Sources
This work was supported by funding from the University of Pennsylvania, Searle Scholars Program (10-SSP-214 to EJP), and the National Science Foundation (NSF CHE-1020205 to EJP). Instruments supported by the National Science Foundation and National Institutes of Health include: HRMS (NIH RR-023444) and MALDI-MS (NSF MRI-0820996).
ABBREVIATIONS
- Cnf or F*
p-cyanophenylalanine
- αS
α-synuclein
- SPPS
solid phase peptide synthesis
- NCL
native chemical ligation
- Fmoc
9-fluorenylmethoxycarbonyl
- Boc
tert-Butoxycarbonyl
- TFA
trifluoroacetic acid
- TFE
trifluoroethanol
- PyBOP
benzotriazol-1-yl-oxytripyrrolidinophosphonium hexafluorophosphate
- TCEP
tris-carboxyethylphosphine
- DTT
dithiothreitol
- HPLC
high pressure liquid chromatography
- MALDI MS
matrix-assisted laser desorption/ionization mass spectrometry
- MPAA
4-mercaptophenylacetic acid
- Thz or Cz
1,3-thiazolidine-4-carboxylic acid
- Csb or Cb
S-tert-Butyl-l-cysteine
- Gla or Go
glycolic acid
- Dkp
diketopiperazine
- Gdn
guanidinium; thioamide residues are represented by the one or three letter code of the equivalent oxoamide amino acids with a prime symbol (e.g. L' or Leu' represent thioleucine)
- BME
β-mercaptoethanol
Footnotes
ASSOCIATED CONTENT
Supporting Information. Procedures for peptide synthesis, protein expression, thioester activation, and ligation reactions; analytical HPLC traces and MALDI MS data; schemes illustrating proposed side reaction mechanisms; primary aggregtion data, including Trp fluorescence, ThT fluorescence, Congo Red absorbance, and PAGE gel analysis. This material is available free of charge via the Internet at http://pubs.acs.org. Primary data used to generate figures and tables have been digitally archived and can be obtained by emailing the corresponding author.
REFERENCES
- 1.Muir TW. Ann. Rev. Biochem. 2003;72:249–289. doi: 10.1146/annurev.biochem.72.121801.161900. [DOI] [PubMed] [Google Scholar]
- 2.Wang L, Schultz PG. Angew. Chem. Int. Ed. 2005;44:34–66. doi: 10.1002/anie.200460627. [DOI] [PubMed] [Google Scholar]
- 3.Choudhary A, Raines RT. 2011;12:1801–1807. doi: 10.1002/cbic.201100272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clausen K, Spatola AF, Lemieux C, Schiller PW, Lawesson SO. Biochem. Biophys. Res. Commun. 1984;120:305–310. doi: 10.1016/0006-291x(84)91449-9. [DOI] [PubMed] [Google Scholar]
- 5.Deechongkit S, Nguyen H, Powers ET, Dawson PE, Gruebele M, Kelly JW. Nature. 2004;430:101–105. doi: 10.1038/nature02611. [DOI] [PubMed] [Google Scholar]
- 6.Powers ET, Deechongkit S, Kelly JW. Peptide Solvation and H-Bonds. 2006;Vol. 72:39–78. [Google Scholar]
- 7.Woll MG, Gellman SH. J. Am. Chem. Soc. 2004;126:11172–11174. doi: 10.1021/ja046891i. [DOI] [PubMed] [Google Scholar]
- 8.Bachmann A, Wildemann D, Praetorius F, Fischer G, Kiefhaber T. Proc. Natl. Acad. Sci. USA. 108:3952–3957. doi: 10.1073/pnas.1012668108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baca M, Kent SBH. Tetrahedron. 2000;56:9503–9513. [Google Scholar]
- 10.Schnolzer M, Kent SBH. Science. 1992;256:221–225. doi: 10.1126/science.1566069. [DOI] [PubMed] [Google Scholar]
- 11.Bartlett PA, Spear KL, Jacobsen NE. Biochemistry. 1982;21:1608–1611. doi: 10.1021/bi00536a022. [DOI] [PubMed] [Google Scholar]
- 12.Frank R, Jakob M, Thunecke F, Fischer G, Schutkowski M. Angew. Chem. Int. Ed. 2000;39:1120–1122. [PubMed] [Google Scholar]
- 13.Hollosi M, Kollat E, Kajtar J, Kajtar M, Fasman GD. Biopolymers. 1990;30:1061–1072. [Google Scholar]
- 14.Goldberg JM, Batjargal S, Petersson EJ. J. Am. Chem. Soc. 2010;132:14718–14720. doi: 10.1021/ja1044924. [DOI] [PubMed] [Google Scholar]
- 15.Goldberg JM, Wissner RF, Klein AM, Petersson EJ. Chem. Commun. 2012;48:1550–1552. doi: 10.1039/c1cc14708k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bardi R, Piazzesi AM, Toniolo C, Jensen OE, Omar RS, Senning A. Biopolymers. 1988;27:747–761. [Google Scholar]
- 17.Bondi A. J. Phys. Chem. 1964;68:441–451. [Google Scholar]
- 18.Lacour TFM, Hansen HAS, Clausen K, Lawesson SO. Int. J. Peptide Protein Res. 1983;22:509–512. doi: 10.1111/j.1399-3011.1983.tb02122.x. [DOI] [PubMed] [Google Scholar]
- 19.Truter MR. J. Chem. Soc. 1960:997–1007. [Google Scholar]
- 20.Dudek EP, Dudek G. J. Org. Chem. 1967;32:823–824. [Google Scholar]
- 21.Hollosi M, Majer Z, Zewdu M, Ruff F, Kajtar M, Kover KE. Tetrahedron. 1988;44:195–202. [Google Scholar]
- 22.Hollosi M, Zewdu M, Kollat E, Majer Z, Kajtar M, Batta G, Kover K, Sandor P. Int. J. Peptide Protein Res. 1990;36:173–181. doi: 10.1111/j.1399-3011.1990.tb00963.x. [DOI] [PubMed] [Google Scholar]
- 23.Laurence C, Berthelot M, Lequestel JY, Elghomari MJ. J. Chem. Soc. Perkin Trans. 1995:2075–2079. [Google Scholar]
- 24.Edman P, Begg G. Eur. J. Biochem. 1967;1:80–91. doi: 10.1007/978-3-662-25813-2_14. [DOI] [PubMed] [Google Scholar]
- 25.Wipf P, Hayes GB. Tetrahedron. 1998;54:6987–6998. [Google Scholar]
- 26.Miwa JH, Pallivathucal L, Gowda S, Lee KE. Org. Lett. 2002;4:4655–4657. doi: 10.1021/ol027056d. [DOI] [PubMed] [Google Scholar]
- 27.Miwa JH, Patel AK, Vivatrat N, Popek SM, Meyer AM. Org. Lett. 2001;3:3373–3375. doi: 10.1021/ol0166092. [DOI] [PubMed] [Google Scholar]
- 28.Reiner A, Wildemann D, Fischer G, Kiefhaber T. J. Am. Chem. Soc. 2008;130:8079–8084. doi: 10.1021/ja8015044. [DOI] [PubMed] [Google Scholar]
- 29.Helbing J, Bregy H, Bredenbeck J, Pfister R, Hamm P, Huber R, Wachtveitl J, De Vico L, Olivucci M. J. Am. Chem. Soc. 2004;126:8823–8834. doi: 10.1021/ja049227a. [DOI] [PubMed] [Google Scholar]
- 30.El-Agnaf OMA, Salem SA, Paleologou KE, Curran MD, Gibson MJ, Court JA, Schlossmacher MG, Allsop D. FASEB J. 2006;20:419–425. doi: 10.1096/fj.03-1449com. [DOI] [PubMed] [Google Scholar]
- 31.Giehm L, Svergun DI, Otzen DE, Vestergaard B. Proc. Natl. Acad. Sci. USA. 2011;108:3246–3251. doi: 10.1073/pnas.1013225108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, Glabe CG. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 33.Lesne S, Koh MT, Kotilinek L, Kayed R, Glabe CG, Yang A, Gallagher M, Ashe KH. Nature. 2006;440:352–357. doi: 10.1038/nature04533. [DOI] [PubMed] [Google Scholar]
- 34.Roychaudhuri R, Yang M, Hoshi MM, Teplow DB. J. Biol. Chem. 2009;284:4749–4753. doi: 10.1074/jbc.R800036200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walsh DM, Hartley DM, Kusumoto Y, Fezoui Y, Condron MM, Lomakin A, Benedek GB, Selkoe DJ, Teplow DB. J. Biol. Chem. 1999;274:25945–25952. doi: 10.1074/jbc.274.36.25945. [DOI] [PubMed] [Google Scholar]
- 36.Yu LP, Edalji R, Harlan JE, Holzman TF, Lopez AP, Labkovsky B, Hillen H, Barghorn S, Ebert U, Richardson PL, Miesbauer L, Solomon L, Bartley D, Walter K, Johnson RW, Hajduk PJ, Olejniczak ET. Biochemistry. 2009;48:1870–1877. doi: 10.1021/bi802046n. [DOI] [PubMed] [Google Scholar]
- 37.Bieschke J, Siegel SJ, Fu YW, Kelly JW. Biochemistry. 2008;47:50–59. doi: 10.1021/bi701757v. [DOI] [PubMed] [Google Scholar]
- 38.Deechongkit S, Powers ET, You SL, Kelly JW. J. Am. Chem. Soc. 2005;127:8562–8570. doi: 10.1021/ja050558c. [DOI] [PubMed] [Google Scholar]
- 39.Fu YW, Gao JM, Bieschke J, Dendle MA, Kelly JW. J. Am. Chem. Soc. 2006;128:15948–15949. doi: 10.1021/ja065303t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hackenberger CPR, Schwarzer D. Angew. Chem. Int. Ed. 2008;47:10030–10074. doi: 10.1002/anie.200801313. [DOI] [PubMed] [Google Scholar]
- 41.Hejjaoui M, Haj-Yahya M, Kumar KSA, Brik A, Lashuel HA. Angew. Chem. Int. Ed. 2011;50:405–409. doi: 10.1002/anie.201005546. [DOI] [PubMed] [Google Scholar]
- 42.Wildemann D, Schiene-Fischer C, Aumuller T, Bachmann A, Kiefhaber T, Lucke C, Fischer G. J. Am. Chem. Soc. 2007;129:4910–4918. doi: 10.1021/ja069048o. [DOI] [PubMed] [Google Scholar]
- 43.Alewood P, Engelhard M, Kent SBH. J. Peptide Sci. 2010;16:513–513. doi: 10.1002/psc.1291. [DOI] [PubMed] [Google Scholar]
- 44.Chae MY, Czarnik AW. J. Am. Chem. Soc. 1992;114:9704–9705. [Google Scholar]
- 45.Corsaro A, Pistara V. Tetrahedron. 1998;54:15027–15062. [Google Scholar]
- 46.Camarero JA, Mitchell AR. Protein Peptide Lett. 2005;12:723–728. doi: 10.2174/0929866054864166. [DOI] [PubMed] [Google Scholar]
- 47.Flemer S. J. Peptide Sci. 2009;15:693–696. doi: 10.1002/psc.1181. [DOI] [PubMed] [Google Scholar]
- 48.Burns JA, Butler JC, Moran J, Whitesides GM. J. Org. Chem. 1991;56:2648–2650. [Google Scholar]
- 49.Dmitrenko O, Thorpe C, Bach RD. J. Org. Chem. 2007;72:8298–8307. doi: 10.1021/jo071271w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu PR, O'Mara BW, Warrack BM, Wu W, Huang YP, Zhang YH, Zhao RL, Lin M, Ackerman MS, Hocknell PK, Chen GD, Tao L, Rieble S, Wang J, Wang-Iverson DB, Tymiak AA, Grace MJ, Russell RJ. J. Am. Soc. Mass Spectrom. 2010;21:837–844. doi: 10.1016/j.jasms.2010.01.016. [DOI] [PubMed] [Google Scholar]
- 51.Valiyaveetil FI, Sekedat M, Muir TW, MacKinnon R. Angew. Chem. Int. Ed. 2004;43:2504–2507. doi: 10.1002/anie.200453849. [DOI] [PubMed] [Google Scholar]
- 52.Bang D, Pentelute BL, Kent SBH. Angew. Chem. Int. Ed. 2006;45:3985–3988. doi: 10.1002/anie.200600702. [DOI] [PubMed] [Google Scholar]
- 53.Boerema DJ, Tereshko VA, Kent SBH. Biopolymers. 2008;90:278–286. doi: 10.1002/bip.20800. [DOI] [PubMed] [Google Scholar]
- 54.Blanco-Canosa JB, Dawson PE. Angew. Chem. Int. Ed. 2008;47:6851–6855. doi: 10.1002/anie.200705471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ingenito R, Bianchi E, Fattori D, Pessi A. J. Am. Chem. Soc. 1999;121:11369–11374. [Google Scholar]
- 56.Mende F, Seitz O. Angew. Chem. Int. Ed. 2011;50:1232–1240. doi: 10.1002/anie.201005180. [DOI] [PubMed] [Google Scholar]
- 57.Sharma I, Crich D. J. Org. Chem. 2011;76:6518–6524. doi: 10.1021/jo200497j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Botti P, Villain M, Manganiello S, Gaertner H. Org. Lett. 2004;6:4861–4864. doi: 10.1021/ol0481028. [DOI] [PubMed] [Google Scholar]
- 59.Chen G, Warren JD, Chen JH, Wu B, Wan Q, Danishefsky SJ. J. Am. Chem. Soc. 2006;128:7460–7462. doi: 10.1021/ja061588y. [DOI] [PubMed] [Google Scholar]
- 60.George EA, Novick RP, Muir TW. J. Am. Chem. Soc. 2008;130:4914–4924. doi: 10.1021/ja711126e. [DOI] [PubMed] [Google Scholar]
- 61.Macmillan D. Angew. Chem. Int. Ed. 2006;45:7668–7672. doi: 10.1002/anie.200602945. [DOI] [PubMed] [Google Scholar]
- 62.Kawakami T, Shimizu S, Aimoto S. J. Peptide Sci. 2010;16:50–50. [Google Scholar]
- 63.Kang J, Macmillan D. Org. Biomol. Chem. 2010;8:1993–2002. doi: 10.1039/b925075a. [DOI] [PubMed] [Google Scholar]
- 64.Jenny RJ, Mann KG, Lundblad RL. Protein Expression Purif. 2003;31:1–11. doi: 10.1016/s1046-5928(03)00168-2. [DOI] [PubMed] [Google Scholar]
- 65.Trexler AJ, Rhoades E. Biochemistry. 2009;48:2304–2306. doi: 10.1021/bi900114z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jiang CT, Chang JY. Biochemistry. 2007;46:602–609. doi: 10.1021/bi062068i. [DOI] [PubMed] [Google Scholar]
- 67.Suk JE, Lokappa SB, Ulmer TS. Biochemistry. 2010;49:1533–1540. doi: 10.1021/bi901753h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Luk KC, Hyde EG, Trojanowski JQ, Lee VMY. Biochemistry. 2007;46:12522–12529. doi: 10.1021/bi701128c. [DOI] [PubMed] [Google Scholar]
- 69.Auluck PK, Caraveo G, Lindquist S. Ann. Rev. Cell Dev. Biol. 2010;26:211–233. doi: 10.1146/annurev.cellbio.042308.113313. [DOI] [PubMed] [Google Scholar]
- 70.Lee JC, Langen R, Hummel PA, Gray HB, Winkler JR. Proc. Natl. Acad. Sci. USA. 2004;101:16466–16471. doi: 10.1073/pnas.0407307101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kaylor J, Bodner N, Edridge S, Yamin G, Hong DP, Fink AL. J. Mol. Biol. 2005;353:357–372. doi: 10.1016/j.jmb.2005.08.046. [DOI] [PubMed] [Google Scholar]
- 72.Klucken J, Outeiro TF, Nguyen P, McLean PJ, Hyman BT. FASEB J. 2006;20:2050–2057. doi: 10.1096/fj.05-5422com. [DOI] [PubMed] [Google Scholar]
- 73.Thirunavukkuarasu S, Jares-Erijman EA, Jovin TM. J. Mol. Biol. 2008;378:1064–1073. doi: 10.1016/j.jmb.2008.03.034. [DOI] [PubMed] [Google Scholar]
- 74.van Rooijen BD, van Leijenhorst-Groener KA, Claessens M, Subramaniam V. J. Mol. Biol. 2009;394:826–833. doi: 10.1016/j.jmb.2009.10.021. [DOI] [PubMed] [Google Scholar]
- 75.Veldhuis G, Segers-Nolten I, Ferlemann E, Subramaniam V. Chembiochem. 2009;10:436–439. doi: 10.1002/cbic.200800644. [DOI] [PubMed] [Google Scholar]
- 76.Middleton ER, Rhoades E. Biophysical Journal. 2010;99:2279–2288. doi: 10.1016/j.bpj.2010.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nath S, Meuvis J, Hendrix J, Carl SA, Engelborghs Y. Biophysical Journal. 2010;98:1302–1311. doi: 10.1016/j.bpj.2009.12.4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.van Ham TJ, Esposito A, Kumita JR, Hsu STD, Schierle GSK, Kaminsk CF, Dobson CM, Nollen EAA, Bertoncini CW. J. Mol. Biol. 2010;395:627–642. doi: 10.1016/j.jmb.2009.10.066. [DOI] [PubMed] [Google Scholar]
- 79.Yushchenko DA, Fauerbach JA, Thirunavukkuarasu S, Jares-Erijman EA, Jovin TM. J. Am. Chem. Soc. 2010;132:7860–7861. doi: 10.1021/ja102838n. [DOI] [PubMed] [Google Scholar]
- 80.Schierle GSK, Bertoncini CW, Chan FTS, van der Goot AT, Schwedler S, Skepper J, Schlachter S, van Ham T, Esposito A, Kumita JR, Nollen EAA, Dobson CM, Kaminski CF. Chemphyschem. 2011;12:673–680. doi: 10.1002/cphc.201000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yap TL, Pfefferkorn CM, Lee JC. Biochemistry. 2011;50:1963–1965. doi: 10.1021/bi2000824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen Y, Barkley MD. Biochemistry. 1998;37:9976–9982. doi: 10.1021/bi980274n. [DOI] [PubMed] [Google Scholar]
- 83.Goldberg JM, Speight LC, Fegley MW, Petersson EJ. doi: 10.1021/ja3005094. Unpublished. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brun MP, Bischoff L, Garbay C. Angew. Chem. Int. Ed. 2004;43:3432–3436. doi: 10.1002/anie.200454116. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.