Abstract
Bacterial two-component systems (TCSs) are signaling pathways composed of two proteins, a histidine kinase (HK) and a response regulator (RR). Upon stimulation, the HK autophosphorylates at a conserved histidine. The phosphoryl group is subsequently transferred to an aspartate on a RR, eliciting an adaptive response, often up- or downregulation of gene expression. TCS signaling controls many functions in bacteria including development, virulence and antibiotic resistance, making the proteins involved in these systems potential therapeutic targets. Efficient methods for the profiling of HKs are currently lacking. For direct readout of HK activity, we sought to design a probe that enables detection of the phosphotransfer event; however, analysis of the phosphohistidine species is made difficult by the instability of the P-N bond. We anticipated that use of a γ-thiophosphorylated ATP analog, which would yield a thiophosphorylated histidine intermediate, could overcome this challenge. We determined that the fluorophore-conjugated probe, ATPγS-BODIPY, labels active HK proteins and is competitive for the ATP-binding site. This activity-based probe provides a new strategy for analysis of TCSs and other HK-mediated processes and will facilitate both functional studies and inhibitor identification.
Two-component systems (TCSs) are stimulus-and-response signaling pathways composed of a histidine kinase (HK) and response regulator (RR). TCSs exist primarily in bacteria, but have also been identified in some lower eukaryotes.1,2 They contribute to many critical bacterial functions including virulence, resistance mechanisms and survival, making the TCS proteins potential therapeutic targets.3–5 For example, TCS signaling has been implicated in pneumonia, toxic shock syndrome, tuberculosis and a plethora of other acute and chronic infections.6 Moreover, TCSs have been correlated to antibiotic resistance in multiple Gram-negative and Gram-positive bacterial systems.1,4,7 HKs are known to be regulated by a number of stimuli such as temperature, peptides and metal ions, which are sensed by an extracellular input domain.1,6,8 HK activation promotes conformational changes in the catalytic and ATP-binding (CA) domain as well as the dimerization and His phosphotransfer (DHp) domain (Figure 1A). Once the protein is in the activated state, a conserved His in the DHp domain attacks the γ-phosphate of ATP resulting in autophosphorylation (Figure 1B).3,9 The phosphoryl group is subsequently transferred to an aspartate (Asp) on a cognate RR eliciting an adaptive response, often up- or downregulation of gene expression.3,9 Most bacteria express 20–30 or more TCSs,10 many of which may have redundant or overlapping functions. Thus, development of facile methods to facilitate their study is an important goal.
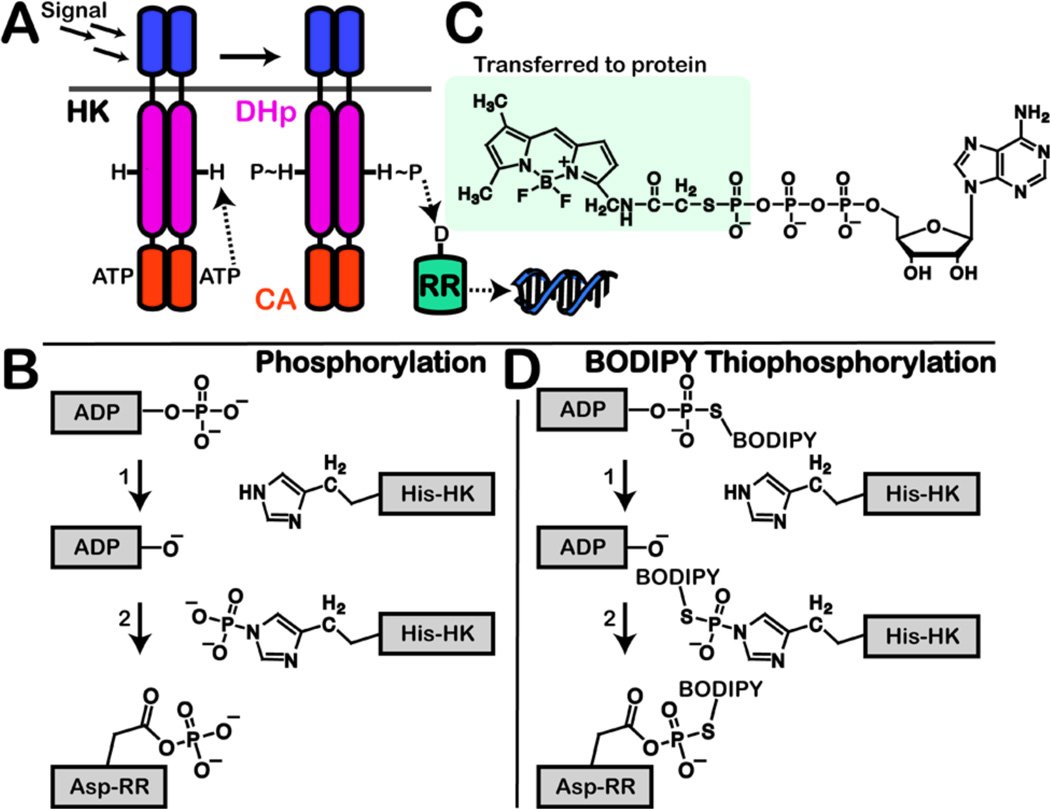
Figure 1.
(A) HK is activated by an extracellular signal, which results in phosphorylation of a conserved histidine by ATP. The phosphoryl group is transferred to the aspartate on a RR. Activated RR then triggers a response, typically acting as a transcription factor. Domains: DHp, dimerization and histidine phosphotransfer; CA, catalytic and ATP-binding. (B) Mechanisms of HK autophosphorylation and phosphotransfer to RR. (C) Structure of B-ATPγS, highlighting the thiophosphate-BODIPY moiety that is transferred during autophosphorylation. (D) Proposed mechanism of B-ATPγS labeling of active HK and cognate RR.
Generation of straightforward methods for the profiling of HK activity has been a long-standing challenge due to the instability of the phosphohistidine (pHis) species.11 Unlike the study of phosphorylation at serine, threonine and tyrosine that has enjoyed great advancement in the last decade,12,13 detection of phosphorylated histidine residues has lagged far behind. Some success has been achieved by application of Phos-tag, a phosphate-binding molecule embedded in a polyacrylamide gel matrix, to facilitate the separation and quantitation of phosphorylated from non-phosphorylated HKs.14,15 However, this technique has not seen wide application. The use of LC-MS-based techniques for detecting pHis-containing proteins/peptides is also severely hindered by the acid lability and short half-life of the P-N bond.16,17 Careful optimization can enable detection of specific phosphorylated HKs, but a global strategy does not exist. pHis instability is also likely responsible for the failure to generate an antibody for detection of phosphorylated histidine-containing proteins to date. Promisingly, some recent progress has been made using stable synthetic pHis analogs.18 Due to the described difficulties, most TCS studies still depend upon the use of radioactivity-based assays or analysis of the isostable phosphorylated RR species.
For direct readout of HK activity, we sought to design a probe that enables detection of the phosphotransfer event; however, it was clear that a strategy for generating a more stable intermediate species would be required. We anticipated that use of a γ-thiophosphorylated ATP analog, which would yield a thio-pHis, could overcome this challenge given the increased stability that the sulfur confers on the P-N bond.16,17 Several labs have demonstrated that ATPγS can act as a substrate for HKs.19,20 Recently, Shokat and Marletta used this alternative substrate to facilitate immunodetection of HK activity20 in a manner analogous to that previously developed for Ser/Thr kinases.21 Their studies demonstrated that ATPγS is a widely accepted substrate analog being turned over by five of six tested enzymes. Taking advantage of the ATPγS scaffold, we sought to devise a direct-readout strategy that could detect HK activity in one step and be visualized immediately.
With this goal in mind, we examined a fluorescently labeled ATP analog, BODIPY-FL-ATPγS (B-ATPγS), for its utility as an activity-based probe (ABP) for HKs (Figure 1C).22 ABPs are commonly substrate-like scaffolds that covalently modify their enzyme target (s) to facilitate detection of the activity state.23 Although B-ATPγS is generally considered to be non-hydrolyzable, we hypothesized that it might be accepted as a substrate of HKs and could serve as an ABP given that autothiophosphorylation with this probe would yield a fluorescently labeled histidine conjugate (Figure 1D). We examined this possibility with HK853, a histidine kinase protein from Thermotoga maritima.24,25
Initial experiments demonstrated that HK853 has good affinity for this probe, Kdapp=474±40 nM as measured using a fluorescence polarization-based assay, which is in stark contrast to reported ATP affinity values (~14–500 µM; Supporting Figure 1).26,27 Next, we assessed the ability of the enzyme to autophosphorylate with the γ-thiophosphate-BODIPY group. HK853 was incubated with B-ATPγS (2 µM) at 25 °C for 1 h prior to running sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) for in-gel fluorescence detection. Addition of the probe resulted in labeling of HK853 (Figure 2A). Examination of the concentration dependence of the B-ATPγS reaction revealed that half-maximal labeling occurs at ~2 µM (Figure 2B and Supporting Figure 2). Labeling was also observed with VicK from Streptococcus pneumoniae (Supporting Figure 3).28 In addition, when B-ATPγS was incubated with an HK construct that contains the ATP-binding domain but lacks the DHp domain, rendering it inactive, no labeling was observed further demonstrating the activity dependence of probe labeling (PhoQ from Salmonella typhimurium;29 Supporting Figure 3).
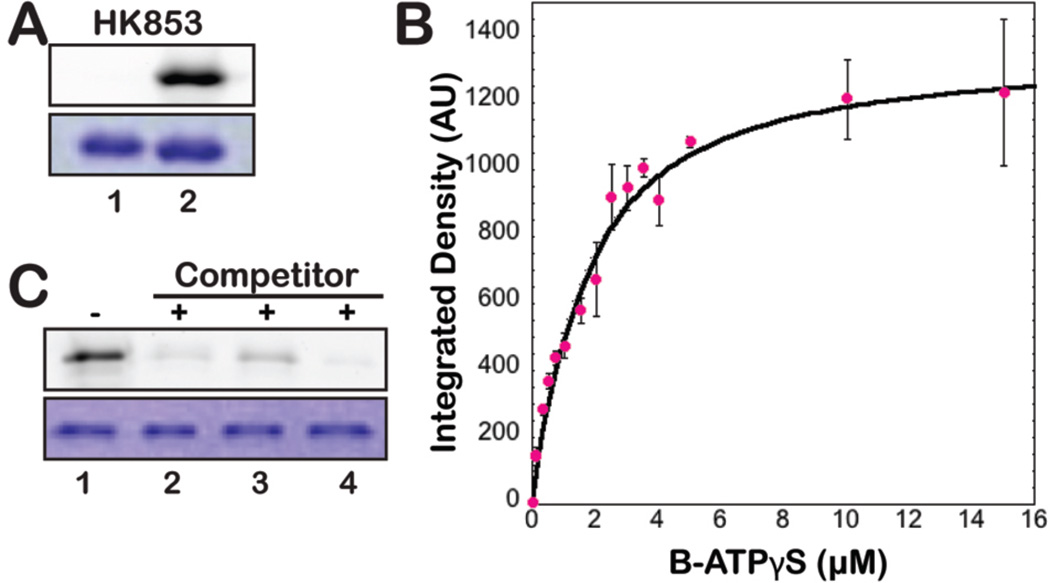
Figure 2.
(A) B-ATPγS used as an ABP to show in vitro thiophosphorylation of active HK853 (T. maritima). Top: in-gel fluorescence detection of the BODIPY fluorophore shows signal only with addition of probe (2 µM). Bottom: Coomassie staining reveals even protein loading. Lanes: 1, no probe; 2, B-ATPγS added. (B) Concentration dependence of HK853 labeling by B-ATPγS. Labeling was saturated at ~10 µM and was measured by in-gel fluorescence scanning (integrated band intensities given in arbitrary units). (C) B-ATPγS labeling of HK853 was inhibited when ATP analogs (200 µM, 10 min) were added prior to B-ATPγS (2 µM). Lanes: 1, no inhibitor; 2, ATP; 3, AMP-PNP; 4, ATPγS.
To provide further evidence that protein labeling occurs in an activity-dependent fashion, we utilized various ATP-analogs as competitive substrates/inhibitors (ATP, AMP-PNP, ATPγS). Upon incubation with each of these compounds (200 µM, 10 min) followed by addition of the probe, a dramatic decrease in labeling was detected indicating that the ABP is binding in the ATP-binding pocket (Figure 2C). We also examined the ability of B-ATPγS to compete with [γ-33P]-ATP and observed the expected decrease in the amount of radioactive protein upon addition of the probe (Supporting Figure 4). Finally, although the sulfur atom in our probe imparts added stability to the formed thiophosphoramidate, these species are still heat sensitive.16,17 To confirm that labeling occurs by formation of such a bond, we heat treated probe-labeled protein, resulting in immediate disappearance of the labeled band (95 °C, 5–30 min; Supporting Figure 5).
We have strong evidence for the thiophosphotransfer of BODIPY-γ-thiophosphate to the conserved histidine of HK853. However, during TCS signaling, this is not the end of the pathway: the phosphoryl group undergoes another transfer event to RR468, its cognate RR (Figure 1B). Thus, we hypothesized that the fluorescently tagged thiophosphate group could be transferred to the aspartic acid on the response regulator (Figure 1D), but it was unclear whether or not the bulky BODIPY group would prevent recognition between the HK and RR. To test this, we first incubated HK853 with B-ATPγS for 1 h and then added the cognate RR468. The reaction was quenched in hour intervals and run on an SDS-PAGE gel. Gratifyingly, the band corresponding to RR468 grew increasingly fluorescent over time, which indicates successful transfer of the BODIPY-γ-thiophosphate from the HK to the RR (Figure 3). As a control, B-ATPγS was incubated with RR468 alone, and no labeling of the RR was observed indicating that HK-dependent transfer of the BODIPY-γ-thiophosphate to RR468 was occurring.
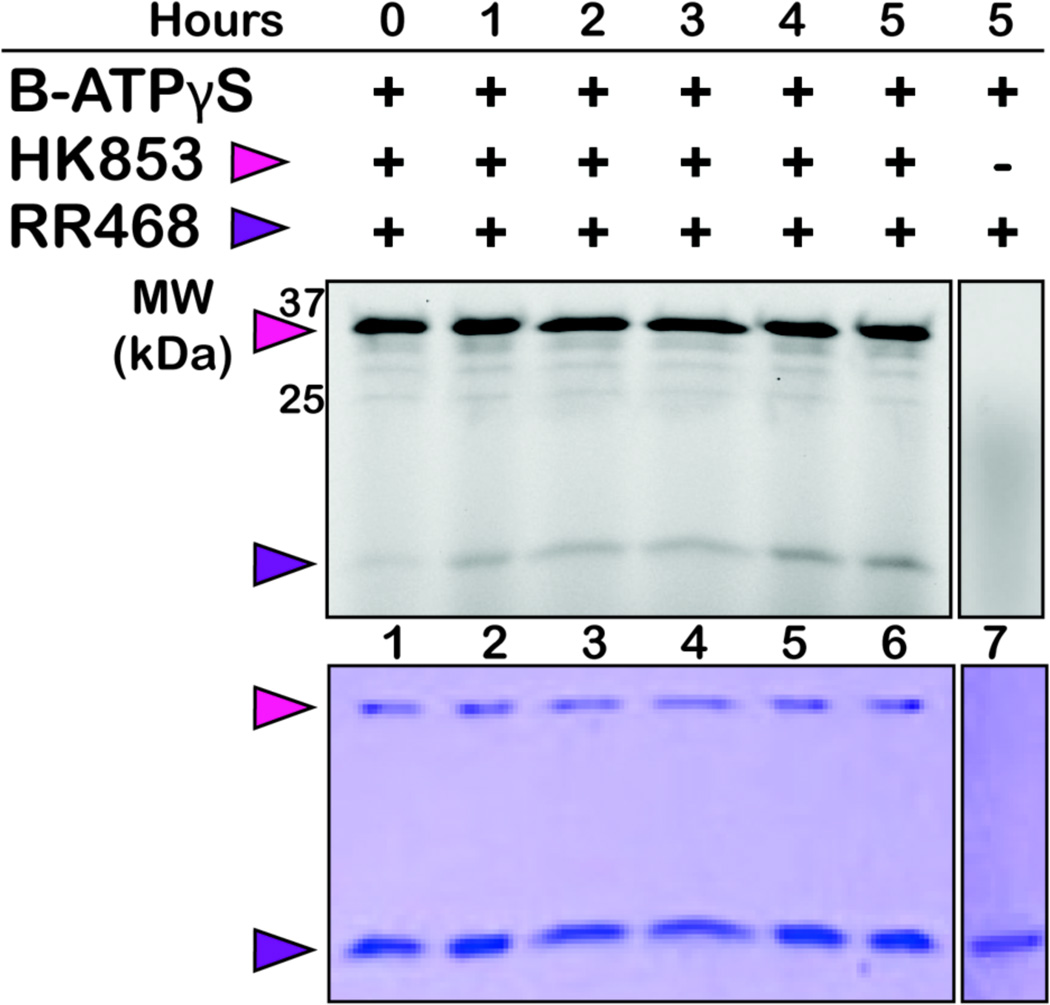
Figure 3.
BODIPY-thiophosphate transfers to RR468 (16 kDa) over time only when HK853 (32 kDa) is present. HK853 was first incubated with the probe for 1 h at 25 °C. The RR468 was then added at 7-fold the concentration of HK853. Top: In-gel fluorescence detection of the BODIPY fluorophore. Bottom: Coomassie staining of the same gel reveals even protein loading. The control lane (7) reveals that RR labeling is HK dependent.
Transfer of the phosphate group between an HK and RR is usually an exceptionally fast process (seconds to minutes). The exchange that we observed was dramatically slower (hrs). This slow RR thiophosphorylation may be due to altered transfer kinetics because of the oxygen-to-sulfur substitution. Furthermore, the size of the BODIPY molecule may hinder HK/RR recognition. In either case, the delayed transfer of the thiophosphate to the RR makes B-ATPγS an excellent tool to visualize the often fleeting phosphorylated HK species, while at the same time still enabling the examination of their cognate RR partners.
The activity of any enzyme is ideally studied in vivo. Currently, in vivo HK regulation (i.e. phosphorylation) has been examined only indirectly, likely due to inherently low protein concentrations, minimal basal activity levels and the instability of the phosphorylated species. Most common in the literature are β-galactosidase reporter assays in which the lacZ gene is engineered into the regulon target of phosphorylated RRs. β-galactosidase activity denotes RR-dependent transcription in cells, and thus, stimulated HK autophosphorylation. 30 While well accepted, this technique is an indirect measure of HK activity.
In vitro study of HKs that have been incorporated into proteoliposomes represents the most complex system in which HK phosphorylation has been directly measured. Because liposomes mimic the native membrane lipid environment, they have been exploited in the reconstitution of transmembrane (TM) proteins for both solubility and in vitro studies.31 Use of a liposomal scaffold provides the two-dimensional landscape and motional organization native to membrane proteins.32 Even for proteins lacking a TM domain, the association of His-tagged proteins to nickel-conjugated liposomes has effectively imparted the influence of the membrane on coordinated protein(s). In particular, template-directed assembly of multiple chemotaxis proteins in liposomes utilizing Ni-NTA-bearing lipids has been utilized to study the CheA-mediated TCS of E. coli.32
Given the importance of the study of HKs in liposomal environments, we sought to test the efficacy of our probe with membrane-associated HK853. For ease of preparation and use, our construct does not contain the transmembrane domain. In place of this domain is the His-tag used for protein purification, which could also be utilized to anchor this HK to the liposome (Supporting Figure 6). It is thought that the coiled-coil portion of HK853 plays an important role in transducing conformational changes from the membrane domain to the cytoplasmic domains that affect catalytic activity.24 Because the His-tag on HK853 is located at the N-terminus directly adjacent to the coiled-coil region, we hypothesized that the incorporation of HK853 into liposomes may confer a native structural impact and poise the coiled-coil domain for greater activity, which could be detected by labeling with B-ATPγS.
The efficacy of B-ATPγS for the activity readout of HK-proteoliposomes was analyzed by comparison of the labeling of HK853 free in solution with protein bound to liposomes. The bare liposomes were mixed with protein to facilitate protein association. Unbound protein (free) was then removed by ultracentrifugation yielding purified proteoliposomes (bound; Figure 4A). We compared the activity of HK853 prior to mixing with the liposomes (stock), from the “free” fraction and the protein associated with the liposomes (bound; Figure 4B). Labeling of all three protein samples was observed and was ATP competitive, indicating that the liposome does not interfere with probe labeling. In addition, although the protein concentrations of the free and bound samples were roughly equivalent (as judged by coomassie staining), there is a noticeable difference in the intensity of the fluorescent signal, with the bound protein showing increased activity. This trend was also noted when liposome-bound HK was phosphorylated with [γ-33P]-ATP indicating that liposome association is likely influencing the activity state of HK853 (Supporting Figure 7). These results confirm B-ATPγS to be akin to other phosphorylation read-out strategies while at the same time boasting superior bond stability and a highly fluorescent signal that readily distinguishes differences in activity.
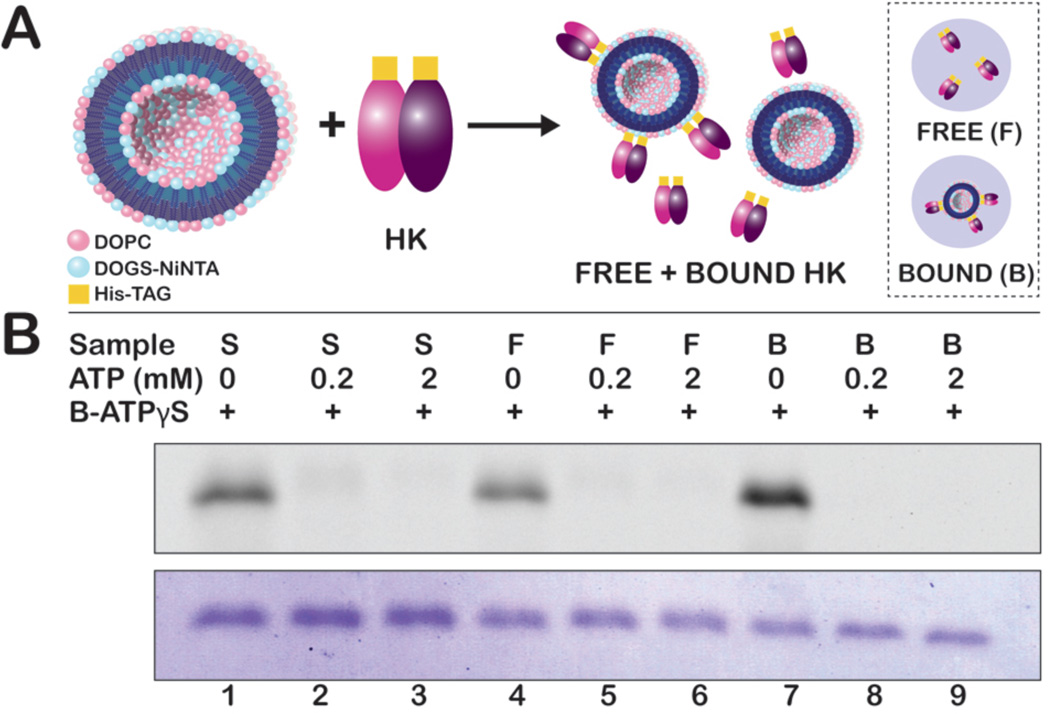
Figure 4.
(A) His-tagged HK853 was conjugated to NiNTA-bearing liposomes and separated into free and bound HK853. (B) 600 ng of HK853 from purified stock solution (S), free HK853 after ultracentrifugation (F), and liposome-bound HK853 (B) was labeled with 2 µM B-ATPγS. Inhibition of fluorescent labeling was achieved by the addition of ATP prior to B-ATPγS. Use of B-ATPγS reveals that HK853 bound to liposomes is more active than that free in solution.
We anticipated that use of a γ-thiophosphorylated ATP analog, which would yield a thiophosphorylated histidine species, could enable detection of the phosphotransfer event. We have identified that BODIPY-FL-ATP-γ-S shows specific labeling of active HK proteins and is competitive for the ATP-binding site. Surprisingly, the fluorophore is also transferred to the RR. Application of the activity-based probe, B-ATPγS, exemplifies the most straightforward and rapid way of visualizing not only HK activity, but also RR phosphotransfer events and will facilitate both functional studies of HKs and inhibitor identification.
Supplementary Material
ACKNOWLEDGMENT
We thank C. Dann for helpful advice and use of a FPLC purification system, M. Winkler for the VicK construct, D. Giedroc for use of a fluorometer, C. Kao for use of an ultracentrifuge and F. Hardin, Jr. for generating HK853 and VicK protein. This work was supported by NIH R00GM82983, NIH DP2OD008592, a Pew Biomedical Scholar Award (E.E.C.) and an Indiana University Quantitative and Chemical Biology training fellowship (K.E.W.).
Footnotes
ASSOCIATED CONTENT
Supporting Information. Methods and results for the following: Protein overexpression and purification, [γ-33P]-ATP assays, fluorescence polarization, ATP analog information, B-ATPγS assay conditions, integration of gel bands, liposome preparation. This material is available free of charge via the Internet at http://pubs.acs.org.
Notes
The authors declare no competing financial interest.
REFERENCES
- 1.Stock AM, Robinson VL, Goudreau PN. Annu. Rev. Biochem. 2000;69:183. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 2.Schaller GE, Shiu SH, Armitage JP. Curr. Biol. 2011;21:R320. doi: 10.1016/j.cub.2011.02.045. [DOI] [PubMed] [Google Scholar]
- 3.Stock JB, Stock AM, Mottonen JM. Nature. 1990;344:395. doi: 10.1038/344395a0. [DOI] [PubMed] [Google Scholar]
- 4.Stephenson K, Hoch JA. Pharmacol. Therapeut. 2002;93:293. doi: 10.1016/s0163-7258(02)00198-5. [DOI] [PubMed] [Google Scholar]
- 5.Wu HJ, Wang AH, Jennings MP. Curr. Opin. Chem. Biol. 2008;12:93. doi: 10.1016/j.cbpa.2008.01.023. [DOI] [PubMed] [Google Scholar]
- 6.Gotoh Y, Eguchi Y, Watanabe T, Okamoto S, Doi A, Utsumi R. Curr. Opin. Microbiol. 2010;13:232. doi: 10.1016/j.mib.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 7.Gooderham WJ, Hancock RE. FEMS Microbiol. Rev. 2009;33:279. doi: 10.1111/j.1574-6976.2008.00135.x. [DOI] [PubMed] [Google Scholar]
- 8.Krell T, Lacal J, Busch A, Silva-Jiménez H, Guazzaroni M, Ramos JL. Annu Rev Microbiol. 2010;64:539. doi: 10.1146/annurev.micro.112408.134054. [DOI] [PubMed] [Google Scholar]
- 9.Laub MT, Goulian M. Annu. Rev. Genet. 2007;41:121. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- 10.Trajtenberg F, Grana M, Ruetalo N, Botti H, Buschiazzo A. J. Biol. Chem. 2010;285:24892. doi: 10.1074/jbc.M110.147843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kee J-M, Muir TW. ACS Chem. Biol. 2012;7:44. doi: 10.1021/cb200445w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green KD, Pflum MKH. J. Am. Chem. Soc. 2007;129:10. doi: 10.1021/ja066828o. [DOI] [PubMed] [Google Scholar]
- 13.Lu CHS, Liu K, Tan LP, Yao SQ. Chem. Eur. J. 2012;18:28. doi: 10.1002/chem.201103206. [DOI] [PubMed] [Google Scholar]
- 14.Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T. Mol. Cell. Proteomics. 2006;5:749. doi: 10.1074/mcp.T500024-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Yamada S, Nakamura H, Kinoshita E, Kinoshita-Kikuta E, Koike T, Shiro Y. Anal. Biochem. 2007;360:160. doi: 10.1016/j.ab.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 16.Attwood PV, Piggott MJ, Zu XL, Besant PG. Amino Acids. 2007;32:145. doi: 10.1007/s00726-006-0443-6. [DOI] [PubMed] [Google Scholar]
- 17.Besant PG, Attwood PV. Mol. Cell Biochem. 2009;329:93. doi: 10.1007/s11010-009-0117-2. [DOI] [PubMed] [Google Scholar]
- 18.Kee J-M, Villani B, Carpenter LR, Muir TW. J. Am. Chem. Soc. 2010;132:14327. doi: 10.1021/ja104393t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lasker M, Bui CD, Besant PG, Sugawara K, Thai P, Medzihradszky G, Turck CW. Prot. Sci. 1999;8:2177. doi: 10.1110/ps.8.10.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carlson HK, Plate L, Price MS, Allen JJ, Shokat KM, Marletta MA. Anal. Biochem. 2010;397:139. doi: 10.1016/j.ab.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allen JJ, Li M, Brinkworth CS, Paulson JL, Wang D, Hubner A, Chou WH, Davis RJ, Burlingame AL, Messing RO, Katayama CD, Hedrick SM, Shokat KM. Nat. Methods. 2007;4:511. doi: 10.1038/nmeth1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Draganescu A, Hodawadekar SC, Gee KR, Brenner C. J. Biol. Chem. 2008;275:4555. doi: 10.1074/jbc.275.7.4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cravatt BF, Wright AT, Kozarich JW. Annu. Rev. Biochem. 2008;77:383. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- 24.Marina A, Waldburger CD, Hendrickson WA. EMBO J. 2005;24:4247. doi: 10.1038/sj.emboj.7600886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Casino P, Rubio V, Marina A. Cell. 2009;139:325. doi: 10.1016/j.cell.2009.08.032. [DOI] [PubMed] [Google Scholar]
- 26.Bilwes AM, Quezada CM, Croal LR, Crane BR, Simon MI. Nat. Struct. Biol. 2001;8:353. doi: 10.1038/86243. [DOI] [PubMed] [Google Scholar]
- 27.Guarnieri MT, Blagg BS, Zhao R. Assay Drug Dev. Technol. 2011;9:174. doi: 10.1089/adt.2010.0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gutu AD, Wayne KJ, Sham LT, Winkler ME. J. Bacteriol. 2010;192:2346. doi: 10.1128/JB.01690-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guarnieri MT, Zhang L, Shen J, Zhao R. J. Mol. Biol. 2008;379:82. doi: 10.1016/j.jmb.2008.03.036. [DOI] [PubMed] [Google Scholar]
- 30.Steinmoen H, Knutsen E, Havarstein LS. Proc. Nat. Acad. Sci. USA. 2002;99:7681. doi: 10.1073/pnas.112464599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moker N, Kramer J, Unden G, Kramer R, Morbach S. J. Bacteriol. 2007;189:3645. doi: 10.1128/JB.01920-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shrout AL, Montefusco DJ, Weis RM. Biochemistry. 2003;42:13379. doi: 10.1021/bi0352769. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.